In this study, Xiao et al. set out to identify molecular mechanisms and genetic pathways that regulate the cellular makeup of a tissue. Using mammalian cardiac fibroblast as a model and unbiased genomic approaches, including high throughput single-cell transcriptomics and enhancer connectome profiling, the authors show that the Hippo signaling pathway is essential in maintaining cardiac fibroblasts in a homeostatic condition.
Keywords: cell state transitions, epigenomics, fibrosis, Hippo signaling, macrophages, myofibroblast, tissue homeostasis, myocardial infarction, single-cell RNA-seq
Abstract
Cardiac fibroblasts (CFs) respond to injury by transitioning through multiple cell states, including resting CFs, activated CFs, and myofibroblasts. We report here that Hippo signaling cell-autonomously regulates CF fate transitions and proliferation, and non-cell-autonomously regulates both myeloid and CF activation in the heart. Conditional deletion of Hippo pathway kinases, Lats1 and Lats2, in uninjured CFs initiated a self-perpetuating fibrotic response in the adult heart that was exacerbated by myocardial infarction (MI). Single cell transcriptomics showed that uninjured Lats1/2 mutant CFs spontaneously transitioned to a myofibroblast cell state. Through gene regulatory network reconstruction, we found that Hippo-deficient myofibroblasts deployed a network of transcriptional regulators of endoplasmic reticulum (ER) stress, and the unfolded protein response (UPR) consistent with elevated secretory activity. We observed an expansion of myeloid cell heterogeneity in uninjured Lats1/2 CKO hearts with similarity to cells recovered from control hearts post-MI. Integrated genome-wide analysis of Yap chromatin occupancy revealed that Yap directly activates myofibroblast cell identity genes, the proto-oncogene Myc, and an array of genes encoding pro-inflammatory factors through enhancer–promoter looping. Our data indicate that Lats1/2 maintain the resting CF cell state through restricting the Yap-induced injury response.
In regenerative organs such as the skin, fibrosis typically resolves as the wound healing process is completed (Shook et al. 2018). In nonregenerative organs, such as the heart, fibrosis persists and serves to both acutely repair the organ and maintain essential long-term organ function (Tallquist and Molkentin 2017). In the acute post-MI period, CFs become activated and proliferate for 2–4 d post-MI. Subsequently, the myofibroblast cell state is induced and becomes maximized at 4–7 d post-MI. At 7 d post-MI, through poorly understood mechanisms, proliferation is extinguished. Lastly, the myofibroblasts transition to matrifibrocytes at 10 d post-MI. It is thought that matrifibrocytes are required to maintain the long term cardiac scar that permits continued pumping function of the injured heart post-MI (Fu et al. 2018).
The Hippo pathway, a kinase cascade, inhibits adult cardiomyocyte (CM) renewal and is considered to be a therapeutic target to treat heart failure (Halder and Johnson 2011; Leach et al. 2017). In contrast to CMs, the Hippo pathway has not been directly investigated in adult resting CFs. The core Hippo pathway components include the Mst and Lats family kinases that are activated by physiologic inputs, such as cell density and matrix stiffness (Halder and Johnson 2011). When activated, Lats1 and Lats2 phosphorylate the downstream effector Yap, a transcriptional coactivator, excluding the modified protein from the nucleus.
While Hippo signaling in adult resting CFs under physiologic conditions has not been investigated, its function has been evaluated in other tissue-specific fibroblasts. Depending on the organ, the Hippo pathway is thought to have distinct roles in fibroblast development. For example, Lats1 and Lats2 deletion in the developing kidney results in increased fibrosis with more myofibroblasts, indicating that the Hippo pathway is anti-fibrotic in kidney development (McNeill and Reginensi 2017). In contrast, in the developing heart, Lats1 and Lats2 deletion inhibits CF development from epicardial progenitors (Xiao et al. 2018). In the absence of Lats1/2, cardiac epicardial progenitors arrest at an intermediate developmental state resulting in a deficiency in CF maturation (Xiao et al. 2018). In the adult, the available studies lead to conflicting conclusions regarding Hippo-Yap function in cardiac fibrosis. Germline inactivation of Mst2, resulting in Mst2 deletion in all cells of the body, results in reduced cardiac fibrosis after trans-aortic constriction (TAC) suggesting that Mst kinases are pro-fibrotic (Zi et al. 2014). A separate study came to the opposite conclusion. Germline deletion of Rassf1a, which normally enhances Mst function, results in more cardiac fibrosis after TAC, suggesting an antifibrotic role for Hippo pathway in cardiac fibrosis (Del Re et al. 2010).
