In this study, Kim et al. set out to elucidate the molecular details of how PIWI proteins in planarian flatworms contribute to stem cell function and regeneration. Using novel biochemical tools, such as IP-seq, ribodepletion, HITS-CLIP, and SHAPE-MaP, the authors show that PIWI proteins enable planarians to repurpose piRNAs for critical roles in neoblast mRNA turnover.
Keywords: PIWI proteins, RNA-binding proteins, S. mediterranea, mRNA surveillance, mRNA turnover, piRNAs, planarians, posttranscriptional gene silencing, regeneration, stem cells
Abstract
PIWI proteins utilize small RNAs called piRNAs to silence transposable elements, thereby protecting germline integrity. In planarian flatworms, PIWI proteins are essential for regeneration, which requires adult stem cells termed neoblasts. Here, we characterize planarian piRNAs and examine the roles of PIWI proteins in neoblast biology. We find that the planarian PIWI proteins SMEDWI-2 and SMEDWI-3 cooperate to degrade active transposons via the ping-pong cycle. Unexpectedly, we discover that SMEDWI-3 plays an additional role in planarian mRNA surveillance. While SMEDWI-3 degrades numerous neoblast mRNAs in a homotypic ping-pong cycle, it is also guided to another subset of neoblast mRNAs by antisense piRNAs and binds these without degrading them. Mechanistically, the distinct activities of SMEDWI-3 are primarily dictated by the degree of complementarity between target mRNAs and antisense piRNAs. Thus, PIWI proteins enable planarians to repurpose piRNAs for potentially critical roles in neoblast mRNA turnover.
Small non-coding RNAs direct cell differentiation, ensure genome integrity, and orchestrate developmental timing in metazoans. In contrast to microRNAs that are ubiquitously expressed and bound by Argonaute proteins to direct post-transcriptional gene regulation (Bartel 2018), piRNAs are mainly found in the germline (Aravin et al. 2006; Vagin et al. 2006). There, they bind to PIWI proteins (Cox et al. 1998), a subclass of the Argonaute protein family, whose major function is to degrade transposable elements. Because transposons are highly active in germ cells, loss of PIWI activity often leads to impaired germ cell function and sterility (Ku and Lin 2014). PIWI proteins degrade active transposons post-transcriptionally via the so-called ping-pong cycle (Brennecke et al. 2007; Gunawardane et al. 2007). The ping-pong cycle consists of sequential nucleolytic cleavages by two different PIWI proteins (heterotypic ping-pong) or twice by the same PIWI protein (homotypic ping-pong) during the degradation of transposable elements with the help of antisense piRNAs. The sequential cleavages thereby result in a characteristic 10-nt overlap of mature piRNAs, a signature indicative of an active ping-pong cycle. Furthermore, piRNAs suppress transposon activity in the nucleus by directing heterochromatin formation and thus preventing their transcription (Sienski et al. 2012 Le Thomas et al. 2013). In addition to transposon control, piRNAs also have transposon-independent functions. For instance, the murine PIWI protein Miwi was found to inhibit the precocious translation of mRNAs not needed until later stages of spermiogenesis (Vourekas et al. 2012). In D. melanogaster, piRNAs were found to direct the degradation of certain maternal mRNAs in the early embryo (Rouget et al. 2010). Finally, in the C. elegans germline piRNAs were demonstrated to establish a transgenerational epigenetic memory of self and non-self transcripts (Ashe et al. 2012; Shirayama et al. 2012).
Intriguingly, all three PIWI proteins present in the planarian flatworm Schmidtea mediterranea, termed SMEDWI-1, SMEDWI -2, and SMEDWI -3, are highly expressed in neoblasts, adult stem cells that constitute ∼20% of all cells in adult worms (Kang and Sánchez Alvarado 2009). The abundance of neoblasts equips planarians with remarkable regenerative abilities, making them an excellent model for studying whole-organ and body regeneration (Elliott and Sánchez Alvarado 2013). SMEDWI-2 and SMEDWI-3 are essential for planarian regeneration and tissue homeostasis (Reddien et al. 2005; Palakodeti et al. 2008;). Their knockdown abolishes the regenerative capacity of planarians and causes a characteristic phenotype that resembles that of irradiated animals, leading to the lysis of the animals on day 12 or day 22 for SMEDWI-2 and SMEDWI-3, respectively. Since an earlier report found that many planarian piRNAs map to exons (Friedländer et al. 2009), we set out to characterize the piRNA pathway in S. mediterranea. We were guided by our interest in deciphering the role of PIWI proteins in stem cell differentiation and development in an organism, where piRNA function can be studied in adult stem cells that give rise to all other tissues (Hayashi et al. 2006).
To unravel why PIWI proteins are essential for planarian stem cell function, we established immunoprecipitation protocols for all planarian PIWI proteins. Deep-sequencing of cobound piRNAs in conjunction with a sequenced genome (Grohme et al. 2018) allowed us to describe the principles of piRNA biogenesis in planarians. Our results provide strong evidence that active transposons are degraded via the ping-pong cycle by SMEDWI-2 and SMEDWI-3. Unexpectedly, a combination of HITS-CLIP, CLASH, small RNA-seq, Degradome-seq and RNA-seq also uncovered additional roles of SMEDWI-3 in mRNA turnover in planarians. By focusing on SMEDWI-3 targets, we found that two distinct modes of PIWI-mediated mRNA regulation are at play in planarian neoblasts. Specifically, one class of mRNAs is overwhelmingly degraded by SMEDWI-3 via a homotypic ping-pong cycle, in analogy to the degradation of transposable elements. In contrast, SMEDWI-3 binds another class of targets in at least partially unstructured regions without degrading them. It is guided to these transcripts by antisense piRNAs that bind their targets with non-perfect complementarity. Our results establish that planarians have repurposed parts of their abundant and complex piRNA reservoir to perform critical functions in mRNA turnover, which may contribute to the stem cell dysfunction phenotypes associated with SMEDWI-2 and -3 knockdowns.
Results
Immunoprecipitation of SMEDWI-bound planarian piRNAs
To unravel the role of PIWI proteins in planarians, we developed immunoprecipitation protocols for all three planarian PIWI proteins (Fig. 1A). Whereas the anti-SMEDWI-1 antibody used here was previously described (Guo et al. 2006), we raised polyclonal antibodies against SMEDWI-2 and SMEDWI-3 and confirmed their specificity for each PIWI protein (Supplemental Fig. S1A). Whole-mount immunofluorescence confirmed that SMEDWI-2 is enriched in the nuclei of neoblasts and differentiated cells (Zeng et al. 2013), while SMEDWI-1 and SMEDWI-3 are predominantly cytoplasmic and enriched in neoblasts (Supplemental Figs. S1B,C). SMEDWI-3 signal also overlapped with Y12 antibody staining, a marker of planarian chromatoid bodies, as shown previously (Rouhana et al. 2012). Despite the abundance of piRNAs in planarians (Supplemental Fig. S2A), the low osmolality of planarian tissues renders planarian PIWI proteins highly sensitive to ionic strength. As a result, we found that only HEPES in its hemisodium form sufficiently stabilizes planarian PIWI proteins in lysates (Supplemental Fig. S2B). In exploiting this finding, we successfully immunoprecipitated each individual PIWI protein with co-bound piRNAs, allowing for the construction and deep sequencing of small RNA libraries.
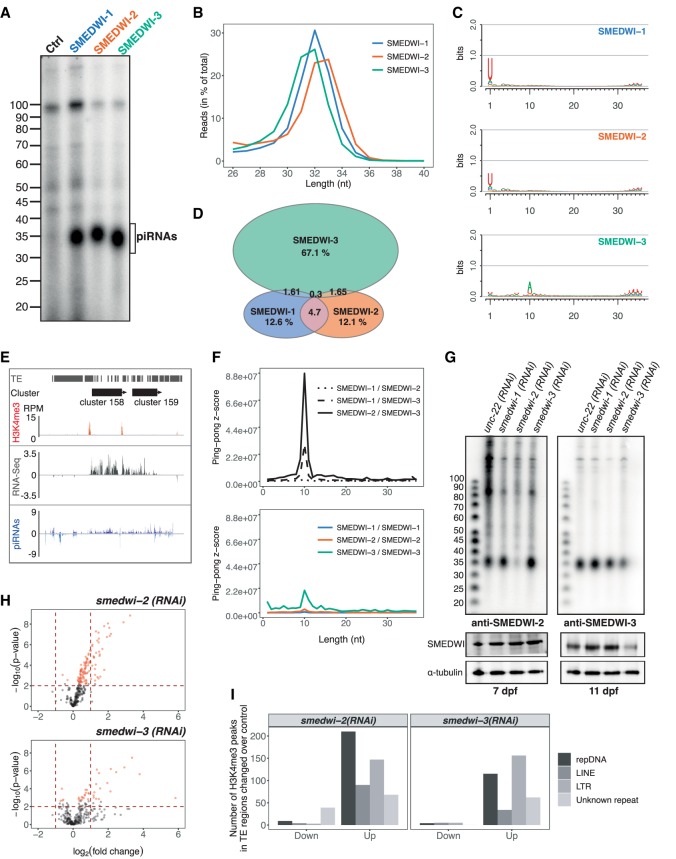
Figure 1.
