Abstract
Well-controlled 2D cell culture systems advance basic investigations in cell biology and provide innovative platforms for drug development, toxicity testing, and diagnostic assays. These cell culture systems have become more advanced in order to provide and to quantify the appropriate biomechanical and biochemical cues that mimic the milieu of conditions present in vivo. Here we present an innovative 2D cell culture system to investigate human stem cell-derived cardiomyocytes, the muscle cells of the heart responsible for pumping blood throughout the body. We designed our 2D cell culture platform to control intracellular features to produce adult-like cardiomyocyte organization with connectivity and anisotropic conduction comparable to the native heart, and combined it with optical microscopy to quantify cell-cell and cell-substrate mechanical interactions. We show the measurement of forces and displacements that occur within individual cells, between neighboring cells, and between cells and their surrounding matrix. This system has broad potential to expand our understanding of tissue physiology, with particular advantages for the study of the mechanically active heart. Furthermore, this technique should prove valuable in screening potential drugs for efficacy and testing for toxicity.
Keywords: Cardiomyocyte, Cell culture systems, Micropatterning, Traction force microscopy, Digital image correlation
INTRODUCTION
It is estimated that more than 92 million American adults have at least one form of cardiovascular disease (CVD) [1]. Fortunately, the mortality of CVD has decreased over the last several decades due to improvements in evidence-based medical therapies and a reduction in the prevalence of risk factors [2]. In spite of these promising developments, CVD continues to be the leading cause of death in the United States. Annually, in the United States alone, there are an estimated 610,000 new myocardial infarctions (MI), commonly referred to as a heart attack, and 325,000 recurrent MIs causing approximately 114,000 deaths [3,4].
One of the reasons for the extremely high mortality rate following an MI is the human heart’s inability to regenerate. Unlike cells present in several other human tissues [5] or the hearts of some other species [6], the proliferation rate of human cardiomyocytes is extremely low. As few as 1% of human cardiomyocytes replicate annually in adults at 25 years of age, further declining to 0.45% by the age of 75 [7]. Carbon-dating studies have shown that less than 50% of cardiomyocytes are regenerated throughout an entire typical lifespan [7]. In the absence of ability to regenerate lost cardiomyocytes, the myocardial response to infarction triggers a complement cascade that produces a large region of collagen-dense scar tissue to replace the damaged myocardium [8]. This scar tissue is profoundly different from the surrounding myocardium in both mechanical [9] and electrophysiological [10] properties. This heterogeneity and loss of functional muscle contributes to the often relentless progressive heart failure following a MI.
Early stage research is investigating the introduction of stem cells and cardiac progenitor cells to the heart to aid in repair after myocardial infarction [11]. There are also a number of ways in which stem cell-derived cardiomyocytes are already being used in disease modeling, drug screening, toxicity testing, and myocardial tissue repair [12,13]. As this new field continues to develop, more robust and reproducible approaches are needed to promote the physiologic maturation of stem-cell derived CMs and to reduce batch-to-batch variability. Additionally, more information is needed pertaining to how these cells function mechanically, and how their local environment influences their biophysical properties.
Numerous investigators have explored a variety of engineered highly-controlled 2D and 3D constructs that generate reproducible in vitro culture platforms to investigate basic cell-cell and cell-extracellular matrix (ECM) interactions [14–24]. 3D culture methods allow cell-cell interactions to occur in a setting that is often more relevant to native biology, which is particularly important when observing phenomena such as organogenesis. Contraction in 3D tissues can be quantified by using force transducers, post deflection, and optical tracking, for example [25–29]. Whereas these methods quantify contraction of the entire tissue, they preclude detailed assessment of single cell function, and they are of relatively low throughput. Advances in lightsheet microscopy [30,31], expansion microscopy [32], and single-cell sequencing [33,34] have begun to address these challenges, but these are still low-throughput in comparison to 2D culture systems.
2D culture methods continue to be a mainstay in the field because they can provide a highly controlled and easily manipulable environment, scalable features with high precision and fidelity, and ease of use for imaging and analysis. 2D systems have advanced basic investigations in cell biology, providing details of cellular architecture, molecular structures, protein interactions, and signal transduction pathways. 2D systems have also provided tools for disease modeling, and high-throughput platforms for drug development and toxicity testing. However, 2D systems do have drawbacks due to limited morphogenesis and differences in cell migration patterns compared to 3D cultures and the in vivo environment. A major limitation to traditional 2D cardiomyocyte cultures has been the disorganization of the cells’ contractile apparatus, which is in marked contrast to the highly aligned and organized features of in vivo cardiac tissue. For detailed contractile physiology studies, a consistently organized, integrated multicellular system is needed. Here we present techniques for studying stem cell derived cardiomyocytes in a 2D cell culture platforms that recapitulated in vivo cellular alignment as well as other key markers of maturity. When combined with optical microscopy, this platform enables mechanics experiments to quantify cell-cell and cell-substrate mechanical interactions.
BACKGROUND
The Cardiomyocyte
Blood flow in the body results from collective force generated by each cardiomyocyte (CM) of the heart. CM contraction is a complex, choreographed event that involves an action potential change in voltage across the cell membrane, calcium (Ca2+) release within the cell, and mechanical contraction of the cell. The contraction is produced by a highly organized apparatus, formed by numerous cytoskeletal structural, motor, and regulatory proteins that enable the cell to produce force in response to high Ca2+ concentration within the cytoplasm inside the cell. The individual repeating units of the contractile apparatus are the sarcomeres, which align end-to-end to produce long myofibrils that span the length of the CM and, when provided the appropriate environmental cues, align laterally adjacent myofibrils. Through numerous connections with neighboring cells, the summation of sarcomere contractile events contributes to the overall contraction and relaxation of the cardiac tissue within a chamber of the heart so that blood can be pumped to distant tissues. Inefficient and aberrant contraction in heart disease can be caused by abnormalities in myofibrillar structure, inefficiencies in contractile force generation and relaxation, and disruption of the normal temporal and spatial variation of Ca2+ flux during excitation-contraction [35–39].
Human Stem Cell-Derived Cardiomyocytes
Scientific study of human cardiac physiology would ideally be conducted using well characterized human cells and tissues in which as many variables as possible can be controlled. In many studies of the human heart, animal models are required because of limited availability of human cells and tissue. Unfortunately, animal models for the human heart inherently have significant drawbacks due to fundamental physiologic differences between species [40,41]. For example, the hearts of small laboratory rodents have different action potentials, roughly five times the resting heart rate of humans, and their force-frequency relation is opposite that of human hearts [42]. Large animal models match human heart characteristics more closely, but they are substantially more expensive than small animal models [43]. Access to human heart tissue for research purposes is exceedingly challenging. Additionally, human heart tissue available for research is often obtained only at a significant state of advanced disease. Appropriate healthy controls are frequently not available, because the demand for heart transplants currently exceeds supply. The emergence of stem cell technologies and the ability to produce large numbers of human stem cell-derived CMs provides a promising new avenue for cardiac research. Stem cell-derived CMs have the potential to support groundbreaking discoveries, particularly if human adult-like CMs in vitro can produced reliably and cultured to maturity.
Human embryonic stem cells (hESCs), which are pluripotent cells capable of long-term proliferation, were first successfully isolated from in vitro fertilization embryos in 1998 at the University of Wisconsin–Madison [44]. This discovery has led to further breakthroughs in the fields of embryology, cellular biology, tissue engineering, and regenerative medicine. Following this extremely promising discovery, researchers focused their efforts on developing methods for producing specific cell types through a wide variety of methods. As a result, we are now capable of producing a wide array of specialized cell types, including cardiomyocytes [45]. Additionally, the ability to generate induced pluripotent stem cells (iPSCs) from adult cells from any individual [46,47] provides added opportunities for creating disease models, improved drug screens, and clinical therapies that evade autoimmune rejection.
Recent investigations have developed increasingly effective means of producing cardiomyocytes from pluripotent stem cells, which have increased production efficiencies of up to 98% [48,49]. Although these cells have committed to the cardiac lineage, they are immature and only begin to show some markers of maturation after many months in standard 2D culture [50].