In parallel experiments, we conditionally deleted Lats1 and Lats2 (Lats1/2) in CFs and subjected these mice to sham or MI surgery to investigate CF cell state transitions. Using single cell RNA sequencing (scRNA-seq), we found that Lats1/2 mutant resting CFs spontaneously transitioned to a myofibroblast-like cell state. Lats1/2 deletion led to a relentless pro-fibrotic and pro-inflammatory cascade that ultimately resulted in organ failure. Reducing levels of Hippo pathway effectors Yap/Taz in Lats1/2 mutant CFs attenuated the lethal fibrotic phenotype after infarction. Thus, Hippo signaling cell-autonomously regulates CF fate transitions and proliferation, and non-cell-autonomously regulates both myeloid and mesenchymal cell polarization.
Results
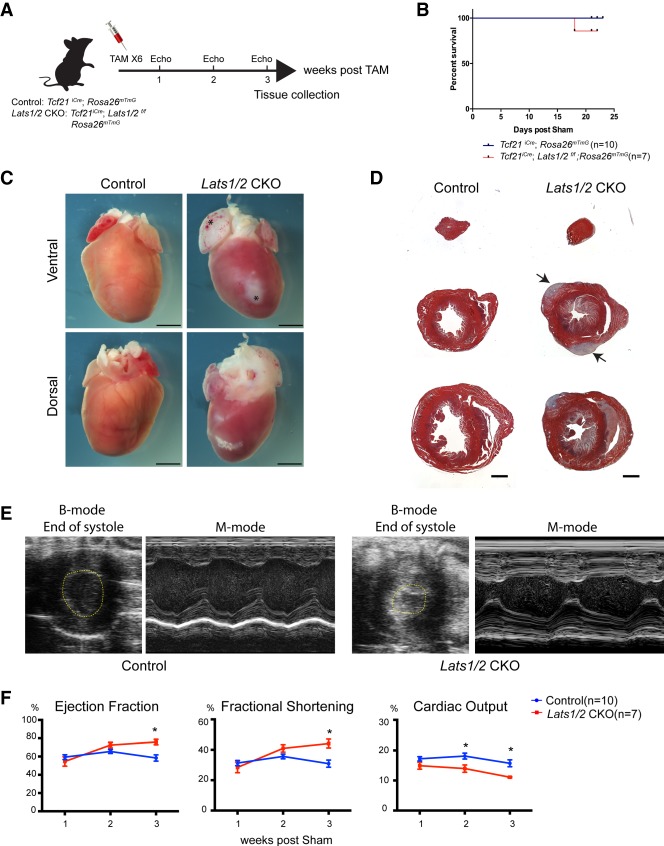
Lats1 and Lats2 inactivation in the Tcf21 lineage results in spontaneous cardiac fibrosis
We set out to characterize Hippo pathway function in adult CFs with and without MI. To label CFs, we used a CF lineage tracing model with Tcf21-iCre;mTmG mice that contain a tamoxifen-inducible Cre recombinase (MerCreMer) knocked into the endogenous transcription factor 21 (Tcf21) locus (Acharya et al. 2011), as well as the mTmG double-fluorescent Cre reporter (Muzumdar et al. 2007). We determined Yap subcellular localization in CFs using confocal microscopy on immunofluorescent (IF) stained tissue sections. GFP positive CFs showed increased nuclear Yap at 3 d post-MI (dPMI) (Supplemental Fig. S1A). These results suggest that the Hippo pathway kinases are active in resting CFs.
Next, we deleted Lats1 and Lats2 in CFs using Tcf21iCre; Lats1fl/fl; Lats2fl/fl; mTmG mice, referred to as Lats1/2 CKO mice (Fig. 1A). Most Lats 1/2 CKO sham mice survived at least 3 wk after inducing Cre activity, and exhibited fibrosis at the gross and histologic levels (Fig. 1B–D). Fibrosis in Lats1/2 CKO sham hearts was primarily localized to subepicardial and subendocardial regions of the ventricle (Fig. 1D; Supplemental Fig. S1B). Three weeks after tamoxifen injection, Lats1/2 CKO shams had increased ejection fraction (EF) and fractional shortening (FS) and reduced cardiac output, consistent with heart failure (Fig. 1E,F). These data indicate that Lats1/2 deletion in adult resting CFs results in spontaneous activation of cardiac fibrosis.