Characterization of piRNAs bound to different SMEDWI proteins. (A) piRNAs were immunoprecipitated by use of anti-SMEDWI-1, anti-SMEDWI-2, and anti-SMEDWI-3 antibodies from total worm lysate, labeled with [γ-32P]-ATP and separated on a 10% Urea PAGE. Pre-immune serum served as negative control. (B) Size distribution of piRNAs bound to different planarian PIWI proteins. (C) Nucleotide composition of immunoprecipitated piRNAs plotted in the form of a position weight matrix with seqLogo. (D) Percentage of unique sequences bound by different SMEDWI proteins. Only genome-mapped piRNAs that appeared in at least two replicates were counted. (E) Coverage profile of H3K4me3 CHIP-seq, RNA-seq, and total piRNAs on unidirectional piRNA clusters 158 and 159. (F) Local Z-score for the occurrence of homotypic and heterotypic ping-pong pairs. (G) piRNAs coimmunoprecipitated from whole-worm lysates of control (unc-22 RNAi) or knockdown (smedwi-1,-2,-3 RNAi) animals using antibodies against SMEDWI-2 or -3. (Below) Western blot showing SMEDWI-2, SMEDWI-3 protein levels in different RNAi knockdown backgrounds. (H) Differentially expressed transposable element families upon RNAi knockdown of smedwi-2 or smedwi-3 (in red). The dashed horizontal red line represents the P-value threshold (P-value <0.01). The two dashed vertical lines illustrate the threshold of log2 fold changes >1 or < −1. (I) Numbers of up- and down-regulated H3K4me3 peaks mapping to transposable elements upon smedwi-2(RNAi) and smedwi-3(RNAi).
Subsequent analysis of piRNA sequences revealed that SMEDWI-1 binds piRNAs with a distribution maximum of 32 nt (Fig. 1B). In contrast, the length distribution of SMEDWI-2- and SMEDWI-3-bound piRNAs lies within a range of 32–33 and 31–32 nt, respectively. Sequence conservation analyses revealed that SMEDWI-1 and SMEDWI-2 bind piRNAs with a strong preference for a 5′-terminal uridine (78% and 67%, respectively) (Fig. 1C). In contrast, SMEDWI-3 binds piRNAs that show a +10 A bias (63%) and a slight conservation of a 5′-terminal uridine (41%). To assess the complexity of planarian piRNAs and to visualize potential sequence overlap between different PIWI proteins, we analyzed unique piRNAs bound by all three PIWI proteins that appeared in at least two replicates (Fig. 1D). Intriguingly, while SMEDWI-1 and SMEDWI-2 showed significant overlap in uniquely mapped piRNAs, SMEDWI-3 binds a distinct pool of piRNAs. Moreover, we detected 71% of SMEDWI-3-bound piRNAs only once, indicating that this piRNA pool is even more complex than we could resolve with our sequencing depth (Supplemental Fig. S2C). Altogether, our findings suggest that SMEDWI-3 has a distinct role in planarians that sets it apart from the two other PIWI proteins.
To examine how planarians are able to produce a highly complex and highly abundant piRNA repertoire, we determined the genomic location of planarian piRNA clusters using proTRAC (Rosenkranz and Zischler 2012). We were able to assign 270 piRNA clusters and found 268 of them to be unidirectional (Supplemental Table S1). Moreover, 14% of planarian piRNA clusters carry histone H3K4me3 marks, suggesting their euchromatic localization (Fig. 1E; Supplemental Fig. S2D). This confirms an earlier report (Friedländer et al. 2009) and resembles murine pachytene clusters and piRNA clusters in ovarian follicle cells in Drosophila. In both cases piRNA clusters show a strong strand bias (Aravin et al. 2006; Czech and Hannon 2016).
SMEDWI-2 and SMEDWI-3 cooperate to degrade planarian transposons
The recognition of an active transposon by antisense piRNAs can initiate the amplification of transposon silencing in the ping-pong cycle (Brennecke et al. 2007; Gunawardane et al. 2007). When analyzing planarian piRNAs for the presence of such signatures, we found that SMEDWI-2 and SMEDWI-3 are the major ping-pong players in planarians, accounting for the majority of ping-pong pairs detected in our piRNA libraries (Fig. 1F). We also observed a less pronounced ping-pong signature for SMEDWI-1 and SMEDWI-3, indicating that those two PIWI proteins also work together in degrading transposable elements. Additionally, SMEDWI-3-bound piRNAs show a homotypic ping-pong signature, whereas piRNAs bound to the other PIWI proteins do not (Fig. 1F).
Since the degradation of transposable elements is the primary function of PIWI proteins in other organisms, we set out to study transposon levels in planarians upon loss of SMEDWI-2 and SMEDWI-3. As both PIWI proteins are essential for planarian homeostasis, we devised an RNA interference (RNAi) knockdown strategy for SMEDWI-2 and SMEDWI-3 without depleting neoblasts. Optimal time points for RNAi knockdown experiments were defined by achieving the maximum possible depletion of SMEDWI-2 or SMEDWI-3 protein levels with only minor accompanying effects on SMEDWI-1, a marker of neoblast maintenance (Supplemental Fig. S2E). Whereas we observed a significant reduction in SMEDWI-3 protein levels upon smedwi-3(RNAi) on day 11 post feeding without affecting SMEDWI-1 expression (Fig. 1G, Supplemental Figs. S2F,G, S3), SMEDWI-2 largely persisted in both neoblasts and differentiated cells on day 7 post feeding. However, as the amount of coimmunoprecipitated piRNAs after SMEDWI-2 immunoprecipitation was drastically reduced upon smedwi-2(RNAi) (Fig. 1G), we speculate that at least part of the persisting pool of SMEDWI-2 is not loaded with piRNAs. Overall, we found the levels of piRNAs coimmunoprecipitated with SMEDWI-3 not altered under smedwi-2(RNAi) conditions and vice versa.
Following RNAi, worms were dissociated and separated by FACS into neoblasts (X1) and differentiated cells (Xins). In the asexual S. mediterranea, neoblasts are the only dividing cells and hence can be separated from differentiated cells by sorting for DNA content (Reddien et al. 2005; Hayashi et al. 2006). Total RNA from 100,000 FACS-isolated neoblasts was prepared and subjected to a custom ribosomal RNA (rRNA) depletion procedure. We developed this procedure to retain noncoding transcripts and those lacking long poly(A) tails (Supplemental Fig. S4A). Our protocol is robust and removes nearly 99% of the rRNA (Supplemental Fig. S4B). Following ribodepletion, we prepared strand-specific libraries for knockdown and control conditions as described (Zhang et al. 2012). Transposon levels after both SMEDWI-2 and SMEDWI-3 knockdown were analyzed by measuring the abundance of 316 consensus transposon families (Bao et al. 2015). Upon SMEDWI-2 knockdown, 109 transposon families were up-regulated, whereas the SMEDWI-3 knockdown increased the expression of only 38 families (Fig. 1H; Supplemental Fig. S4C).
As epigenetic silencing of transposable elements by PIWI proteins has been reported in other organisms (Sienski et al. 2012; Yang et al. 2016), we asked whether this is also the case in planarians. We performed CHIP-seq experiments for trimethylated histone H3 Lysine 4 (H3K4me3), a chromatin mark associated with active transcription (Howe et al. 2017). We found a considerable increase of H3K4me3 peaks at transposable elements upon smedwi-2(RNAi) and smedwi-3(RNAi) (Supplemental Fig. S4D). Out of 515 up-regulated peaks overlapping with transposable elements upon smedwi-2(RNAi), 40% were located at DNA transposons and 28% at long terminal repeat elements (LTRs) (Fig. 1I). Smedwi-3(RNAi) also led to an increase in the active transcription mark at DNA transposons (115 out of 367 up-regulated peaks mapped to transposable elements) and LTRs (156 peaks) (Fig. 1I). Altogether, the enrichment of SMEDWI-2 in the nucleus (Supplemental Fig. S1B) and the observed increase in the expression of transposable elements upon SMEDWI-2 and SMEDWI-3 knockdown (Supplemental Fig. S4C), indicate that SMEDWI-2 is likely directly involved in epigenetic silencing of transposable elements. Why a knockdown of SMEDWI-3 also leads to an increase in H3K4me3 peaks remains to be investigated.
Genic piRNAs bound to SMEDWI-3 are degradation products of planarian mRNAs
To decipher which genomic targets apart from transposable elements are subjected to piRNA-mediated regulation, we mapped all immunoprecipitated piRNAs to the planarian genome (Supplemental Methods). By allowing no mismatches and retaining multiple alignments we achieved mapping rates of 52%, 56%, and 43% for SMEDWI-1, -2, and -3-bound piRNAs, respectively. The mapping rates we obtained are rather low, likely due to the high degree of genome heterozygosity in planarians (Nishimura et al. 2015; Guo et al. 2017). Nonetheless, the alignment of SMEDWI-1, -2, and -3-bound piRNAs resulted in 2.8, 3.0, and 9.9 million uniquely mapped sequences, respectively. As expected, we found that the majority of planarian piRNAs mapped to unannotated regions and repetitive loci associated with transposable elements (Fig. 2A). However, a significant fraction of SMEDWI-3-bound piRNAs mapped to genic features, especially to coding regions, also displaying a strong sense bias and comparatively little multimapping (Fig. 2A,B; Supplemental Fig. S5A,B). What is more, SMEDWI-3-bound genic piRNAs do not map to annotated transposable elements, further strengthening our conclusion that these piRNAs are directly derived from mRNA transcripts (Supplemental Fig. S5C).