2D Patterned Substrates
One of the most attractive features of 2D systems is the relative ease with which images can be captured of the cells and their substrates through bright field and florescent microscopy. Cells cultured on a micropatterned substrate can be treated with immunostaining techniques to visualize cellular components of interest. Bright field observations and live imaging techniques can be used in concert with image tracking or digital image correlation software. With the incorporation of tracking particles within the substrate, techniques such as traction force microscopy can be employed. As their role is to produce force, cardiomyocytes in particular lend themselves to biomechanics analysis.
Muscle cells respond to certain micropatterns in 2D culture systems with improved cell-cell junctions, increased contractile force, and greater sarcomere organization [51–53]. These micropatterns also influence cell shape and cell attachment [54,55]. The majority of studies involving micropatterned CMs have used rat neonatal ventricular CMs. Deriving human CMs from pluripotent stem cells is a more recent achievement, so researchers have only begun to examine how engineered substrate conditions influence behavior of human CMs. For example, the human CM excitation-contraction coupling and sarcomere structure have been improved by culturing cell populations of CMs onto mesh-like micropatterns for tissue engineering applications [14]. Culturing human CMs in micropatterned arrays also allows for high-throughput screening [56]. Micropatterning has also been used to demonstrate that cell shape and substrate stiffness impact shortening of sarcomeres and the translation of sarcomere activity to contractility [57].
Although glass and tissue culture plastic are commonly used substrates in cell culture, a wide range of polymeric materials can be employed to more closely mimic the mechanical properties of the in vivo environment. In addition to the type and pattern of proteins available to the cells for attachment, the mechanical stiffness of the system is important and allows the cell to display more mature behavior [58]. Cardiac tissue in vivo has a Young’s modulus of 4 to 50 kPa, with the lowest values being that of healthy myocardium [59]. The desired stiffness can be created with substrates composed of polydimethylsiloxane (PDMS) with tunable stiffness [60–63], polyacrylamide (PA), which can be made with a range of moduli spanning from <1 kPa to >100 kPa [64–66], or alginate, which has also been shown to have a high level of mechanical property controllability through its ionic and covalent crosslinking [67] and can be blended at different ratios with gelatin to modify its mechanical properties for CM culture [68].
In this work we focus on 2D cell culture systems and use micropatterning techniques to create ECM patterns on substrates ranging from glass [69] to polymeric materials such as PA and PDMS in order to examine how patterning impacts the organization and alignment of a CM’s contractile apparatus and its ability to enhance maturation of the CMs. When direct microcontact printing is not feasible, we use sacrificial films [60] to transfer ECM patterns to substrates. Importantly, the polymeric substrates can be easily modified to incorporate fluorescent particles that enable techniques such as traction force microscopy to be utilized [70,71,62].
MATERIALS AND METHODS
Stem Cell Differentiation
In the research reported here, immature CMs were differentiated from pluripotent stem cells using a modified version of the small molecule Wnt-agonist method [46]. Briefly, on day −3 the cTnT-GFP stem cells were seeded onto Matrigel coated (BD Biosciences) 12-well plates at a density of 400,000 cells per well and fed with E8 medium supplemented with 5 μM ROCK inhibitor (Tocris). On day −1, the cells were again fed with E8 medium. On day 0 (exactly 72 hours after initial seeding), cells were treated with RPMI (Life Technologies) supplemented with B27-insulin (Life Technologies), 12 μM of the Wnt agonist CHIR 99021 (Tocris) and 1 μg/ml of insulin (Sigma). On day 1, cells were fed with RPMI supplemented with B27-insulin. On day 3 for hESCs and when confluent for iPSCs, the cells were treated with RPMI supplemented with B27-insulin and 10 μM of the Wnt inhibitor IWP4 (SemGent). On day 5, cells were fed with RPMI containing B27-insulin and then treated with RPMI containing B27-complete on days 7, 9, and every other day until day 15.
On day 15 the media was switched to lactate media to begin CM purification[72]. The lactate media consisted of RPMI-glucose (Life Technologies), B27-complete (Life Technologies) and 5mM lactate (Sigma). The cells were exposed to lactate for 10 days and then the media was changed back to RPMI containing B27-complete on day 25. Spent medium was replaced with fresh RPMI+B27 every three days thereafter. Purified stem cell-derived CM cultures were washed with PBS and a 10–15 minute exposure to TrypLE (Life Technologies) was then conducted to dissociate and singularize CMs before seeding them onto micropatterned substrates in a 10% serum-containing media.
Engineered Substrate System
The results reported below were produced on polymeric substrates of PDMS or PA. PDMS of desired stiffness was created by blending Sylgard 184 with Sylgard 527 (Dow Corning) [73]. Sylgard 184 was made by mixing ten parts base to one part curing agent, and Sylgard 527 was prepared by mixing equal weights of part A and part B. Both Sylgard 184 and 527 were mixed for 5 min with a glass stir rod. After each blend was properly mixed, they were then combined in different mass ratios of Sylgard 184:527 and again mixed for 5 min. The mass ratio of 1:10 produced a substrate with Young’s modulus of approximately 10 kPa [60]. Once mixed, the PDMS was poured into 100 mm diameter petri dishes and cured overnight at 60°C. The following day the samples were cut into the correct shape with a razor blade and UV sterilized prior to patterning with extracellular matrix proteins.
PA is used for the Traction Force Microscopy results presented below. A solution of 3% acrylamide and 0.1% bisacrylamide was prepared to give a Young’s modulus of 1 kPa upon polymerization. Simply mixing fluorescent particles into the prepolymerized solution results in the particles being dispersed throughout the bulk of the substrate. As tractions applied by the cells to the substrate occur only on the top surface of the PA gel, more accuracy is obtained by embedding the particles only at the top surface. For this, we centrifuge the gels upside down during polymerization. The centrifugation brings the fluorescent particles to the top surface of the gel, thereby allowing for imaging with a common wide field fluorescent microscope. We use a high concentration (0.07% volume/volume) of 0.5 μm particles (Fluospheres, Life Technologies), which together produce high contrast bright and dark spots within the image. To avoid leaking of the prepolymerized gel solution during centrifugation, we sometimes use a 2-step method as follows. A 100 μm thick polyacrylamide gel is fully polymerized with no particles. Then, a second gel ~10 μm thick with particles is polymerized on the first and centrifuged upside down during polymerization. Although not used in this study, other authors report the fluorescent particles can also be incorporated in PDMS[74] [75].
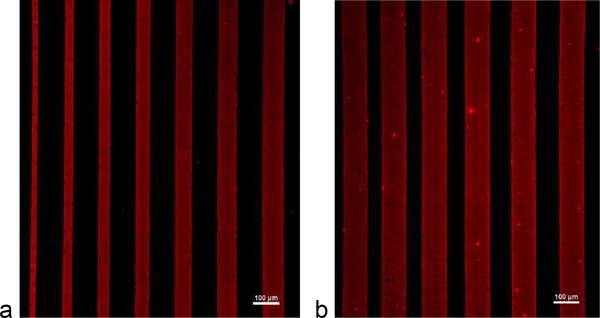
To accomplish the micropatterning of cells in lanes on soft substrates, reusable microcontact printing stamps were fabricated on a silicon wafer (FlowJEM, Toronto, ON, Canada) with the specific micropatterns designed for study. The PDMS (Sylgard 184) was poured on top of the patterned silicon wafer and cured at 60°C overnight to produce a reverse replica of the silicon wafer. After removal from the silicon wafer, the PDMS was cut into individual stamps which were coated with ECM proteins overnight (83 μg/mL Matrigel). Using microcontact printing, the ECM-coated PDMS stamps were used to transfer lanes of ECM to the substrate. Fig. 1 illustrates how the micropatterning can be used to transfer Matrigel onto various substrates, so that experiments can be conducted on glass or on a substrate with a more physiologically relevant stiffness.
Fig. 1.