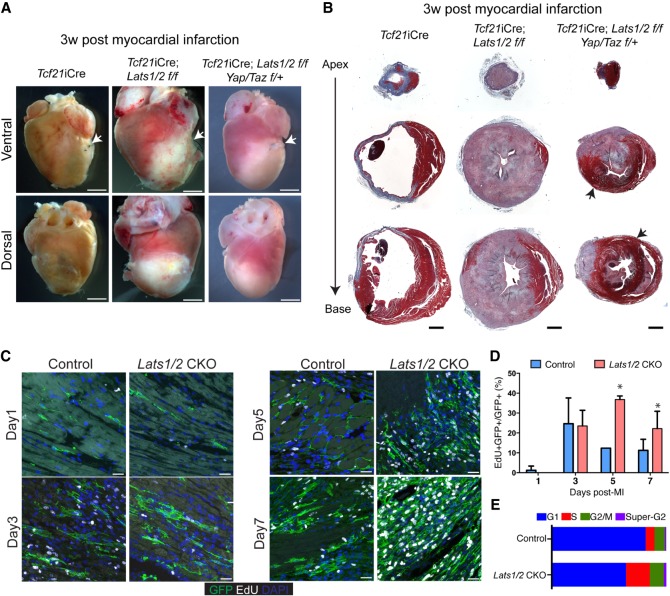
Figure 1.
Lats1/2 deletion in uninjured cardiac fibroblasts results in pervasive myocardial fibrosis. (A) Experimental strategy for cardiac fibroblast Cre activation and tissue collection. (B) Survival curve of control mice (Tcf21iCre/+; Rosa26mTmG/+) and Lats1/2 CKO (Tcf21iCre/+; Lats1/2flox/flox; Rosa26mTmG/+) mice by 3 wk after Cre activation and sham. (C) Representative gross heart morphology at 3 wk after Cre activation. Atria were stiffened and enlarged (asterisks). Scale bar, 200 µm. (D) Representative Masson's trichrome serial sections of control and Lats1/2 CKO hearts. Lats1/2 CKO hearts possessed expansive and aggregated (arrows) cardiac fibrosis (stained blue) within the myocardium (stained red). Scale bar, 1000 µm. (E) Representative M-mode and B-mode echocardiographic images of control and Lats1/2 CKO mouse hearts 3 wk after tamoxifen induction. (F) Percent ejection fraction, fractional shortening, and cardiac output as determined by echocardiography. Control, n = 10; Lats1/2 CKO, n = 7. Statistical significance was determined by t-test, P-value <0.05.
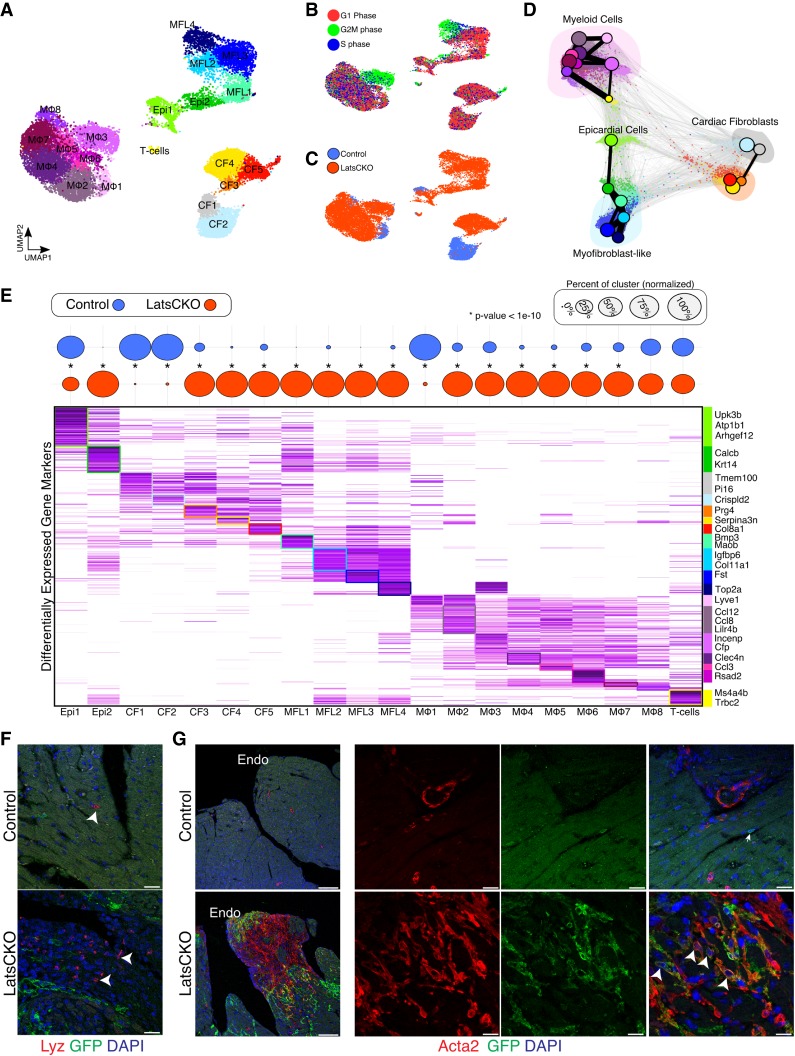
Lats1 and Lats2 deletion in cardiac fibroblasts disrupts cardiac tissue composition
To determine if Lats1/2 deletion in resting CFs promotes their differentiation and/or elicits an injury response we performed Drop-seq (Macosko et al. 2015) 3 wk following Tcf21-iCre induction. After computational processing, we captured 17,501 noncardiomyocytes that separated into 20 distinct clusters (Fig. 2A; Supplemental Fig. S2A; Supplemental Table S1). Overall, we detected two epicardial clusters (Epi1-2), five CF clusters (CF1-5), four clusters of myofibroblast-like cells (MFL1-4), eight monocytes/macrophages clusters (Mϕ1-8), and one T lymphocyte cluster (T-cells). Many cells from Lats1/2 CKO sham hearts appeared to be actively undergoing mitosis, with MFLs, CF5, and Mϕ3 being among the most proliferative clusters (Fig. 2B,C). To further dissect the topology of these cells, we applied partition-based graph abstraction (PAGA), an algorithm that maps discrete connected and continuous connected cell-to-cell variation (Wolf et al. 2019). Importantly, the resulting PAGA graphs were consistent with our UMAP results (Fig. 2D).