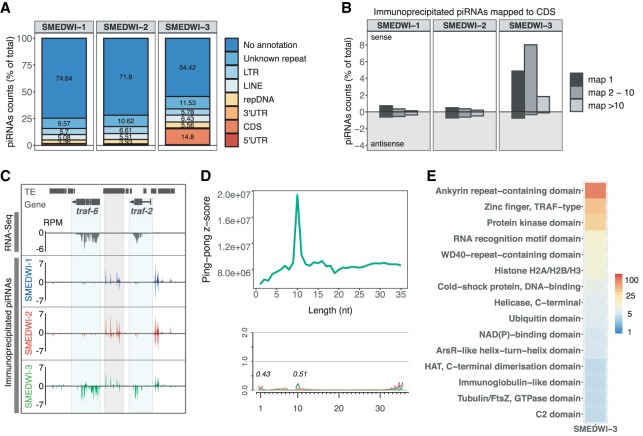
Figure 2.
SMEDWI-3 binds a diverse class of genic piRNAs. (A) Mapping of immunoprecipitated piRNAs to the planarian genome uncovers a distinct class of genic piRNAs bound to SMEDWI-3. piRNA counts weighted by the number of places they map to were used for the genomic annotation of piRNAs. (B) SMEDWI-3-bound piRNAs mapped to gene-coding regions show a strong sense bias. (C) Coverage profile of RNA-seq and immunoprecipitated piRNAs for traf-6 (SMESG000000371.1). The gene loci are highlighted in blue. A neighboring transposable element is highlighted in gray. (D) Ping-pong analysis of transcripts producing sense piRNAs. Nucleotide composition analysis of total piRNAs mapped to processed transcripts confirms the involvement of the ping-pong cycle. (E) Heatmap showing the enrichment of predicted InterPro protein domains in transcripts with mapped genic piRNAs.
We then focused our attention on mRNAs to which SMEDWI-3-bound piRNAs mapped with an average of 10 reads per million (RPM) in all three immunoprecipitation replicates. Using this cutoff, we identified 925 transcripts that are processed into SMEDWI-3 bound piRNAs (Supplemental Table S2). Figure 2C and Supplemental Figure S5D show two representative genes, traf-6 (SMESG000000371.1) and npk1-like (SMESG000017261.1), both of which are degraded into great numbers of SMEDWI-3 bound piRNAs. The resulting genic piRNAs map in a sense orientation exclusively to coding regions of the targeted genes. In contrast, we only detected negligible amounts of traf-6 and npk1-like piRNAs bound to SMEDWI-1 and SMEDWI-2. Neither of the two proteins showed the strong sense bias of mapped piRNAs that we find to be characteristic for SMEDWI-3. Furthermore, transposon-related elements near genes are not degraded into strand-specific SMEDWI-3-bound piRNAs, yet can be detected bound by all three PIWI proteins (Fig. 2C). We also detected significant ping-pong signatures and the characteristic 5′ 1U and 10A nucleotide biases that accompany ping-pong amplification for our list of genic SMEDWI-3 bound piRNAs (Fig. 2D). These data strongly suggest that the piRNA-mediated mRNA turnover results in genic piRNAs specifically bound to SMEDWI-3. Notably, the large number of targeted mRNAs contains ankyrin-repeat-containing domain, zinc-finger TRAF-type domain, histone fold, etc. (Fig. 2E), suggesting the possibility that piRNAs might regulate the expression levels of entire protein domain families.
Crosslinking immunoprecipitation confirms that SMEDWI-3 targets hundreds of planarian transcripts
To gain direct evidence for the involvement of SMEDWI-3 in planarian mRNA decay, we established a crosslinking immunoprecipitation (CLIP) protocol for SMEDWI-3 in planarians (Vourekas and Mourelatos 2014). Since UV irradiation does not penetrate tissues efficiently, we first rapidly dissociated planarians into a single cell suspension and then crosslinked the dissociated cells on ice at λ = 254 nm (Fig. 3A). Crosslinked protein–RNA complexes were isolated using the anti-SMEDWI-3 antibody and resolved by PAGE. The average size of RNAs crosslinked to SMEDWI-3 was below 200 nt, hinting at strong nucleolytic activity in planarian lysates (Supplemental Fig. S6A,B). We also used a modified CLASH (cross-linking ligation and sequencing hybrids) protocol to capture chimeric piRNA-mRNAs reads to gain direct experimental evidence of piRNA-target interactions (Fig. 3B,C; Helwak et al. 2013; Shen et al. 2018). Both HITS-CLIP and CLASH libraries were prepared from two crosslinked RNA species that were separately isolated: a shorter, very prominent species crosslinked to SMEDWI-3 (labeled “down”), and a smear of longer RNA fragments (labeled “up”) (Fig. 3B,C; Supplemental Fig. S6C).
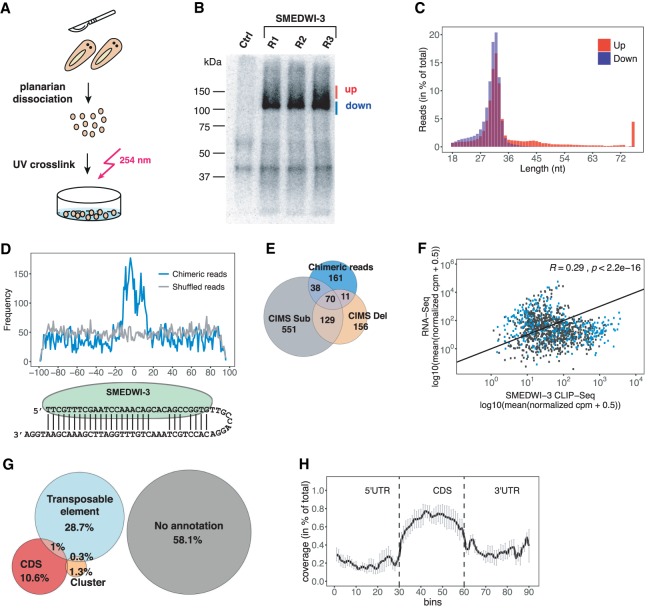
Figure 3.
Identification of SMEDWI-3-targeted transcripts by HITS-CLIP. (A) Schematic representation of HITS-CLIP in planarians. (B) Immunoprecipitated crosslinked SMEDWI-3–RNA complexes were radioactively labeled, separated on an 8% Bis-Tris gel, and blotted onto a nitrocellulose membrane. CLIP libraries were prepared from RNA extracted separately from the upper and lower bands. Pre-immune serum served as negative control. (C) Length distribution of CLIP reads from the upper and lower bands. (D) Alignment of the piRNAs parts of chimeric reads onto the ±100-nt extended midpoint of mRNA fragments. An alignment of piRNAs to random mRNA fragments was used as a negative control. (E) Venn diagram representing the identified SMEDWI-3 CLIP targets carrying deletions, substitutions, and chimeric reads. (F) Scatter plot comparing log2 normalized CPM (counts per million) of mapped reads for CLIP-seq and RNA-seq data. Transcripts carrying chimeric reads are highlighted in blue. The Spearman's correlation coefficient is indicated. (G) The annotation of CLIP fragments to the planarian genome confirms the involvement of SMEDWI-3 in the posttranscriptional regulation of transposable elements and coding transcripts. (H) Density of the CLIP fragments mapping to 5′-UTRs, coding regions, and 3′-UTRs of coding genes. Each feature was divided into 30 bins. The mean density of CLIP fragments was calculated over the corresponding features. Error bars represent SE.
In total, we analyzed nine CLIP replicates. After removal of PCR duplicates and rRNA filtering (Supplemental Fig. S6D), fragments from “up” and “down” libraries were pooled and jointly analyzed. CLIP reads were mapped to the planarian genome, resulting in a mapping rate of 55%–65%. This corresponds to between 0.4 and 6.6 million uniquely assigned reads per replicate (Supplemental Fig. S7A). Next, we applied crosslinking induced mutation site analysis (CIMS) to define CLIP sites in planarian mRNAs (Zhang and Darnell 2011; Shah et al. 2017). As reported for human Argonaute-2 and others, mutation-induced base substitutions were most frequent in our data (present in 56% of CLIP tags), while deletions accounted only for 13% of all mutations (Supplemental Figs. S7B,C; Moore et al. 2014). We focused our analysis on CLIP tags that carry both deletions (FDR ≤ 0.001) and substitutions (FDR ≤ 0.001) and that appear in at least two replicates, since both types of mutations were evenly distributed throughout the length of the CLIP read (Supplemental Fig. S7D). We detected 745 deletions in 366 mRNAs and 1629 substitutions in 788 mRNAs. All final CLIP tags comprise >30 unique reads in at least two replicates per site of mutation. Using the CLIP-chimaeric pipeline, our data also revealed thousands of chimeric piRNA-mRNA reads (Supplemental Fig. S7A; Vourekas et al. 2016; Alexiou et al. 2018). The piRNA parts of these chimeric reads were mapped on the extended mRNA fragments, and chimeric reads with alignments of piRNA parts within ±20 bp around the midpoint of the mRNA fragments were kept for the subsequent analysis (Fig. 3D; Supplemental Fig. S7E). In total, we discovered chimeric reads for 280 planarian mRNAs (Supplemental Table S3). The mRNAs carrying chimeric reads were also detected by our CIMS analysis (Fig. 3E).
After combining chimeric reads and CIMS-analyzed CLIP tags we were able to define 1116 transcripts as SMEDWI-3 targets (Fig. 3E; Supplemental Table S4). To compare the distribution of the reads mapping to genes in CLIP-seq and RNA-seq experiments, we calculated the Spearman correlation coefficient between both types of experiments. It revealed that the correlation between the gene expression and CLIP tag abundance is low (R = 0.29, P < 2.2 × 10−16) strengthening the specificity of our CLIP experiment (Fig. 3F). Moreover, we note that about 50% of all chimeric reads are derived from the top 25% of highly abundant transcripts in the CLIP libraries (Supplemental Fig. S7F). As expected, a significant fraction of CLIP reads (10%) could be traced back to coding regions of genes and not UTRs (Fig. 3G,H; Supplemental Fig. S7G). They preferentially map in sense orientation, confirming that SMEDWI-3 indeed targets coding transcripts (Supplemental Fig. S7G). In addition, 28% of our CLIP reads are derived from transposable elements, a fact that supports SMEDWI-3's central role in the ping-pong cycle (Figs. 1F, 3G).