Immunofluorescence images of different patterns of Matrigel (red), visualized with an anti-laminin antibody, on (a) glass and (b) 5 kPa PDMS substrates. Scale bar = 100 μm
For patterning on compliant substrates, such as in Fig. 1b, stamping of ECM proteins was conducted on a sacrificial polyvinyl alcohol (PVA) film which was then brought into conformal contact with the substrate [76]. To make a sacrificial film, 0.5 g of PVA (Sigma Aldrich) was dissolved in 10 mL of deionized water and dried overnight in a petri dish. Once dry, the PVA film was removed from the petri dish and cut into rectangles slightly larger than the PDMS stamps. The PDMS stamps were coated with Matrigel overnight and then placed on top of the PVA films. A glass slide and 50 g weight was added to ensure even pressure distribution and better pattern transfer from the PDMS stamp to the PVA film. After one hour, the PVA film was removed from the PDMS stamp and then brought into conformal contact with the soft substrate for 30 min. Afterward, the substrate was washed to dissolve the PVA film, leaving behind the patterned proteins which were then seeded with cells. The remaining areas of the substrate without ECM proteins were then backfilled with Pluronic F127 (Sigma Aldrich) to prevent cell outgrowth. Cells must be seeded onto the patterns with a density high enough to ensure complete pattern coverage by the cells, and low enough to allow a moderate amount of normal growth of each cell. In these experiments, cells were seeded onto patterns in the range of 2000–3000 cells per mm2 of ECM area available. This is higher than the 1400–1500 cells/mm2 in monolayer control samples, even though the ultimate seeding density after attachment is equivalent by a count of nuclei.
Immunofluorescent Staining
For immunofluorescent staining, cells were cultured in the 2D substrate systems, stained at various time points, and then imaged. Staining began with a PBS wash and then exposure to 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) for 15-min at room temperature. The cells were washed again with PBS and then the cell membranes were permeabilized with 0.1% Triton (Sigma) for 6 min at room temperature. After another PBS wash they were treated for 30 min with a blocking solution consisting of PBS, 2% FBS, 0.1% Triton, 11.2 mg/mL glycine, and 50 mg/mL BSA. The following primary anitbodies were left on the samples overnight at 4°C: rabbit anti-α-actinin (1:200 dilution, Abcam) and mouse anti-N-cadherin (1:250 dilution, BD Biosciences). The following day, cells were washed once with PBS for 5 minutes, and treated for 1 hour with blocking solution containing the secondary antibody, Alexa Fluor 488 goat anti-mouse and/or Alexa Fluor 647 goat anti-rabbit (1:1000 dilution). When desired, cells were washed once more, and phalloidin conjugated to tetramethylrhodamine B isothiocyanate (TRITC) (Sigma) was applied at a 50μg/mL concentration to label actin filaments, and/or DAPI was applied at a 1:10,000 dilution to label nuclei. Finally, cells were washed with PBS and transferred to coverslips and mounted with ProLong Gold Antifade (Life Technologies). Samples were imaged on a Nikon A1RSi Confocal Microscope. Widefield images were also acquired with an attached Photometrics CoolSNAP HQ2 camera. Nis-D Elements – Advanced Research v.3.22 software was used for image acquisition and analysis. Objectives for this microscope included a CFI Plan Apochromat Lambda 40× NA 0.95 air objective and a CFI Plan Apochromat Lambda 60x NA 1.4 oil immersion objective.
To visualize the transfer of Matrigel from the patterned PDMS stamp to the substrate, the primary antibody rabbit anti-laminin (1:500, Sigma-Aldrich) was incubated with the substrate overnight at 4 °C. The next day the substrate was washed with PBS for 5 min and then treated for 1 h with blocking solution containing the secondary antibody, Cy3-conjugated mouse anti-rabbit (1:1000 dilution, Abcam).
Surface Feature Tracking and Calcium Imaging
Contraction and relaxation were quantified using surface feature tracking of stem cell-derived CMs grown on micropattened lanes. Using the IonOptix myocyte calcium and contractility system (Westwood, MA), with the micropatterned slide containing the stem cell-derived CMs placed in a custom open perfusion chamber, detailed cell shortening data was collected. Additionally, the relationship between the calcium transient and contraction was interrogated by introducing the cells to calcium ratiometric dye Fura-2-AM (Invitrogen). Using the IonOptix calcium imaging system with its HyperSwitch in conjunction with the perfusion chamber placed on the flat stage of an appropriate microscope (Nikon TI-U inverted microscope with 40X fluorescence objective), intracellular calcium release and reuptake and cell shortening were simultaneously recorded.
Digital Image Correlation
Certainly when observing a collection of CMs, but even within an individual CM, the deformation taking place is not uniform across the field of view. The details of the full-field behavior are often critical to developing a deeper understanding of the biomechanics at play. To measure full-field displacements, we used digital image correlation (DIC) [77]. DIC requires an image having a random high-contrast pattern, which in this case was a phase contrast image of the cells obtained with a Nikon Eclipse Ti microscope with a Plan Fluor 10× NA 0.3 objective and Nikon DS-QiMc camera. In our preliminary analyses, we found the results to be the same with two different free software packages, Ncorr (https://github.com/justinblaber/ncorr_2D_matlab) [78] and Fast Iterative Digital Image Correlation (FIDIC, https://github.com/FranckLab/FIDIC) [79]. Results presented here used FIDIC.
Traction Force Microscopy
In addition to measuring displacements and strains of the cells, we measured forces applied by the cells to the substrate using traction force microscopy (TFM), which computes the traction (force per area) at the interface between the cells and the compliant substrate [70,71,80–83]. This required markers to be embedded in to the substrate of our cell culture platform. It is crucial to achieve a high contrast image of a single plane of particles at the top surface of the substrate. As cell-induced deformations cause particle displacements throughout the substrate, the exact location of the particles must be known for accurate calculations. Often measurements are assumed to be taken from particles at the top surface of the substrate nearest the cells. Unfortunately, this assumption is often incorrect, resulting in an image that shows beads for various positions through the bulk of the substrate. To address this issue, we make PA substrates with particles located at the cell-substrate interface as described above.
The cells and fluorescent particles were imaged with a widefield florescent microscope (Nikon Ti-E) with a 10× NA 0.3 objective during an experiment. After the experiment, the cells were released from the substrate with trypsin, which eliminated the tractions applied by the cells to the substrate and allowed the substrate to recover to a stress-free state. A final image of the particles was acquired in that stress-free state; this final image was used as a reference to compute cell-induced displacements of the particles using DIC. After displacements were measured, the tractions were computed by applying equations of force equilibrium to the substrate [70,71,80–83]. There are multiple different ways to implement the equations; we used an approach based on the Fourier transform [71] with corrections for a substrate of finite thickness [82,83].
RESULTS AND DISCUSSION
Cell Culture System
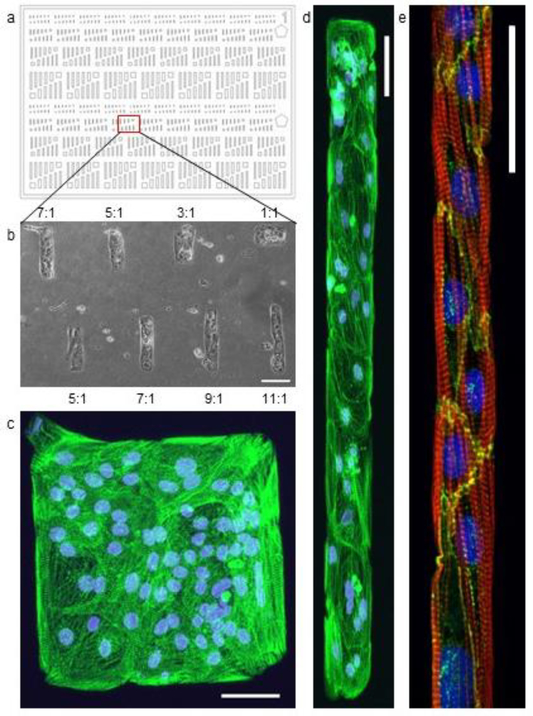
Fig. 2a illustrates a pattern designed to investigate the influence of the pattern aspect ratio on CM morphology. Adult human CMs are rod-shaped with a minimum length-to-width ratio of 7:1 and lengths in the 100 μm range [84]. This characteristic elongated shape of mature CMs produces a highly organized internal cytoskeletal network consisting of myofibrils running parallel to the length of the cell body. In these experiments immature cardiomyocytes differentiated from induced pluripotent stem cells (iPSCs) (19–9-11 cell line, WiCell, Madison, WI) were seeded onto PDMS substrate patterned with Matrigel. By fixing and staining the cells after they have grown in the pattern for a period of time, the internal structure of the cell was explored with immunofluorescence microscopy. As Fig. 2c and d show, the internal sarcomere structure of the iPSC-CMs is significantly more organized along the length of the 11:1 feature compared to the disordered central region of the 1:1 feature after just 6 days of being cultured on the patterns.