Figure 2.
Lats1/2 prevent the differentiation of resting cardiac fibroblasts to immunostimulatory myofibroblasts. (A) UMAP plot of combined control and Lats1/2 CKO Drop-seq. MFL, myofibroblast-like cells; CF, cardiac fibroblasts; Mϕ, macrophages and monocytes; Epi, epicardial cells; T-cells, T lymphocyte. (B) Cell cycle phase analysis of single cardiac cells projected onto UMAP plot. (C) UMAP plot showing the genotype for each individual transcriptome. (D) Force-directed graph of PAGA-initialized embedding with PAGA graph superimposed. Colored according to Figure 1A. (E, top) Cellular composition of each cluster in control and Lats1/2 CKO hearts. Dot plot displays the relative proportion of cells from control and mutant hearts within each cluster. Dot size represents the percentage of cell origin within each cluster. The statistical difference of cell origin composition within each cluster were analyzed by Chi-square analysis (*P < 10−10). (Bottom) Average differential expression heat map for the top marker genes (n = 784), with genes as rows and clusters as columns. Colors for each cluster match those in A. (F) Representative immunofluorescence confocal images from control and Lats1/2 CKO hearts 3 wk following Tcf21-iCre activation showing Lyz (white arrowheads), a myeloid cell marker, expression (red), cardiac fibroblast fate mapping (green), and nuclei labeling (blue). Scale bar, 25 µm. (G) Representative low magnification (left) and high magnification (right) immunofluorescence confocal images from control and Lats1/2 CKO hearts 3 wk following Tcf21-iCre induction displaying lineage traced GFP+ cardiac fibroblasts (green) and Acta2 (αSMA) expression (red). Acta2 labels Lats1/2 CKO CFs (white arrowheads). Nuclei are stained with DAPI (blue). Endo, endocardium of left ventricle. Scale bar, 100 µm (left), and 25 µm (right).