SMEDWI-3 degrades planarian mRNAs using the ping-pong cycle and binds other mRNAs without degrading them
To dissect how planarian mRNAs are converted into piRNAs, we analyzed the abundance of SMEDWI-3-bound genic piRNAs in the identified CLIP regions. piRNAs were mapped to CLIP regions allowing one mismatch to account for planarian heterozygosity (Nishimura et al. 2015; Guo et al. 2017). Next, we computed the piRNA density per bp in all CLIP regions by normalizing weighted piRNA counts to the length of the corresponding CLIP regions. Unexpectedly, not all CLIP regions were densely covered with genic piRNAs. In contrast, our analysis revealed two distinct modes of SMEDWI-3 transcript targeting. A first class of transcripts displayed significant CLIP peaks across their coding regions and showed SMEDWI-3-bound piRNA coverage in the sense direction (Fig. 4A; Supplemental Fig. S8A). A second class of transcripts showed CLIP signals, yet hardly any piRNA coverage (<0.5 piRNA counts per bp of CLIP region) (Fig. 4B; Supplemental Fig. S8B).
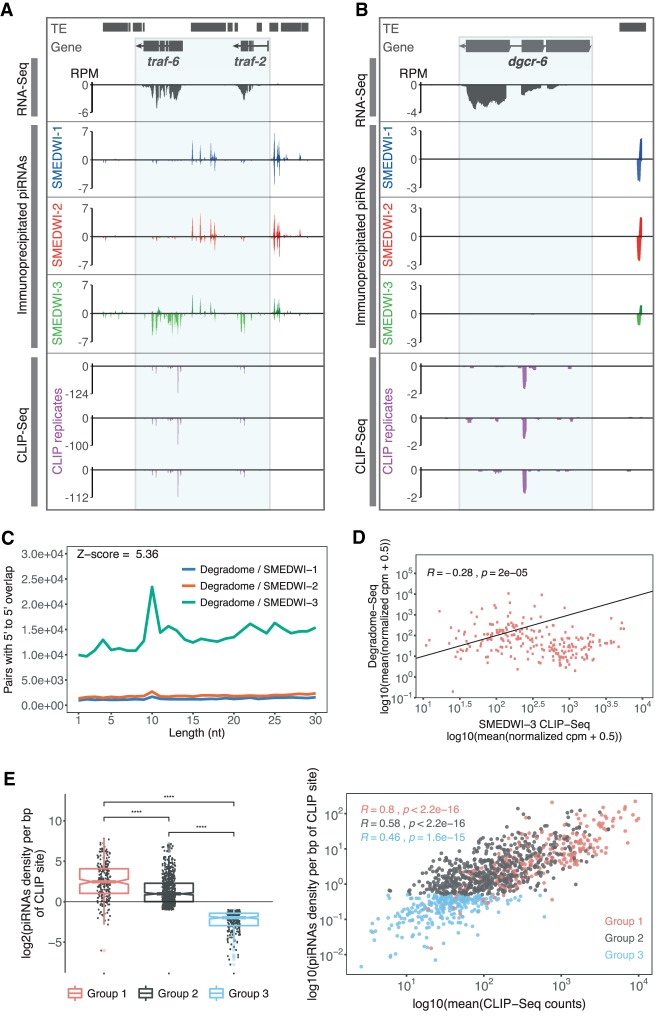
Figure 4.
The dual role of SMEDWI-3 in neoblast mRNA surveillance. (A) RNA-seq, piRNA-seq, and HITS-CLIP coverage profiles for traf-6 (SMESG000000371.1). The presence of both genic SMEDWI-3-bound piRNAs and CLIP fragments suggests their piRNA-mediated degradation by SMEDWI-3. (B) The presence of CLIP fragments along with the simultaneous absence of sense piRNAs for dgcr-6 (SMESG000021088.1) indicates that those transcripts are regulated by SMEDWI-3 in a cleavage-independent manner. (C) Number of unique pairs with 5′ to 5′ 10-nt overlaps between degradome sequences and piRNAs immunoprecipitated with anti-SMEDWI antibodies. (D) Correlation of normalized CPMs of CLIP-seq and Degradome-seq data. Each dot represents a transcript with ping-pong degradation signature. Spearman's correlation coefficient is indicated. (E, left) Boxplots of three groups of CLIP transcripts that are defined based on SMEDWI-3-bound genic piRNA density per bp of CLIP site and the presence of a piRNA-dependent degradation signature. The unpaired t-test was applied to estimate significant differences between groups. P-value ≤0.0001 is marked with (****). (Right) Scatter plot showing the cumulative density of genic piRNAs across the CLIP sites of SMEDWI-3 targets and the corresponding abundance of CLIP counts assigned to the transcript. Group 1 includes transcripts that display a ping-pong degradation pattern along with high genic piRNAs density. Group 2 comprises transcripts with piRNAs density ≥0.5/bp CLIP and without a piRNA-dependent degradation signature. Transcripts with a piRNA density <0.5/bp CLIP are sorted in group 3. Spearman's correlation coefficients for all three groups are indicated in different colors.
The presence of genic piRNAs in CLIP regions suggested that these targets might be cleaved and processed into piRNAs. To confirm that the degradation of SMEDWI-3 CLIP targets is indeed piRNA-dependent, we performed 5′-monophosphate-dependent cloning of mRNA fragments (Degradome-seq) in sorted planarian neoblasts (X1 cells). We focused on X1 cells since SMEDWI-3 is predominantly present in these cells (Supplemental Fig. S1C). Degradome-seq allows the capture of RNA cleavage products that result from piRNA targeting (Reuter et al. 2011; Wang et al. 2014). In total, we were able to map 10,230,560, 10,346,869, and 10,372,010 unique degradome reads to the planarian genome for three biological replicates, respectively. Approximately 9% of all degradome reads mapped to gene coding regions, with 1% possessing a 5′-5′ 10-nt overlap with immunoprecipitated piRNAs (Fig. 4C; Supplemental Fig. S8C). We noticed that the vast majority of ping-pong pairs between degradome reads and PIWI-bound piRNAs are formed with piRNAs bound by SMEDWI-3 (Z-score = 5.36). This suggests that SMEDWI-3 plays a leading role in degrading planarian transcripts that are not transposable elements.
Next, we intersected the list of genes showing piRNA-dependent degradation with our list of 1116 CLIP targets (Supplemental Fig. S8D). Using at least five unique ping-pong pairs per transcript as cutoff, we identified 232 SMEDWI-3 CLIP targets to be processed into genic piRNAs (Fig. 4D; Supplemental Table S5). CLIP targets with stronger degradation signal were slightly less abundant in our CLIP-seq libraries (R = −0.28, P = 2 × 10−5), a fact likely attributed to their efficient degradation into genic piRNAs. In agreement with the latter, the majority of degraded transcripts showed a high density of genic piRNAs across their CLIP sites (Fig. 4E). We therefore combined these SMEDWI-3 targets into a first group of targets (group 1). Subsequently, we assigned 621 transcripts to a second group of SMEDWI-3 targets (group 2). Group 2 targets exhibit substantial density of genic piRNAs (>0.5 piRNAs/bp of CLIP region); however, we did not detect any ping-pong degradation pairs for these transcripts in our degradome data (Fig. 4E). Most likely, group 2 transcripts are degraded in the ping-pong cycle as well; however, their degradation might be triggered by piRNAs that exhibit non-perfect base-pairing for target recognition. As we only considered piRNAs mapping to the genome with a single mismatch, we have no means to detect these cases. Combining all SMEDWI-3 CLIP targets with significant genic piRNA density (0.5 piRNAs/CLIP bp) left 263 transcripts. For these group 3 transcripts we detected significant SMEDWI-3 binding, yet very little piRNA mapping (<0.5 piRNAs/CLIP bp) (Fig. 4E; Supplemental Table S6). The correlation coefficients for the three groups of transcripts support our classification, as the positive correlation between the number of CLIP reads and the density of genic piRNAs decreases from group 1 through group 3. We therefore conclude that SMEDWI-3 binds group 3 transcripts, but does not trigger their degradation into significant numbers of genic piRNAs.