Fig. 2.
(a) Pattern consisting of rectangles with a range of aspect rations between 1:1 and 11:1. (b) A bright field image of 19–9-11 stem cell derived cardiomyocytes cultured on a portion of the pattern created with a Matrigel coated stamp using this pattern design. Aspect ratio is noted for each feature detailed above and below the image. Scale bar: 100 μm. (c, d) Fluorescent microscopy image of a 1:1 and a 9:1 feature. The nuclei are stained with DAPI (blue) and the protein α-actinin (green) highlights the sarcomere structure within the cells. (a-d) Scale bar: 50 μm. (e) Micropatterned lane with hESC-CMs cultured in Matrigel lanes on a 5kPa stiffness PDMS substrate for 18 days. Cell alignment and sarcomere organization shown in lanes of 20μm width. The nucleus is stained with DAPI (blue), the protein actin (red) highlights the internal organization of the cell, and the protein N-Cadherin (green) highlights the cell-cell interfaces. Scale bar 50 μm.
Although the aspect ratio of the pattern has interesting implications, we found that the pattern width is more important than aspect ratio in controlling the internal organization of the CMs within a multicellular 2D construct [69]. Fig. 2e shows the cell alignment and sarcomere organization within hESC-CMs (H9 hESC line containing a cardiac troponin T GFP promoter [85]) that have been cultured for 18 days in a 20μm wide Matrigel lane on a PDMS substrate with Young’s modulus of 5 kPa (measured by tensile testing). Rather than the internal disarray observed in CMs grown in unpatterned monolayer conditions, the lane guides the cells to create an organized internal structure similar to that of adult CMs in vivo. As micropatterning provides control over these intracellular features, it reduces some of the heterogeneity common in biology, thereby providing a means to design well-controlled, repeatable experiments.
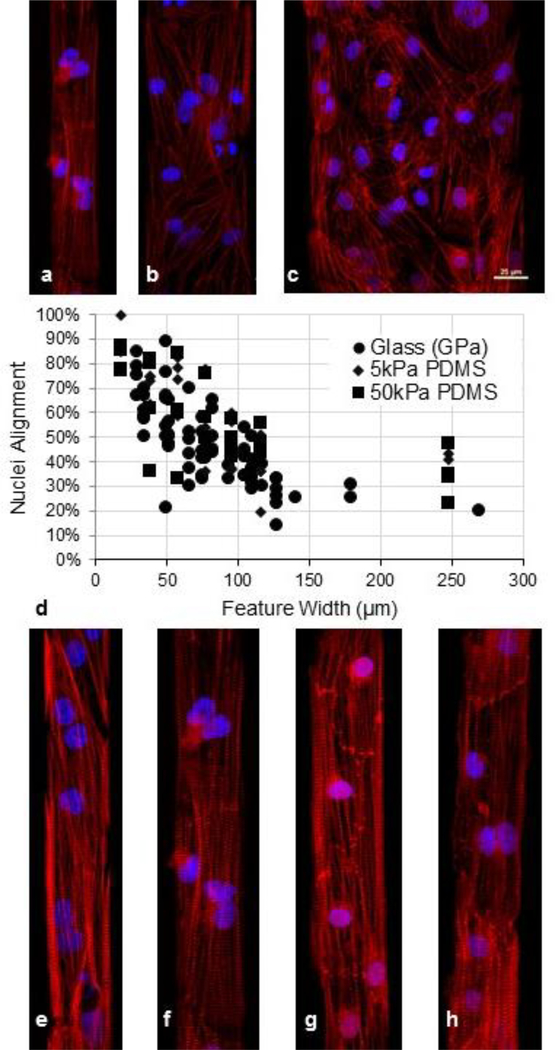
Experiments using varying pattern shapes have shown that patterns with widths less than 100 μm induce an increase in nuclear alignment for CMs on substrates of different stiffnesses. Patterns with widths of ~40 μm were found to be ideal for producing connected, highly structured, one-cell-wide lanes of hESC-CMs [69]. Using our culture system, we find that CMs cultured in lanes of various width on PDMS substrates display a similar trend in organization to that found on glass (Fig. 3a-d).
Fig. 3.
(a-c) Sarcomere organization in hESC-CMs seeded on different lane widths of Matrigel on 1.72 MPa PDMS, including (a) 37 μm, (b) 75 μm, and (c) 150 μm. Red = actin, blue = DAPI. Scale bar is 25 μm. (d) Nuclei alignment for hESC-CMs grown on a range of lane widths spanning 30 to 270 μm using glass and PDMS substrates. (e-f) hESC-CMs grown on substrates of different Young’s moduli, each having a lane width of 37 μm: (e) Glass, (f) 1.72 MPa PDMS, (g) 50 kPa PDMS, and 5 kPa PDMS. Red = actin, blue = DAPI
We explored more than seven orders of magnitude in Young’s modulus with these techniques, from glass to low stiffness PDMS. Fig. 3e-h illustrates the results seen in stem cell-derived CMs under this range of conditions. The actin stain shows not only elements of the sarcomere organization but also stress fibers, which are least prevalent in cells cultured on the 5 kPa substrate. Analysis of nuclear alignment with the DAPI stain was used to determine the directionality of the cells populating lanes The cells were defined as aligned if their major axis was within 20° of the lane direction. As Fig. 3d shows, alignment improves with decreasing lane width and is relatively unaffected by substrate stiffness.
Contraction Displacement and Calcium Imaging
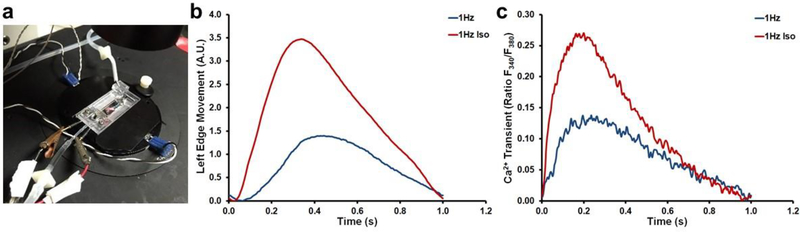
We simultaneously measured contraction amplitude, and intracellular calcium transients in micropatterned hESC-CMs that were cultured in 30 μm lanes on 5 kPa PDMS for 6 days prior to testing. A major advantage of using our micropatterned lanes rather than conventional monolayer culture of hESC-CMs is that the micropatterning produced organization in the CMs with most sarcomeres aligned parallel to the long axis of the cell, allowing us to assess contraction along a known direction. Contraction was assessed by surface feature tracking, while calcium transients were assessed by ratiometric Fura2 fluorescence using an IonOptix myocyte calcium and contractility system. During testing, micropatterned hESC-CMs were subjected to electrical pacing at 1Hz by field stimulation, allowing us to gather detailed kinetic data of contraction (CM shortening), relaxation (CM re-lengthening), calcium release, and calcium sequestration both in the absence and presence of 1 μM isoproterenol (Fig. 4). β–adrenergic stimulation with isoproterenol treatment of the micropatterned hESC-CMs increased both contraction amplitude and calcium transient amplitudes while also increasing the rates of contraction, relaxation, calcium release, and sequestration (Fig. 4) This combination of outcomes indicates a mature functional status of the CMs in these culture conditions [86,50].
Fig. 4.