In addition to being expressed in adult CFs, Tcf21 is also detected at low levels in the uninjured adult epicardium (Braitsch et al. 2013). Hence, epicardial Cre-recombinase activity is expected. In Lats1/2 CKO sham hearts, the control Epi1 cluster was depleted and the Epi2 cluster expanded (Fig. 2C,E). Epi2 expressed genes that are signatures of Wnt and Tgfβ-signaling, such as Rspondin1 and Tgfbr2. These findings suggest that Hippo signaling plays an important role in maintaining adult epicardial cell identity.
Among the five CF clusters, we discovered two control heart-enriched subpopulations of fibroblasts, CF1 and CF2 (Fig. 2E). The CF1 cluster expressed Tmem100, Sema3c and Cd248 (Fig. 2E; Supplemental Fig. S2B). The CF2 cluster expressed ECM genes such as Mgp and Fbln5, and the lipid metabolism gene, Apoe (Supplemental Fig. S2B). The CF3 and CF4 clusters were primarily derived from Lats1/2 CKO sham hearts. Both of these mutant clusters exhibited gene signatures consistent with an inflammatory phenotype, highlighted by the expression of Cxcl1, and Ccl7, which encode secreted immunostimulatory factors (Supplemental Fig. S2C). The third mutant CF cluster, CF5, expressed Postn, a marker of activated and pro-fibrotic CFs (Supplemental Fig. S2D; Tallquist and Molkentin 2017).
The dramatic shifts in cellular composition, and fibroblast activation observed in Lats1/2 CKO sham hearts resembled an injured heart. Indeed, the phenotypic expansion of myeloid cells in Lats1/2 CKO sham hearts is consistent with recent work which uncovered a unique cluster of injury-induced interferon-inducible cells (IFNICs) (King et al. 2017). We found that the cluster we annotated as Mϕ6 were Rsad2+ and Ifit1-3+ (Fig. 2E; Supplemental Fig. S2F) and resembled IFNICs. IF with the macrophage marker Lyz confirmed the increased composition of macrophages in Lats1/2 CKO sham hearts (Fig. 2F). Thus, in the absence of injury, Lats1/2 CKO sham hearts acquired characteristics of an injured heart.
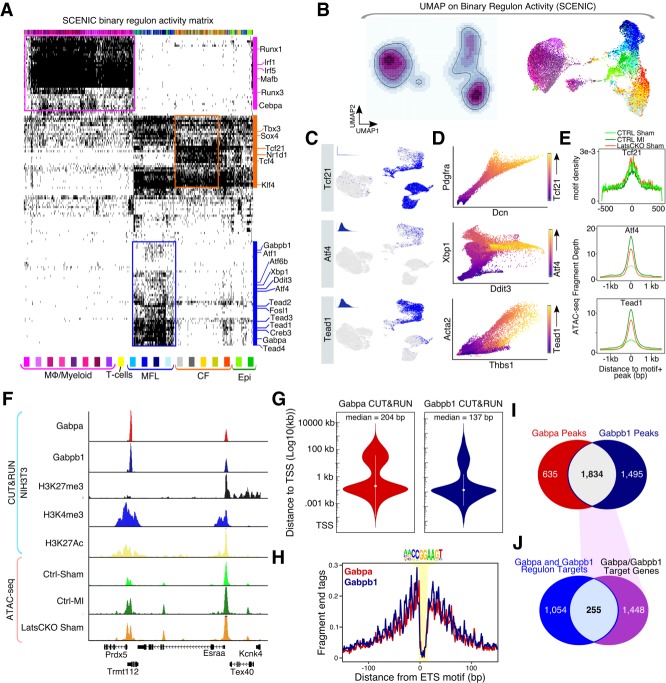
Lats1 and Lats2 regulate cardiac fibroblast cell states both autonomously and nonautonomously
In Lats1/2 CKO sham hearts, in addition to CF3-5, we identified distinct MFL populations, which had strong expression of myofibroblast markers such as Acta2 (Supplemental Fig. S2A). Additionally, MFL1–4 also express several Yap/TEAD targets, including Vgll3 and Amotl2 (Supplemental Fig. S2E). Moreover, the MFLs expressed Egfp (Supplemental Fig. S3A). Thus, we hypothesized that the MFLs represented Lats1/2 CKO CFs. To further validate the presence of MFLs in Lats1/2 CKO sham hearts, we performed IF experiments (Fig. 2G). Importantly, the majority of lineage-traced CFs exhibited Acta2 staining in Lats1/2 CKO sham hearts revealing that Lats1/2 deletion in resting CFs promotes a cell state transition to MFLs characteristic of an injury response (Fig. 2G).