The base-pairing pattern between SMEDWI-3-bound piRNAs and target mRNAs determines whether SMEDWI-3 degrades or merely binds its targets
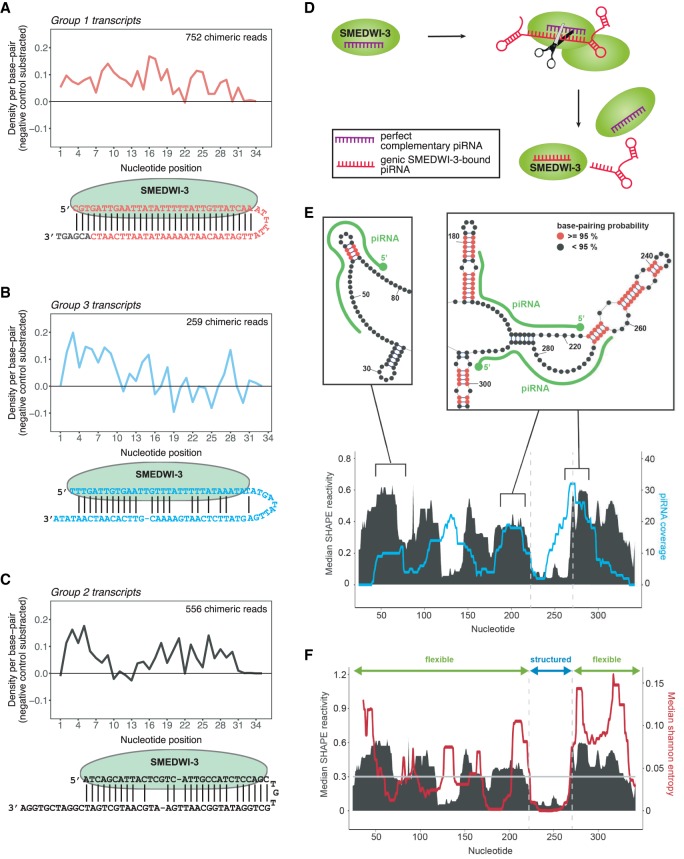
To investigate whether piRNAs are responsible for guiding SMEDWI-3 to group 3 transcripts, we examined the presence of chimeric reads mapping to this group of transcripts. In total, we identified 259 chimeric reads mapping to 75 group 3 transcripts. In addition, our data revealed 752 chimeric reads for 88 group 1 transcripts and 556 for 117 group 2 transcripts. To determine the base-pairing patterns underlying the interaction of SMEDWI-3-bound piRNAs with all three groups of transcripts, the unweighted local read aligner as implemented in the CLIP-chimeric pipeline was used (Vourekas et al. 2016; Alexiou et al. 2018). After subtraction of a randomized control, we observed continuous base pairing over the whole length of antisense piRNAs for group 1 targets (Fig. 5A). This observation confirms that antisense piRNAs need to recognize their targets with perfect or near-perfect complementarity to initiate target cleavage. It also confirms prior knowledge on cleavage-competent Argonaute proteins, which require continuous base-pairing across the scissile phosphate between target nucleotides 10 and 11 (Schirle et al. 2014). Meanwhile, piRNAs that target group 3 transcripts show a significant drop in base-pairing at nucleotides 10 and 11 and a general lack of base-pairing between nucleotide positions 16 and 26 (Fig. 5B). These data argue that group 3 transcripts are recognized by SMEDWI-3 with piRNAs as guides. However, target cleavage by SMEDWI-3 is likely precluded due to the multitude of mismatches between piRNA and mRNA target. In support of this notion, group 3 transcripts show a low density of genic piRNAs across their CLIP sites. Last, when analyzing the base-pairing patterns of group 2 chimeras we noticed almost continuous piRNA-mRNA base-pairing apart from a significant drop thereof between nucleotide positions 10 and 13 (Fig. 5C). Because we detected significant genic piRNAs for this group of transcripts, yet no degradome signal (Fig. 4E), we speculate that this drop may illustrate that efficient target cleavage is impaired for these transcripts due to a lack of base-pairing over the scissile phosphate group. Alternatively, due to limitations of our classification of group 2 transcripts, this group might include transcripts from both group 1 and group 3. Taken together, our results are in agreement with the previously described piRNA targeting rules in C. elegans (Shen et al. 2018; Zhang et al. 2018). Moreover, they are also in line with the rules established for Argonaute targeting and underline the importance of continuous base-pairing over the scissile phosphate bond to initiate target cleavage (Fig. 5D; Schirle et al. 2014).
Figure 5.
The base-pairing patterns between SMEDWI-3-bound piRNAs and target mRNAs. (A) Average density of base-pairing events per nucleotide position of the piRNA parts of chimeric reads. Chimeric piRNA parts were mapped onto targeted mRNA fragments in the vicinity of ±20 nt from the mRNA fragment midpoint. Only unique chimeric reads mapping to transcripts of Group 1 were included from all nine replicates. Random piRNA mapping densities were subtracted as negative control. (Below) An illustration of one exemplary chimeric read from group 1 transcripts is shown. The extended mRNA part of the chimeric read is shown in gray. (B) Same as in A for group 3 SMEDWI-3 targets. (C) Same as in A for group 2 SMEDWI-3 targets. (D) SMEDWI-3 degrades a set of mRNAs in a homotypical ping-pong cycle. It recognizes these transcripts with a high degree of complementarity between the SMEDWI-3-bound piRNA and the target mRNA. (E) Plot showing the correlation between experimentally determined median SHAPE reactivity (in black) and the mapping profile of unique predicted antisense piRNAs (in cyan) for histone H2B (SMESG000052758.1). For piRNA mapping, one mismatch was allowed aside from a mandatory seed match for nucleotides 2–8. Exemplary regions from the SHAPE reactivity-constrained secondary structure model are shown above the plots. Double-stranded regions with base-pairing probabilities >0.95 are colored in red. piRNAs (green) are drawn along the structures. (F) Plot showing the correlation between median SHAPE reactivity (in black) and median Shannon entropy (in red). Transcript regions with median SHAPE reactivity <0.3 and a Shannon entropy of <0.04 were considered structured.
piRNAs guide SMEDWI-3 to at least partially unstructured regions in its targets
Our results indicate that for all instances of target recognition, SMEDWI-3 is guided by a cobound piRNA. This necessitates that the piRNA target sites on SMEDWI-3-targeted mRNAs are available to base-pairing interactions. To characterize the piRNA-target interactions we therefore determined the structures of three SMEDWI-3 CLIP targets using ex vivo SHAPE-MaP (selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling), a powerful technique for RNA secondary structure probing (Smola et al. 2015b). In total, we collected sufficiently deep sequencing data for three SMEDWI-3 CLIP targets: one group 1 target (histone H2B) (SMESG000052758.1), one group 2 target (smedwi-3) (SMESG000081970.1), and one group 3 target (MYH15) (SMESG000056451.1). We used the computed SHAPE reactivities to constrain secondary structure modeling as implemented in RNAstructure (Fig. 5E, Extended Supplemental Data; Reuter and Mathews 2010).
Next, we extracted predicted piRNA-binding events across all three CLIP targets using a custom script (Supplemental Methods). We had to rely on predictions here since our CLASH data only comprised very few chimeric reads for the three mRNAs. Figure 5D shows the median SHAPE reactivity for histone H2B in conjunction with predicted piRNA binding events across the entire mRNA sequence. To analyze a possible correlation between mRNA structure and piRNA binding, we defined structured regions as having a median SHAPE reactivity <0.3 and a Shannon entropy of <0.04 (Mustoe et al. 2018). Using this definition, we found that for histone H2B 77% of piRNA binding sites fall in unstructured regions, whereas the entire histone H2B mRNA is to 74% unstructured (Fig. 5F). Clearly, our data does not support that all piRNA binding sites are obligatory single-stranded. However, in conjunction with the results for smedwi-3 and MYH15 mRNA (Extended Supplemental Data), our data suggest that piRNA target sites need to be at least partially unstructured. This is conceivable because guide RNA-target interactions by Argonaute proteins propagate after stable association of the seed region and thereby might be able to unfold loosely folded structures (Chandradoss et al. 2015). Our data also confirm an earlier report using a synthetic piRNA sensor in C. elegans (Zhang et al. 2018). There, as in our case, piRNA binding sites were at times found in structured regions. Therefore, we propose that only experimentally determined RNA structures in combination with sufficiently deep CLASH data will enable us to fully correlate mRNA structure and piRNAs binding sites.
Transcripts degraded by SMEDWI-3 reveal a piRNA pathway in the planarian epidermis
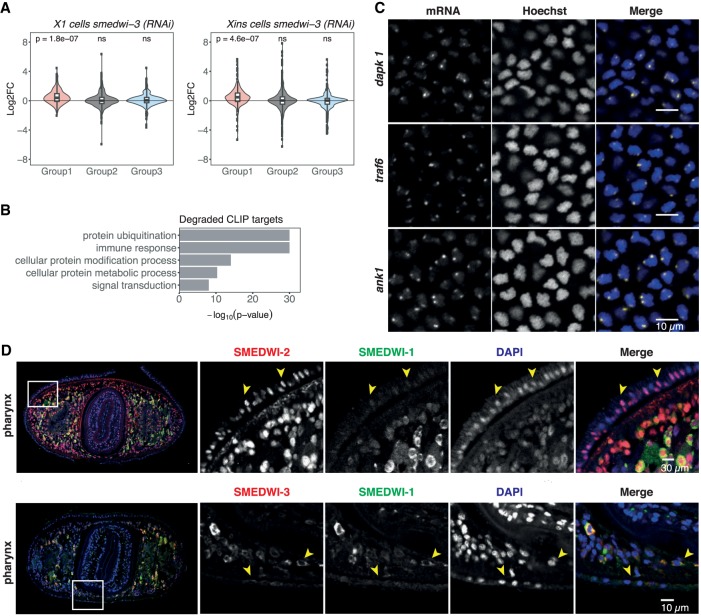
To decipher the impact of SMEDWI-3 on its target mRNAs, we examined the expression levels of transcripts in all three previously defined groups after smedwi-3(RNAi) knockdown. A gene set enrichment analysis (Luo et al. 2009) revealed that the expression of ping-pong degraded group 1 targets increased significantly in both smedwi-3(RNAi) neoblasts and differentiated cells (Fig. 6A). However, we also observed a significant up-regulation of group 1 targets upon smedwi-2(RNAi) (Supplemental Fig. S9A). We therefore speculate that piRNA pathway-dependent factors contribute to the degradation of these mRNAs or that SMEDWI-2 is involved in piRNAs precursor transcription and processing. Alternatively, a knockdown of either PIWI protein might disturb neoblasts fate and lead to the up-regulation of SMEDWI-3 ping-pong targets (group 1). We note here that group 1 targets are enriched in transcripts involved in protein ubiquitination and immune response (Fig. 6B). Overall, group 2 and group 3 targets do not show significant expression changes upon SMEDWI-2 and SMEDWI-3 knockdown. This suggests that SMEDWI-3 binding to these mRNAs does not have an immediate impact on their stability. However, their interaction with SMEDWI-3 might still be important by either licensing these transcripts in a type of surveillance pathway or by regulating their translation (Vourekas et al. 2012; Seth et al. 2018).