(a) IonOptix myocyte calcium and contractility system with custom chamber. β–adrenergic stimulation of micropatterned hESC-CMs. hESC-CM (b) contraction amplitude and (c) rate of Ca2+ transient when paced under different conditions in the presence and absence of isoproterenol. Fura-2-AM ratiometric fluorescent dye (Abcam). Exposure to isoproterenol at 1 μM increased Ca2+-transient amplitude, contraction amplitude, the rate of contraction, and the rate of relaxation with 1 Hz pacing.
Contractile Force
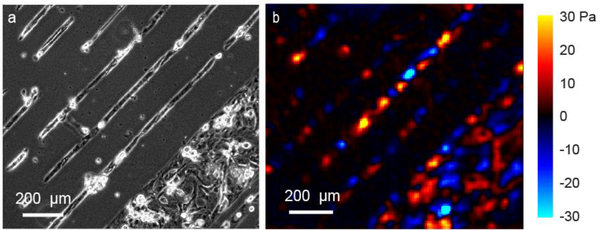
To quantify the forces of CM contraction, we used traction force microscopy. Polyacrylamide substrates embedded with fluorescent particles at the top were generated using the 2-step protocol described in the methods. Matrigel was micropatterned onto the polyacrylamide substrate, and cells were seeded on top (in this case, H9 hESC-CMs). Representative tractions applied by CMs to the substrate are shown in Fig. 5 for unconnected lanes which do not contract in synchrony with neighboring lanes.
Fig. 5.
TFM analysis of micropatterned hESC-CMs seeded within lanes on a 5 kPa polyacrylamide substrate at one moment in time. (a) Phase contrast image of CMs. Narrow lanes are 30 μm in width; wide lane is 350 μm. (b) Tractions applied by cells to substrate in direction of lanes. Positive (warm colors) is up and right; negative (cool colors) is down and left
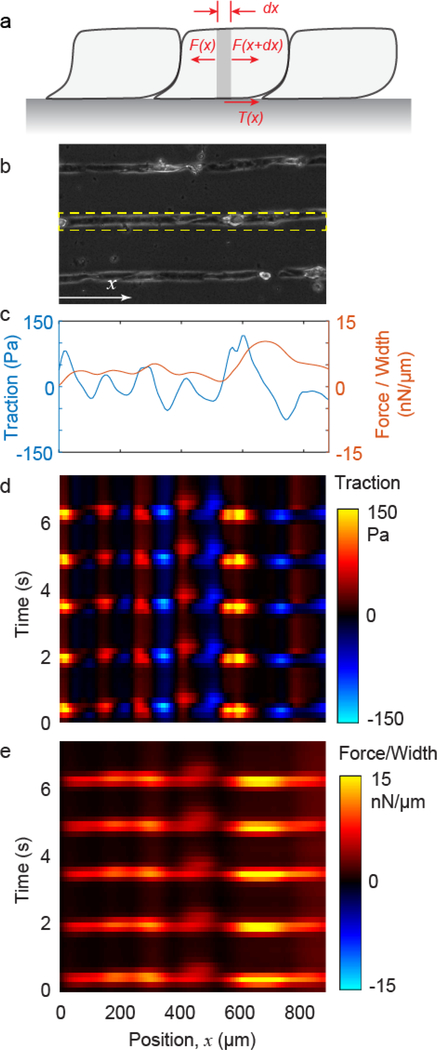
Whereas Fig. 5 shows traction at the interface between the cells and substrate, the contraction within a layer or lane of cells can be quantified as well, using an additional analysis that applies the principle of force equilibrium to the layer of cells [83,87–89]. The analysis begins with a micropatterned lane of cells having width w (Fig. 6a). Cell contraction generates an intracellular force at location x, F(x), which is balanced by a nearby force in the opposite direction F(x+dx) and the traction T(x) having units of force per area. Force equilibrium gives F(x) = F(x+dx) + T(x)wdx, which can be rewritten dF/dx + wT = 0. From this relationship, integrating the traction gives the contractile force scaled by lane width,
Fig. 6.
CM contraction over time. (a) Forces within a lane of cells. (b) Cardiomyocytes patterned into lanes on a polyacrylamide substrate. Tractions are computed for the lane shown by the dashed rectangle. (c) Traction (averaged over lane width) and contractile force (normalized by lane width) is plotted against position x for a single point in time. (d, e) Traction and force are calculated for all time points and plotted over position and time as a kymograph. The periodic bands of bright signal in force indicate periodic cell contraction, i.e., beating.
Integration begins at the endpoint of a cell lane, where force is known to be zero. As the force f is a direct measure of contractile activity within the cell, it is a more physiologically relevant metric than traction. Thus, the micropatterning both produces organized sarcomeres (Fig. 3) and enables mechanics-based quantification of CM contraction. The data show that traction and force increase and decrease periodically over time with repeating magnitude (Fig. 6c-e), indicating collective and consistent cell beating.
CM Contraction on Patterned Substrates
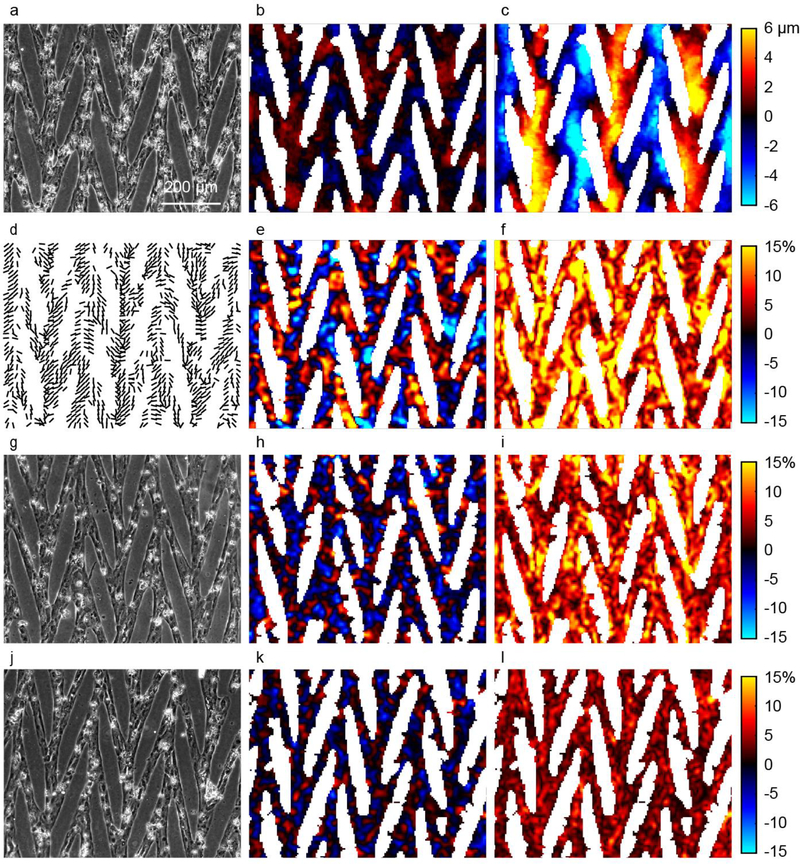
We designed more complex patterns for multicellular studies to produce alignment of the internal myofibrils while providing for cell-cell connections that span a large area. The resulting connected mesh of CMs displayed synchronized spontaneous contractions. We focus here on a 15° chevron pattern with representative data shown in Fig. 7. In this data set, CMs were cultured for 16 days in the pattern prior to imaging. Bright field images were captured over the course of several spontaneous contractions (representative images are shown in Fig. 7a, g, and j). Displacements were computed by correlating images in the relaxed and contracted states. In contrast to an unpatterned monolayer, the contraction is anisotropic and aligned primarily in the vertical direction (Fig. 7c), which matches the orientation of the chevron patterns and corresponds to the general alignment of the cells in the primary direction of the chevron patterns at ±15° from vertical. Minimal displacements occur in the horizontal direction (Fig. 7b).
Fig. 7.
Displacements and strains of a CM array computed with DIC. (a,g,j) Bright field images of CMs micropatterned into a chevron pattern. (b, c) Displacements in horizontal (b) and vertical (c) directions, uh and uv. Red (positive) indicates rightward and upward. Color scale bars for panels (b) and (c) are the same. (d) Lines show orientation of second principal strain, indicating the primary axis of contraction. Lines are shown only in locations for which the magnitude of second principal strain is greater than 2%. (e,h,k) Sum of principal strains, e1 + e2, gives a measure of relative change in area at each location within the CM array. (f,i,l) Difference of principal strains, e1 − e2, quantifies a relative magnitude of shape change at each location within the CM array. (a-f) CMs on a compliant substrate with Young’s modulus of 5 kPa, in comparison to substrates of 10kPa (g-h) and 50 kPa (j-l). Color scale bars for panels (e-f, h-i, and k-l) are the same.
To further quantify the contraction, the strain tensor is computed by taking the gradient of the displacement data; subsequently, the principal strains, defined as the eigenvalues of the strain tensor, are computed. Here, we show the sum and difference of the principal strains (Fig. 7e-f), which correspond to changes in local area and shape, respectively (see supplemental data for video). In Fig. 7e, blue (negative) corresponds to a decrease in area, indicating contraction in the plane of the sample. Given the short time between the images, constant volume of the cells can be assumed such that the out of plane strain is −(e1 + e2). In Fig. 7f, the magnitude of the shape change is illustrated by the difference in principal strains, e1 − e2.
Because the soft substrate is attached to a rigid glass slide below, global macroscopic area and shape changes are not possible. The chevron mesh pattern not only promotes sarcomere alignment through the pattern edges, but also provides the cells with a template for creating a globally self-accommodating structure that permits large coordinated contractions along the chevron orientations of ±15°. The large contractile strains typically reported for CMs in tissue can be observed at the local level and sustained in the chevron mesh pattern of this 2D platform. In contrast when a 90° bridge pattern design was used to connect lanes, CMs were torn apart at junctions by the contractions (data not shown). The primary axis of contraction in the plane of the sample is indicated by the lines in Fig. 7d, which gives the orientations of the second principal strain.
Data in Fig. 7a-f are for CMs on a compliant substrate having Young’s modulus of 5 kPa. Results for cells on substrates of Young’s modulus 10 kPa and 50 kPa are shown in Fig. 7g-i and Fig. 7j-l, respectively. The data show that contraction decreases nonlinearly as substrate stiffness increases. For example, the root-mean-squares of the differences in principal strains, e1 − e2, are 10.9%, 7.7%, and 4.0% for cells on 5 kPa, 10 kPa, and 50 kPa substrates, respectively. The strains diminish with increasing substrate stiffness, because the cardiomyocytes are increasingly constrained in their ability to contract by the underlying substrate. Interestingly, the decrease in contractile strains are not proportional to the increase in substrate stiffness, indicating that the CMs produce different contractile force on substrates of different stiffness.
CONCLUSION
Here we have shown a 2D cell culture platform designed to produce organized collections of cardiomyocytes in vitro, thereby enhancing key maturity markers. We quantified cell alignment and contraction using florescent dyes, image tracking, digital image correlation, and traction force microscopy. We showed that lanes with widths less than 100 μm induce an increase in nuclear alignment for CMs on substrates of Young’s modulus spanning seven orders of magnitude. Calcium transients and contraction amplitude were measured for CMs in lanes with exposure to 1Hz electrical pacing, with and without the presence of isoproterenol. The mature functional status of the CMs exposed to isoproterenol were illustrated by the increases in contraction amplitude, calcium transient amplitudes, rates of contraction, relaxation, calcium release, and sequestration. We also presented data for a connected pattern design (15° chevron) that maintains the feature width restrictions of lanes while producing cell-cell connections that span a large area, recapitulating adult-like cardiomyocyte organization within heart tissue. The connected CMs in the chevron design displayed synchronized spontaneous contractions, with contractile strains of the magnitude typically reported for CMs in vivo. Furthermore, we show that contraction decreases nonlinearly as substrate stiffness increases over range of 5 kPa to 50 kPa which spans the range of moduli reported for healthy and diseased heart tissue. Taken together, these findings demonstrate the value of this platform to enhance maturation in stem cell derived cardiomyocytes and study cardiomyocyte behavior. As mechanical contraction is one of the most critical functions of CMs in vivo, we expect the methods described here will useful for drug development, toxicity testing, and diagnostic assays.
Supplementary Material
ACKNOWLEDGEMENTS
Research reported in this publication was supported in part by the National Science Foundation under grant number 1660703 (JN) and by the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health under award number R01 HL107367 (JCR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation. Support was also provided by the Karen Thompson Medhi Professorship (WCC), the Graduate School (WCC) and the Office of the Vice Chancellor for Research and Graduate Education (WCC, JN) at the University of Wisconsin-Madison. Additional thanks are given to Dr. Timothy Kamp of the University of Wisconsin-Madison for providing the cTnT H9 hESC line used in some of the experiments described above.
REFERENCES
- 1.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jiménez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P (2017) Heart Disease and Stroke Statistics—2017 Update: A Report From the American Heart Association. Circulation. doi: 10.1161/cir.0000000000000485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhyari P, Kamiya H, Haverich A, Karck M, Lichtenberg A (2008) Myocardial tissue engineering: the extracellular matrix. Eur J Cardiothorac Surg 34 (2):229–241 [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Kochanek K, Murphy S, Tejada-Vera B (2010) Deaths: Final Data for 2007. National Vital Statistics Reports, 58 (19). [PubMed] [Google Scholar]
- 4.Xu J, Kochanek KD, Murphy SL, Tejada-Vera B (2016) Deaths: final data for 2014. [PubMed]
- 5.Michalopoulos GK, DeFrances MC (1997) Liver regeneration. Science 276 (5309):60–66 [DOI] [PubMed] [Google Scholar]
- 6.Poss KD, Wilson LG, Keating MT (2002) Heart regeneration in zebrafish. Science 298 (5601):2188–2190 [DOI] [PubMed] [Google Scholar]
- 7.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H (2009) Evidence for cardiomyocyte renewal in humans. Science 324 (5923):98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frangogiannis NG, Smith CW, Entman ML (2002) The inflammatory response in myocardial infarction. Cardiovasc Res 53 (1):31–47 [DOI] [PubMed] [Google Scholar]
- 9.Holmes JW, Borg TK, Covell JW (2005) Structure and mechanics of healing myocardial infarcts. Annu Rev Biomed Eng 7:223–253 [DOI] [PubMed] [Google Scholar]
- 10.Bigger JT, Fleiss JL, Kleiger R, Miller JP, Rolnitzky LM (1984) The relationships among ventricular arrhythmias, left ventricular dysfunction, and mortality in the 2 years after myocardial infarction. Circulation 69 (2):250–258 [DOI] [PubMed] [Google Scholar]
- 11.Wu R, Hu X, Wang Ja (2018) Concise Review: Optimized Strategies for Stem Cell based Therapy in Myocardial Repair: Clinical Translatability and Potential Limitation. Stem Cells [DOI] [PubMed] [Google Scholar]
- 12.Buikema JW, Wu SM (2017) Untangling the Biology of Genetic Cardiomyopathies with Pluripotent Stem Cell Disease Models. Curr Cardiol Rep 19 (4):30. [DOI] [PubMed] [Google Scholar]