Figure 6.
SMEDWI-3 group 1 ping-pong targets form nuclear foci in the planarian epidermis. (A) Boxplots showing the log2 fold differential expression changes of the three distinct SMEDWI-3 target groups upon smedwi-3 knockdown in neoblasts (X1) and differentiated cells (Xins). Statistical significance of differential expression of the gene sets was assessed using generally applicable gene-set enrichment (GAGE) analysis with a two-sample t-test (ns, not significant). (B) GO term enrichment analysis of ping-pong degraded group 1 SMEDWI-3 targets. (C) Expression and localization pattern of three exemplary ping-pong targets of SMEDWI-3 [dapk1 (SMESG000043474.1), traf6 (SMESG000000294.1), ank1 (SMESG000076223.1)] in the planarian epidermis analyzed with whole-mount in situ hybridization (WISH). Nuclei stained with Hoechst are in blue, mRNAs are in yellow. (D) Coimmunostaining of SMEDWI-1, -2, and -3 in the epidermis on a cross-section through the planarian pharynx. SMEDWI-1 is in green, SMEDWI-2 and -3 are in red. Nuclei stained with DAPI are in blue.
Given the significant up-regulation of SMEDWI-3 group 1 targets in neoblasts and differentiated cells, we asked where these targets are expressed in planarians. We carried out whole-mount fluorescent in situ hybridization (WISH) of four group 1 transcripts that we found targeted by the ping-pong cycle [histone H2B (SMESG000052758.1), ank1 (SMESG000076223.1), dapk1 (SMESG000043474.1), traf-6 (SMESG000000294.1)]. Staining for histone H2B confirmed an earlier report that found histone mRNAs to be enriched in chromatoid bodies of neoblasts (Supplemental Fig. S9B; Rouhana et al. 2014). However, we were unable to detect the other three transcripts in neoblasts, a fact likely owed to their low expression levels (average expression level 23 TPM) when compared with histone H2B mRNA (22,677 TPM). Instead, we detected strong punctuated staining for all three transcripts in the epidermal cell layer in planarians (Fig. 6C). This finding was unexpected, since the expression levels of these transcripts in differentiated cells (Xins) is low (ank1 = 2.3 TPM, traf6 = 37 TPM, dapk 1 = 97.7 TPM). To understand the nature of the foci, we quantified their exact localization and found them to be predominantly nuclear. More precisely, they were enriched in the nuclear periphery (Supplemental Fig. S9C). Moreover, using immunostaining on sectioned animals, we could demonstrate that SMEDWI-2 is present in the epidermis, whereas the amount of SMEDWI-3 is negligible (Fig. 6D; Supplemental Fig. S9D). We found the observed nuclear foci to be reminiscent of the accumulation of piRNA precursors in Dot COM structures in somatic cells in Drosophila (Dennis et al. 2013). Thus, we suspect that the three tested transcripts might actually be processed just like piRNA precursors in the epidermis. Taken together, our in situ data on group 1 SMEDWI-3 targets demonstrates that planarians possess an epidermal somatic piRNA pathway involving likely only a single PIWI protein, SMEDWI-2. That is in addition to the piRNA pathway, which operates in neoblasts and requires the presence of at least SMEDWI-2 and SMEDWI-3 (Reddien et al. 2005; Palakodeti et al. 2008).
Discussion
The piRNA pathway was discovered in the germline, where it orchestrates germline development and ensures genome integrity by silencing of transposable elements (Cox et al. 1998; Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006). However, piRNAs were recently found to also have important roles in somatic cells such as neurons, cancer cells, embryonic stem cells, and adult somatic stem cells (van Wolfswinkel 2014; Rojas-Ríos and Simonelig 2018). Somatic piRNAs are widely expressed across arthropods, where they target both transposable elements and protein-coding transcripts. Yet the best-studied arthropod, D. melanogaster, only expresses piRNAs in the germline, thus representing an evolutionary exception in its phylum (Lewis et al. 2018). Therefore, much remains to be understood about the role of piRNAs in somatic tissues of animals. In this study, we developed an arsenal of biochemical tools that will now make it possible to further dissect piRNA function in the planarian S. mediterranea—an organism that neither senesces nor develops stem cell-based diseases like cancer (Pearson and Sanchez Alvarado 2008; Sahu et al. 2017).
Planarian piRNAs may function in cell cycle regulation and immune defense
Embryonic stem cells are transcriptionally hyperactive. This allows for the expression of repetitive sequences and mobile elements as well as the expression of lineage- and tissue-specific genes at low levels (Efroni et al. 2008). In addition to their role as a counteracting force to detrimental transposon activity (Fig. 1H,I), we show that the planarian PIWI protein SMEDWI-3 utilizes a diverse set of piRNAs to degrade, among others, a multitude of traf mRNAs, histone mRNAs, mRNAs coding for E3 ubiquitin ligases, and transcripts containing ankyrin repeats (Supplemental Table S5). These results suggest that piRNA-mediated degradation of SMEDWI-3 group 1 targets might be important for planarian regeneration and neoblasts cell cycle control. In addition, a modulation of the histone mRNA metabolism has the potential to directly control cell cycle progression and cell division (Marzluff and Duronio 2002). Moreover, E2- and E3-ubiquitin ligases are known to regulate the proteolysis of key cell cycle-regulatory proteins, chromosome separation, cytokinesis, and cell differentiation (Teixeira and Reed 2013). As in other organisms, secondary piRNA production operates in chromatoid bodies or the nuage (Lim and Kai 2007); we envision that SMEDWI-3 needs to be localized to chromatoid bodies for the degradation of transposons and mRNAs in the ping-pong cycle (Supplemental Figs. S1B, S9B; Lim and Kai 2007; Rouhana et al. 2014; Kashima et al. 2016).
In addition to the piRNA pathway in neoblasts, we also found an active piRNA pathway in epidermal cells operating with the use of SMEDWI-2 only (Fig. 6C,D). As epidermal cells do not divide and rapidly turn over, we hypothesize that the function of the epidermal piRNA pathway goes beyond the maintenance of genome integrity. We propose that piRNAs in the planarian epidermis might play a role in innate immunity along with the role of the epidermis in phagocytic cell responses, the secretion of antimicrobial mucus (Peiris et al. 2014), and the response to bacterial infection (Arnold et al. 2016). The piRNA pathway also plays an important role in epithelium cell metabolism and development. In support, planarian myb-1, whose mouse homolog initiates pachytene piRNA production in testes (Li et al. 2013), was recently shown to be responsible for the progression of the epidermal lineage, in particular for the specification of the early progeny cell state (prog-1) (Zhu and Pearson 2018). Moreover, we note that smedwi-3(RNAi) treatment leads to skin lesions in planarians on days 13–14 post RNAi, well before the animal lyses (21 d post-RNAi) (Palakodeti et al. 2008). This might be due to reduced SMEDWI-2 protein levels in the epidermis of smedwi-3(RNAi) animals (Supplemental Figs. S3, S9D).
The potential role of SMEDWI-3 in determining neoblast mRNA fate
SMEDWI-3, apart from degrading planarian mRNAs by targeting their coding sequences in the ping-pong cycle, specifically binds numerous planarian mRNAs, including its own. The key to these seemingly opposed SMEDWI-3 activities might lie in the presence or absence of antisense piRNAs that exhibit distinct base-pairing patterns when recognizing their mRNA targets (Fig. 5A–C). We suspect that the piRNA-targeted regions in these transcripts need to be devoid of stably folded structures for efficient targeting (Fig. 5E,F). Moreover, piRNAs render it unnecessary to evolve conserved nucleotide binding motifs for each mRNA to be targeted.
We hypothesize that the degradation-independent interaction of SMEDWI-3 with group 3 transcripts might be a sign of transcript binding by SMEDWI-3, analogous to mRNA surveillance in the C. elegans germline. There, PRG-1 and CSR-1 act out a functional rivalry to achieve the degradation of transcripts that are rated foreign and to establish an epigenetic memory of this distinction (Ashe et al. 2012; Lee et al. 2012; Shirayama et al. 2012; Seth et al. 2013). However, at this point we are not certain whether SMEDWI-3-mediated piRNA–mRNA interactions that do not trigger mRNA degradation persist long enough to have a biological function in planarians. Whether SMEDWI-3 is involved in recognizing and licensing neoblast mRNAs in the planarian S. mediterranea will thus require a thorough study of the piRNA pathway response to the invasion of foreign nucleic acids. Furthermore, to dissect the molecular mechanism of piRNA-mediated mRNA turnover biochemical studies addressing the function and composition of planarian chromatoid bodies as well as a thorough investigation of the role of the planarian epidermis in immune defense are necessary.
Materials and methods
Antibodies
The SMEDWI-2 antibody was generated by immunizing rabbits with the peptide KKDEEGVEKEK. For the production of a polyclonal anti-SMEDWI-3 antibody, the N-terminal 200 amino acids of SMEDWI-3 were fused to GST, expressed in E. coli, and then used for immunization. Obtained SMEDWI-3 antiserum was further purified against the antigen coupled to NHS-activated sepharose.