- 13.Mordwinkin NM, Burridge PW, Wu JC (2013) A review of human pluripotent stem cell-derived cardiomyocytes for high-throughput drug discovery, cardiotoxicity screening, and publication standards. J Cardiovasc Transl Res 6 (1):22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang BY, Xiao Y, Hsieh A, Thavandiran N, Radisic M (2011) Micro- and nanotechnology in cardiovascular tissue engineering. Nanotechnology 22 (49). doi: Artn 494003 [DOI] [PubMed] [Google Scholar]
- 15.Falconnet D, Csucs G, Grandin HM, Textor M (2006) Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials 27 (16):3044–3063. doi: 10.1016/j.biomaterials.2005.12.024 [DOI] [PubMed] [Google Scholar]
- 16.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF (1999) Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials 20 (6):573–588. doi:Doi 10.1016/S0142-9612(98)00209-9 [DOI] [PubMed] [Google Scholar]
- 17.Bettinger CJ, Langer R, Borenstein JT (2009) Engineering Substrate Topography at the Micro- and Nanoscale to Control Cell Function. Angew Chem Int Edit 48 (30):5406–5415. doi: 10.1002/anie.200805179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehnert D, Wehrle-Haller B, David C, Weiland U, Ballestrem C, Imhof BA, Bastmeyer M (2004) Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J Cell Sci 117 (1):41–52. doi:DOI 10.1242/jcs.00836 [DOI] [PubMed] [Google Scholar]
- 19.Parker KK, Brock AL, Brangwynne C, Mannix RJ, Wang N, Ostuni E, Geisse NA, Adams JC, Whitesides GM, Ingber DE (2002) Directional control of lamellipodia extension by constraining cell shape and orienting cell tractional forces. Faseb J 16 (10). doi:UNSP 0892–6638/02/0016–1195 DOI 10.1096/fj.02-0038com [DOI] [PubMed] [Google Scholar]
- 20.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS (2004) Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6 (4):483–495. doi:Doi 10.1016/S1534-5807(04)00075-9 [DOI] [PubMed] [Google Scholar]
- 21.Thomas CH, Collier JH, Sfeir CS, Healy KE (2002) Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci U S A 99 (4):1972–1977. doi: 10.1073/pnas.032668799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilian KA, Bugarija B, Lahn BT, Mrksich M (2010) Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A 107 (11):4872–4877. doi: 10.1073/pnas.0903269107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaji H, Takii Y, Nishizawa M, Matsue T (2003) Pharmacological characterization of micropatterned cardiac myocytes. Biomaterials 24 (23):4239–4244 [DOI] [PubMed] [Google Scholar]
- 24.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE (1997) Geometric control of cell life and death. Science 276 (5317):1425–1428 [DOI] [PubMed] [Google Scholar]
- 25.Legant WR, Pathak A, Yang MT, Deshpande VS, McMeeking RM, Chen CS (2009) Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues. Proceedings of the National Academy of Sciences 106 (25):10097–10102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonard A, Bertero A, Powers JD, Beussman KM, Bhandari S, Regnier M, Murry CE, Sniadecki NJ (2018) Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues. J Mol Cell Cardiol 118:147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroer AK, Shotwell MS, Sidorov VY, Wikswo JP, Merryman WD (2017) I-Wire Heart-on-a-Chip II: Biomechanical analysis of contractile, three-dimensional cardiomyocyte tissue constructs. Acta Biomater 48:79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Lange WJ, Hegge LF, Grimes AC, Tong CW, Brost TM, Moss RL, Ralphe JC (2011) Neonatal mouse-derived engineered cardiac tissue: a novel model system for studying genetic heart disease. Circ Res 109 (1):8–19. doi: 10.1161/CIRCRESAHA.111.242354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shadrin IY, Allen BW, Qian Y, Jackman CP, Carlson AL, Juhas ME, Bursac N (2017) Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nature communications 8 (1):1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH (2004) Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305 (5686):1007–1009 [DOI] [PubMed] [Google Scholar]
- 31.Mertz J (2011) Optical sectioning microscopy with planar or structured illumination. Nat Methods 8 (10):811. [DOI] [PubMed] [Google Scholar]
- 32.Chen F, Tillberg PW, Boyden ES (2015) Expansion microscopy. Science:1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161 (5):1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW (2015) Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161 (5):1187–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes S (2004) The pathology of hypertrophic cardiomyopathy. Histopathology 44 (5):412–427 [DOI] [PubMed] [Google Scholar]
- 36.Hoskins AC, Jacques A, Bardswell SC, McKenna WJ, Tsang V, dos Remedios CG, Ehler E, Adams K, Jalilzadeh S, Avkiran M (2010) Normal passive viscoelasticity but abnormal myofibrillar force generation in human hypertrophic cardiomyopathy. J Mol Cell Cardiol 49 (5):737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fidzianska A, Glinka-Lindner Z, Religa G, Walczak E (2010) Usefulness of the ultrastructural and immunohistochemical analysis of cardiac biopsy in affected heart. Folia Neuropathol 48:57–63 [PubMed] [Google Scholar]
- 38.Zhao Y-T, Valdivia CR, Gurrola GB, Hernández JJ, Valdivia HH (2015) Arrhythmogenic mechanisms in ryanodine receptor channelopathies. Science China Life Sciences 58 (1):54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novak A, Barad L, Zeevi-Levin N, Shick R, Shtrichman R, Lorber A, Itskovitz-Eldor J, Binah O (2012) Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to beta-adrenergic stimulation. J Cell Mol Med 16 (3):468–482. doi: 10.1111/j.1582-4934.2011.01476.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halapas A, Papalois A, Stauropoulou A, Philippou A, Pissimissis N, Chatzigeorgiou A, Kamper E, Koutsilieris M (2008) In vivo models for heart failure research. In Vivo 22 (6):767–780 [PubMed] [Google Scholar]
- 41.Bers DM (2002) Cardiac excitation–contraction coupling. Nature 415 (6868):198–205 [DOI] [PubMed] [Google Scholar]
- 42.Endoh M (2004) Force–frequency relationship in intact mammalian ventricular myocardium: physiological and pathophysiological relevance. Eur J Pharmacol 500 (1):73–86 [DOI] [PubMed] [Google Scholar]
- 43.Dixon JA, Spinale FG (2009) Large animal models of heart failure a critical link in the translation of basic science to clinical practice. Circ Heart Fail 2 (3):262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM (1998) Embryonic stem cell lines derived from human blastocysts. science 282 (5391):1145–1147 [DOI] [PubMed] [Google Scholar]
- 45.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S (2007) Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25 (9):1015–1024 [DOI] [PubMed] [Google Scholar]
- 46.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5):861–872 [DOI] [PubMed] [Google Scholar]
- 47.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318 (5858):1917–1920 [DOI] [PubMed] [Google Scholar]
- 48.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP (2012) Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences 109 (27):E1848–E1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, Cashman J, Mercola M (2011) Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell–derived mesoderm. Circulation research 109 (4):360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang X, Pabon L, Murry CE (2014) Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res 114 (3):511–523. doi: 10.1161/CIRCRESAHA.114.300558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cimetta E, Pizzato S, Bollini S, Serena E, De Coppi P, Elvassore N (2009) Production of arrays of cardiac and skeletal muscle myofibers by micropatterning techniques on a soft substrate. Biomed Microdevices 11 (2):389–400. doi: 10.1007/s10544-008-9245-9 [DOI] [PubMed] [Google Scholar]
- 52.McDevitt TC, Angello JC, Whitney ML, Reinecke H, Hauschka SD, Murry CE, Stayton PS (2002) In vitro generation of differentiated cardiac myofibers on micropatterned laminin surfaces. J Biomed Mater Res 60 (3):472–479 [DOI] [PubMed] [Google Scholar]