Immunoprecipitation of SMEDWI proteins
Approximately 150 worms (7–10 mm) were collected, snap-frozen in liquid nitrogen, and homogenized in 4 mL lysis buffer (30 mM HEPES, pH 7.7 [Sigma, H989], 150 mM NaCl, 10 mM KCl, 4 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, Complete EDTA-free protease inhibitor [Roche]) with a Dounce homogenizer. The lysate was cleared by centrifugation at 50,000g for 30 min at 4°C, followed by filtering it through a 0.20-µm cellulose acetate syringe filter. For each immunoprecipitation experiment 1 mL of cleared lysate (protein concentration 3 mg/mL) was incubated with 10 µg purified antibody for 2 h at 4°C with gentle rotation. Pre-immune serum was used as negative control. Next, 50 µL pre-equilibrated Dynabeads Protein A (Invitrogen) were added to each sample and incubation was continued for an additional 2 h at 4°C with gentle rotation. Beads were washed twice with low-salt buffer (30 mM HEPES, pH 7.7, 150 mM NaCl, 10 mM KCl, 4 mM MgCl2, 5 mM EDTA, 1 mM DTT, 0.1% Triton X-100) and twice with high-salt buffer (30 mM HEPES, pH 7.7, 300 mM NaCl, 10 mM KCl, 4 mM MgCl2, 5 mM EDTA, 1 mM DTT, 0.1% Triton X-100). Next, beads were transferred to a new tube and resuspended in 200 µL Proteinase K buffer (200 mM Tris, pH 7.5, 300 mM NaCl, 25 mM EDTA, 1.5% SDS) containing 120 µg/mL Proteinase K (Roche). To extract piRNAs that coimmunoprecipitated with SMEDWI proteins, beads were incubated with Proteinase K for 20 min at 42°C and 1000 rpm, followed by phenol-chloroform extraction and ethanol precipitation of all isolated nucleic acids.
Small RNA library preparation
Small RNA libraries were constructed as described (Hauptmann et al. 2015) with some modifications. Briefly, RNA coimmunoprecipitated by anti-SMEDWI-1, anti-SMEDWI-2, or anti-SMEDWI-3 antibodies was used for small RNA cloning. Pre-adenylated 3′-adapters were ligated to the 3′-end of all RNAs by a truncated T4 RNA Ligase 2 in the presence of 10% PEG-8000 at 16°C overnight (PEG-8000 considerably improved the ligation reaction due to the modified piRNA 3′-end). In a second ligation step, T4 RNA Ligase 1 was used to add an RNA adapter to the 5′-end of all RNAs at 37°C for 1 h. Deep sequencing was performed on Illumina HiSeq1000 (single-end 50 nt) or Illumina NextSeq 500 (single-end 75 nt) platforms.
rRNA depletion
To deplete planarian rRNA a pool of 88 3′-biotin-labeled DNA probes with a length of 40 nt were synthesized (siTOOLs Biotech, Martinsried, Germany). Total planarian RNA from 100,000 FACS-sorted cells was extracted with Trizol. Next, DNA probes were hybridized to planarian rRNA in hybridization buffer (10 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM EDTA). The RNA-probe hybrids were then separated twice from total RNA by use of Dynabeads MyOne Streptavidin C1 (invitrogen) according to the manufacturer's protocol. Finally, the supernatant was collected, concentrated with Zymo-spin IC columns with size selection ≥200 nt and treated with DNaseI.
SMEDWI-3 HITS-CLIP
SMEDWI-3 HITS-CLIP libraries were prepared as described (Vourekas and Mourelatos 2014) with the following modifications: Approximately 50 worms (7–10 mm) were dissociated in cold calcium- and magnesium-free buffer with 1% BSA (CMFB). Next, cells were irradiated at 254 nm using a Stratalinker 1800 (Stratagene) once with 400 mJ/cm2 and then again after 30 sec at 200 mJ/cm2 (Moore et al. 2014). Then cells were pelleted, snap-frozen in liquid nitrogen, and resuspended in lysis buffer (30 mM HEPES, pH 7.7, 150 mM NaCl, 10 mM KCl, 4 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 0.8 units/µL RNasin [Promega], Complete EDTA-free protease inhibitors). After treatment with DNase I the whole-cell extract was centrifuged at 50,000g for 30 min at 4°C. It was then incubated with 18 µg of anti-SMEDWI-3 antibody for 2 h at 4°C with gentle rotation, followed by incubation with 150 µL Dynabeads Protein A slurry (Invitrogen) for another 2 h at 4°C with gentle rotation. Next, beads were washed twice with low-salt buffer (30 mM HEPES, pH 7.7, 150 mM NaCl, 10 mM KCl, 4 mM MgCl2, 5 mM EDTA, 1 mM DTT, 0.1% Triton X-100), twice with high-salt buffer (30 mM HEPES, pH 7.7, 500 mM NaCl, 10 mM KCl, 4 mM MgCl2, 5 mM EDTA, 1 mM DTT, 0.1% Triton X-100), and twice with 1x PNK buffer (70 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 5 mM DTT, 0.1% Triton). Adapter ligation and library preparation was conducted as previously described (Vourekas and Mourelatos 2014). The resulting libraries were sequenced on an Illumina NextSeq 500 platform (single-end 75 nt).
SMEDWI-3 CLASH
CLASH libraries were prepared as described for SMEDWI-3 HITS-CLIP with slight modifications (Shen et al. 2018). After immunoprecipitation of crosslinked RNA-SMEDWI-3 complexes, samples were treated with RNase T1 (0.1 units in 500 µL of lysis buffer) for 5 min at 20°C. Next, samples were washed once with low-salt buffer, once with high-salt buffer and three times with 1x PNK buffer. To phosphorylate the mRNA 5′-ends for chimera formation and discard possible DNA contaminants, samples were treated with DNase I and PNK 3′-phosphatase minus. Chimera ligation was conducted overnight in the presence of T4 RNA ligase1 (1 unit/µL) and 10% PEG 8,000. Next, 3′-ends of crosslinked transcripts were dephosphorylated with Antarctic phosphatase. Adapter ligation, library preparation, and sequencing were carried out as described for SMEDWI-3 HITS-CLIP.
Degradome-seq library preparation
Degradome libraries (5′-monophosphate-dependent cloning of mRNA fragments) were prepared from 2.5 µg of X1 FACS sorted total RNA as previously described (Wang et al. 2014). Next generation sequencing was carried out on an Illumina Next-Seq 500 platform (single-end 75-nt mode).
CHIP-seq library preparation
CHIP-seq libraries were prepared from X1 FACS-sorted cells as previously described (Duncan et al. 2015).
Ex vivo SHAPE-map
Total RNA was extracted from worms under non-denaturing conditions as described previously (Smola et al. 2015a) with the following modifications: 50 worms (7–10 mm) were dissociated in cold calcium- and magnesium-free buffer with 1% BSA (CMFB). Cells were pelleted and resuspended in 200 µL Proteinase K buffer (40 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1.5% SDS). DNase I (150 units, Roche) was added to the cell suspension to degrade genomic DNA followed by Proteinase K (500 µg/mL) digestion for 30 min at 37°C, 1000 rpm. Next, RNA was purified by phenol-chloroform extraction and treated with 1M7 (1-methyl-7-nitroisatoic anhydride) in DMSO or in DMSO only (negative control). The final concentration of 1M7 in the reaction after both treatments was 20 mM. DNaseI treatment, reverse transcription, and SHAPE-MaP library construction were performed according to the amplicon workflow (Smola et al. 2015b) with the following modifications: For each target mRNA specific RT-primers were used. The size of the reverse-transcribed sequences did not exceed 1 kb. Reverse transcription was performed with SuperScript II in the presence of Mn2+ ions followed by RNA–cDNA hybrid purification with AMPure magnetic beads (Beckman). Deep-sequencing libraries were constructed using the Nextera XT Kit (Illumina) and sequenced on an Illumina NextSeq 500 platform (paired-end 150 nt).
Data availability
All sequencing data have been deposited in the Gene Expression Omnibus (GEO), series GSE122199.
Supplementary Material
Acknowledgments
We thank the Core Unit Systems Medicine at the University of Würzburg for excellent technical support during next-generation sequencing. The anti-SMEDWI-1 antibody was a kind gift of Dr. Kerstin Bartscherer, Hubrecht Institute, Utrecht, Netherlands. We thank Silke Spudeit for technical support, David Rosenkranz for help with piRNA analysis scripts, and Anthony Mustoe for advice on SHAPE-MaP. We thank Olaf Stemmann and Gunter Meister for comments on the manuscript. C.-D.K. gratefully acknowledges the possibility to start this project in the laboratory of Leemor Joshua-Tor at Cold Spring Harbor Laboratory. This work was supported by the Elite Network of Bavaria (N-BM-2013-244), the University of Bayreuth, the German Research Foundation (DFG Grant KU 3514/1-1), and the Paul Ehrlich and Ludwig Darmstaedter Prize for Young Researchers (to C.D.K.). A.S.A. is an investigator of the Howard Hughes Medical Institute and the Stowers Institute for Medical Research. This work was funded in part by National Institutes of Health R37GM057260 to A.S.A.
Author contributions: I.V.K., E.M.D., E.J.R., A.S.A., and C.D.K. conceived and designed the studies; I.V.K., E.M.D., V.G., S.N., and S.A.E. acquired data; I.V.K., E.J.R., and V.G. performed computational analyses; A.S.A. and C.D.K. supervised the study. I.V.K. and C.D.K. drafted the manuscript with input from all authors.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.322776.118.
References
- Alexiou P, Maragkakis M, Mourelatos Z, Vourekas A. 2018. cCLIP-seq: retrieval of chimeric reads from HITS-CLIP (CLIP-seq) libraries. Methods Mol Biol 1680: 87–100. 10.1007/978-1-4939-7339-2_6 [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 431: 343–345. [DOI] [PubMed] [Google Scholar]
- Arnold CP, Merryman MS, Harris-Arnold A, McKinney SA, Seidel CW, Loethen S, Proctor KN, Guo L, Sánchez Alvarado A. 2016. Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. Elife 5: e16793 10.7554/eLife.16793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, Cording AC, Doebley A-L, Goldstein LD, Lehrbach NJ, Le Pen J, et al. 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99. 10.1016/j.cell.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6: 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2018. Metazoan microRNAs. Cell 173: 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C. 2015. A dynamic search process underlies microRNA targeting. Cell 162: 96–107. 10.1016/j.cell.2015.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727. 10.1101/gad.12.23.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci 41: 324–337. 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis C, Zanni V, Brasset E, Eymery A, Zhang L, Mteirek R, Jensen S, Rong YS, Vaury C. 2013. “Dot COM”, a nuclear transit center for the primary piRNA pathway in Drosophila. PLoS ONE 8: e72752 10.1371/journal.pone.0072752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EM, Chitsazan AD, Seidel CW, Sánchez Alvarado A. 2015. Set1 and MLL1/2 target distinct sets of functionally different genomic loci in vivo. Cell Rep 13: 2741–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RDG, Buetow KH, et al. 2008. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2: 437–447. 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott SA, Sánchez Alvarado A. 2013. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip Rev Dev Biol 2: 301–326. 10.1002/wdev.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, Marra M, Nusbaum C, Lee WL, Jenkin JC, et al. 2009. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci 106: 11546–11551. 10.1073/pnas.0905222106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714. 10.1101/gad.1434406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohme MA, Schloissnig S, Rozanski A, Pippel M, Young GR, Winkler S, Brandl H, Henry I, Dahl A, Powell S, et al. 2018. The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature 554: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. 10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- Guo T, Peters AHFM, Newmark PA. 2006. A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell 11: 159–169. 10.1016/j.devcel.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Guo L, Zhang S, Rubinstein B, Ross E, Alvarado AS. 2017. Widespread maintenance of genome heterozygosity in Schmidtea mediterranea. Nat Ecol Evol 1: 19 10.1038/s41559-016-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann J, Schraivogel D, Bruckmann A, Manickavel S, Jakob L, Eichner N, Pfaff J, Urban M, Sprunck S, Hafner M, et al. 2015. Biochemical isolation of Argonaute protein complexes by Ago-APP. Proc Natl Acad Sci 112: 11841–11845. 10.1073/pnas.1506116112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. 2006. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ 48: 371–380. 10.1111/j.1440-169X.2006.00876.x [DOI] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153: 654–665. 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe FS, Fischl H, Murray SC, Mellor J. 2017. Is H3K4me3 instructive for transcription activation? Bioessays 39: 1–12. 10.1002/bies.201670013 [DOI] [PubMed] [Google Scholar]
- Kang H, Sánchez Alvarado A. 2009. Flow cytometry methods for the study of cell-cycle parameters of planarian stem cells. Dev Dyn 238: 1111–1117. 10.1002/dvdy.21928 [DOI] [PubMed] [Google Scholar]
- Kashima M, Kumagai N, Agata K, Shibata N. 2016. Heterogeneity of chromatoid bodies in adult pluripotent stem cells of planarian Dugesia japonica. Dev Growth Differ 58: 225–237. 10.1111/dgd.12268 [DOI] [PubMed] [Google Scholar]
- Ku H-Y, Lin H. 2014. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev 1: 205–218. 10.1093/nsr/nwu014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Gu W, Shirayama M, Youngman E, Conte D, Mello CC. 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150: 78–87. 10.1016/j.cell.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399. 10.1101/gad.209841.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, et al. 2018. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol 2: 174–181. 10.1038/s41559-017-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50: 67–81. 10.1016/j.molcel.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T. 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci 104: 6714–6719. 10.1073/pnas.0701920104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. 2009. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10: 161 10.1186/1471-2105-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Duronio RJ. 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol 14: 692–699. 10.1016/S0955-0674(02)00387-3 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. 2014. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc 9: 263–293. 10.1038/nprot.2014.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe AM, Busan S, Rice GM, Hajdin CE, Peterson BK, Ruda VM, Kubica N, Nutiu R, Baryza JL, Weeks KM. 2018. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell 173: 181–195.e18. 10.1016/j.cell.2018.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura O, Hosoda K, Kawaguchi E, Yazawa S, Hayashi T, Inoue T, Umesono Y, Agata K. 2015. Unusually large number of mutations in asexually reproducing clonal planarian Dugesia japonica. PLoS ONE 10: e0143525 10.1371/journal.pone.0143525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. 2008. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14: 1174–1186. 10.1261/rna.1085008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Sanchez Alvarado A. 2008. Regeneration, stem cells, and the evolution of tumor suppression. Cold Spring Harb Symp Quant Biol 73: 565–572. 10.1101/sqb.2008.73.045 [DOI] [PubMed] [Google Scholar]
- Peiris TH, Hoyer KK, Oviedo NJ. 2014. Innate immune system and tissue regeneration in planarians: an area ripe for exploration. Semin Immunol 26: 295–302. 10.1016/j.smim.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. 2005. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310: 1321330. [DOI] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267. [DOI] [PubMed] [Google Scholar]
- Rojas-Ríos P, Simonelig M. 2018. piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development 145: dev161786 10.1242/dev.161786 [DOI] [PubMed] [Google Scholar]
- Rosenkranz D, Zischler H. 2012. proTRAC–a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinformatics 13: 5 10.1186/1471-2105-13-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier A-C, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. 2010. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467: 1128–1132. 10.1038/nature09465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Vieira AP, Roberts-Galbraith RH, Newmark PA. 2012. PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development 139: 1083–1094. 10.1242/dev.076182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Weiss JA, King RS, Newmark PA. 2014. PIWI homologs mediate Histone H4 mRNA localization to planarian chromatoid bodies. Development 141: 2592–2601. 10.1242/dev.101618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu S, Dattani A, Aboobaker AA. 2017. Secrets from immortal worms: What can we learn about biological ageing from the planarian model system? Semin Cell Dev Biol 70: 108–121. 10.1016/j.semcdb.2017.08.028 [DOI] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, MacRae IJ. 2014. Structural basis for microRNA targeting. Science 346: 608–613. 10.1126/science.1258040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D Jr, Mello CC. 2013. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell 27: 656–663. 10.1016/j.devcel.2013.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Tang W, Shen E-Z, Tu S, Lee H-C, Weng Z, Mello CC. 2018. The coding regions of germline mRNAs confer sensitivity to argonaute regulation in C. elegans. Cell Rep 22: 2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. 2017. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 33: 566–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E-Z, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding Y-H, Dai S-Y, Weng Z, Mello CC. 2018. Identification of piRNA binding sites reveals the argonaute regulatory landscape of the C. elegans germline. Cell 172: 937–951.e18. 10.1016/j.cell.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D Jr, Mello CC. 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. 10.1016/j.cell.2012.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Calabrese M, Weeks KM. 2015a. Detection of RNA–protein interactions in living cells with SHAPE. Biochemistry 54: 6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. 2015b. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10: 1643–1669. 10.1038/nprot.2015.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira LK, Reed SI. 2013. Ubiquitin ligases and cell cycle control. Annu Rev Biochem 82: 387–414. 10.1146/annurev-biochem-060410-105307 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC. 2014. Piwi and potency: PIWI proteins in animal stem cells and regeneration. Integr Comp Biol 54: 700–713. 10.1093/icb/icu084 [DOI] [PubMed] [Google Scholar]
- Vourekas A, Mourelatos Z. 2014. HITS-CLIP (CLIP-seq) for mouse Piwi proteins. Methods Mol Biol 1093: 73–95. 10.1007/978-1-62703-694-8_7 [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19: 773–781. 10.1038/nsmb.2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Alexiou P, Vrettos N, Maragkakis M, Mourelatos Z. 2016. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 531: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yoshikawa M, Han BW, Izumi N, Tomari Y, Weng Z, Zamore PD. 2014. The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs. Mol Cell 56: 708–716. 10.1016/j.molcel.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Chen K-M, Pandey RR, Homolka D, Reuter M, Janeiro BKR, Sachidanandam R, Fauvarque M-O, McCarthy AA, Pillai RS. 2016. PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol Cell 61: 138–152. 10.1016/j.molcel.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A, Li Y-Q, Wang C, Han X-S, Li G, Wang J-Y, Li D-S, Qin Y-W, Shi Y, Brewer G, et al. 2013. Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J Cell Biol 201: 409–425. 10.1083/jcb.201207172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. 2011. Mapping in vivo protein–RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol 29: 607–614. 10.1038/nbt.1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Theurkauf WE, Weng Z, Zamore PD. 2012. Strand-specific libraries for high throughput RNA sequencing (RNA-seq) prepared without poly(A) selection. Silence 3: 9 10.1186/1758-907X-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu W-S, Huang W-C, Weng Z, Lee H-C. 2018. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359: 587–592. 10.1126/science.aao2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Pearson BJ. 2018. Smed-myb-1 specifies early temporal identity during planarian epidermal differentiation. Cell Rep 25: 38–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited in the Gene Expression Omnibus (GEO), series GSE122199.