- 53.Feinberg AW, Alford PW, Jin H, Ripplinger CM, Werdich AA, Sheehy SP, Grosberg A, Parker KK (2012) Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture. Biomaterials 33 (23):5732–5741. doi: 10.1016/j.biomaterials.2012.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bray MA, Sheehy SP, Parker KK (2008) Sarcomere alignment is regulated by myocyte shape. Cell Motil Cytoskel 65 (8):641–651. doi: 10.1002/cm.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chopra A, Patel A, Shieh AC, Janmey PA, Kresh JY (2012) alpha-Catenin Localization and Sarcomere Self-Organization on N-Cadherin Adhesive Patterns Are Myocyte Contractility Driven. Plos One 7 (10). doi:ARTN e47592 10.1371/journal.pone.0047592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serena E, Cimetta E, Zatti S, Zaglia T, Zagallo M, Keller G, Elvassore N (2012) Micro-arrayed human embryonic stem cells-derived cardiomyocytes for in vitro functional assay. PLoS One 7 (11):e48483. doi: 10.1371/journal.pone.0048483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ribeiro AJ, Ang YS, Fu JD, Rivas RN, Mohamed TM, Higgs GC, Srivastava D, Pruitt BL (2015) Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc Natl Acad Sci U S A 112 (41):12705–12710. doi: 10.1073/pnas.1508073112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126 (4):677–689 [DOI] [PubMed] [Google Scholar]
- 59.Bhana B, Iyer RK, Chen WL, Zhao R, Sider KL, Likhitpanichkul M, Simmons CA, Radisic M (2010) Influence of substrate stiffness on the phenotype of heart cells. Biotechnol Bioeng 105 (6):1148–1160. doi: 10.1002/bit.22647 [DOI] [PubMed] [Google Scholar]
- 60.Napiwocki BN, Salick MR, Ashton RS, Crone WC (2017) Controlling hESC-CM Cell Morphology on Patterned Substrates Over a Range of Stiffness In: Mechanics of Biological Systems and Materials, Volume 6 Springer, pp 161–168 [Google Scholar]
- 61.Napiwocki BN, Salick MR, Ashton RS, Crone WC (2016) Polydimethylsiloxane Lanes Enhance Sarcomere Organization in Human ESC-Derived Cardiomyocytes. In: Tekalur SA, Zavattieri P, Korach CS (eds) Mechanics of Biological Systems and Materials, Vol 6. Conference Proceedings of the Society for Experimental Mechanics Series. Springer, New York, pp 105–111. doi: 10.1007/978-3-319-21455-9_12 [DOI] [Google Scholar]
- 62.Notbohm J, Banerjee S, Utuje KJ, Gweon B, Jang H, Park Y, Shin J, Butler JP, Fredberg JJ, Marchetti MC (2016) Cellular contraction and polarization drive collective cellular motion. Biophysical journal 110 (12):2729–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Napiwocki BN, Stempien A, Notbohm J, Ashton RS, Crone W (2018) Two-Dimensional Culture Systems to Investigate Mechanical Interactions of the Cell In: Mechanics of Biological Systems, Materials and other topics in Experimental and Applied Mechanics, Volume 4 Springer, pp 37–39 [Google Scholar]
- 64.Pelham RJ, Wang YL (1997) Cell locomotion and focal adhesions are regulated by substrate flexibility. Proceedings of the National Academy of Sciences of the United States of America 94 (25):13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming WY, Weaver V, Janmey PA (2005) Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskel 60 (1):24–34. doi: 10.1002/cm.20041 [DOI] [PubMed] [Google Scholar]
- 66.Tse JR, Engler AJ (2010) Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol Chapter 10:Unit 10 16. doi: 10.1002/0471143030.cb1016s47 [DOI] [PubMed] [Google Scholar]
- 67.Kong HJ, Wong E, Mooney DJ (2003) Independent control of rigidity and toughness of polymeric hydrogels. Macromolecules 36 (12):4582–4588 [Google Scholar]
- 68.Rosellini E, Cristallini C, Barbani N, Vozzi G, Giusti P (2009) Preparation and characterization of alginate/gelatin blend films for cardiac tissue engineering. J Biomed Mater Res Part A 91 (2):447–453 [DOI] [PubMed] [Google Scholar]
- 69.Salick MR, Napiwocki BN, Sha J, Knight GT, Chindhy SA, Kamp TJ, Ashton RS, Crone WC (2014) Micropattern width dependent sarcomere development in human ESC-derived cardiomyocytes. Biomaterials 35 (15):4454–4464. doi: 10.1016/j.biomaterials.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dembo M, Wang YL (1999) Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J 76 (4):2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ (2002) Traction fields, moments, and strain energy that cells exert on their surroundings. American Journal of Physiology-Cell Physiology 282 (3):C595–C605 [DOI] [PubMed] [Google Scholar]
- 72.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y (2013) Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell stem cell 12 (1):127–137 [DOI] [PubMed] [Google Scholar]
- 73.Palchesko RN, Zhang L, Sun Y, Feinberg AW (2012) Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PloS one 7 (12):e51499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaefer JA, Tranquillo RT (2016) Tissue Contraction Force Microscopy for Optimization of Engineered Cardiac Tissue. Tissue Eng Part C Methods 22 (1):76–83. doi: 10.1089/ten.TEC.2015.0220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soiné JR, Hersch N, Dreissen G, Hampe N, Hoffmann B, Merkel R, Schwarz US (2016) Measuring cellular traction forces on non-planar substrates. Interface focus 6 (5):20160024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu HY, Xiong SJ, Tay CY, Leong WS, Tan LP (2012) A novel and simple microcontact printing technique for tacky, soft substrates and/or complex surfaces in soft tissue engineering. Acta Biomater 8 (3):1267–1272. doi: 10.1016/j.actbio.2011.09.006 [DOI] [PubMed] [Google Scholar]
- 77.Sutton MA, Orteu JJ, Schreier H (2009) Image correlation for shape, motion and deformation measurements: basic concepts, theory and applications. Springer Science & Business Media, [Google Scholar]
- 78.Blaber J, Adair B, Antoniou A (2015) Ncorr: Open-Source 2D Digital Image Correlation Matlab Software. Exp Mech 55 (6):1105–1122. doi: 10.1007/s11340-015-0009-1 [DOI] [Google Scholar]
- 79.Bar-Kochba E, Toyjanova J, Andrews E, Kim KS, Franck C (2015) A Fast Iterative Digital Volume Correlation Algorithm for Large Deformations. Exp Mech 55 (1):261–274. doi: 10.1007/s11340-014-98742 [DOI] [Google Scholar]
- 80.Sabass B, Gardel ML, Waterman CM, Schwarz US (2008) High resolution traction force microscopy based on experimental and computational advances. Biophys J 94 (1):207–220. doi: 10.1529/biophysj.107.113670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B, Safran SA (2002) Calculation of forces at focal adhesions from elastic substrate data: The effect of localized force and the need for regularization. Biophys J 83 (3):1380–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.del Alamo JC, Meili R, Alonso-Latorre B, Rodriguez-Rodriguez J, Aliseda A, Firtel RA, Lasheras JC (2007) Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. P Natl Acad Sci USA 104 (33):13343–13348. doi: 10.1073/pnas.0705815104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ (2009) Physical forces during collective cell migration. Nat Phys 5 (6):426–430. doi: 10.1038/nphys1269 [DOI] [Google Scholar]
- 84.Gerdes AM, Kellerman SE, Moore JA, Muffly KE, Clark LC, Reaves PY, Malec KB, McKeown PP, Schocken DD (1992) Structural remodeling of cardiac myocytes in patients with ischemic cardiomyopathy. Circulation 86 (2):426–430 [DOI] [PubMed] [Google Scholar]
- 85.Wrighton PJ, Klim JR, Hernandez BA, Koonce CH, Kamp TJ, Kiessling LL (2014) Signals from the surface modulate differentiation of human pluripotent stem cells through glycosaminoglycans and integrins. Proceedings of the National Academy of Sciences 111 (51):18126–18131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pillekamp F, Haustein M, Khalil M, Emmelheinz M, Nazzal R, Adelmann R, Nguemo F, Rubenchyk O, Pfannkuche K, Matzkies M (2012) Contractile properties of early human embryonic stem cell-derived cardiomyocytes: beta-adrenergic stimulation induces positive chronotropy and lusitropy but not inotropy. Stem cells and development 21 (12):2111–2121 [DOI] [PubMed] [Google Scholar]
- 87.Maruthamuthu V, Sabass B, Schwarz US, Gardel ML (2011) Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proceedings of the National Academy of Sciences of the United States of America 108 (12):4708–4713. doi: 10.1073/pnas.1011123108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, Zhou EHH, Zaman MH, Butler JP, Weitz DA, Fredberg JJ, Trepat X (2011) Collective cell guidance by cooperative intercellular forces. Nat Mater 10 (6):469–475. doi: 10.1038/nmat3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tambe DT, Croutelle U, Trepat X, Park CY, Kim JH, Millet E, Butler JP, Fredberg JJ (2013) Monolayer stress microscopy: limitations, artifacts, and accuracy of recovered intercellular stresses. Plos One 8 (2):e55172. doi: 10.1371/journal.pone.0055172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.