We hypothesized that clusters CF3–5, which did not express direct Yap target genes, had escaped Tcf21iCre-induced Lats1/2 deletion and were phenotypically distinct from control-specific CF1–2 because of intercellular communication with Lats1/2 CKO MFLs. To map the gene regulatory networks (GRNs) active in these distinct cell clusters, we deployed the single-cell regulatory network inference and clustering (SCENIC) computational pipeline to our scRNA-seq data (Aibar et al. 2017). Briefly, SCENIC determines the activity of transcription factors from scRNA-seq data by integrating coexpression analysis, with transcription factor motif enrichment analysis. An expressed transcription factor and all of its coexpressed target genes are referred to as regulons. We identified 541 active regulons out of the initial 1001 present in our transcription factor coexpression matrix (Fig. 3A; Supplemental Table S2). Next, we performed UMAP on the binary regulon matrix and found that the resulting UMAP had a distribution of cells that closely matched our expression-based UMAP (Fig. 3B). Importantly, all macrophage clusters displayed high levels of myeloid lineage-specific regulons that were not active in CFs (Fig. 3A). All of the CF, Epi, and MFL clusters displayed Tcf21 regulon activity (Fig. 3C). To further assess the transcription factor activity predicted by SCENIC we applied the Markov affinity-based graph imputation of cells (MAGIC) algorithm (van Dijk et al. 2018). Importantly, Tcf21 expression progressively increased in relationship to the expression of two of its predicted target genes, Dcn and Pdgfra, indicating that regulon activity identified by SCENIC is robust (Fig. 3D).
Figure 3.
Tead and ER stress-response-associated transcription factor GRNs are active in myofibroblasts. (A) Binary regulon activity matrix results from SCENIC algorithm carried out on cardiac Drop-seq data. Individual cells are columns, and regulons are rows. (B) Cardiac cell identity UMAP generated from the binary regulon activity matrix. (Left) Regulon activity density plot embedded on UMAP. Light color indicates low cumulative regulon activity, and darker color indicates high cumulative regulon activity. (Right) Binary regulon activity UMAP colored by individual cell cluster identity. (C) Regulon activity for individual cells embedded on the expression based UMAP. Blue highlights active regulon for indicated transcription factor, and gray indicates that a regulon is not active. Insets show the AUCell score histogram for the regulon. (D) MAGIC scatterplots of gene–gene relationships for regulon components. Transcription factor expression encoded by highlighted color gradient. (E) Evaluation of regulon activity using Fast-ATAC. (Top) Global Tcf21 motif enrichment across peaks from each indicated experimental condition. (Bottom) Fast-ATAC read enrichment across motif-containing peaks annotated to genes in either the Atf4 (middle) or Tead1 (bottom) regulons identified in Figure 3A. (F) Genome browser tracks displaying GABP CUT&RUN. (G) Violin plot showing the absolute distance of Gabpa and Gabpb1 peaks to the nearest transcription start site (TSS). (H) CUT&RUN footprint analysis for GABP across the ETS motif. (I) Venn diagram showing intersect of Gabpa and Gabpb1 peaks. (J) Venn diagram showing intersect of GABP peaks (Fig. 3I), with the Gabpa and Gabpb1 regulon target genes (Fig. 3A).
Next, we explored the GRNs distinguishing MFLs from CFs. Importantly, Tead1-4 regulons were highly enriched in MFLs compared with CF1-5 (Fig. 3A,C). Moreover, Tead1 expression was positively correlated with the expression of two of its targets, Acta2 and Thbs1 (Fig. 3D). MFLs also uniquely activated regulons associated with the unfolded protein response (UPR), and endoplasmic reticulum (ER) stress, including Atf4 and Creb3. Next, we performed ATAC-seq on FACS sorted CFs (GFP+) harvested from control sham hearts and Lats1/2 CKO sham hearts. Additionally, we included cells collected from control hearts post-MI (three dpMI). Tcf21 motifs were enriched in accessible regions across all conditions, consistent with Tcf21-iCre activity (Fig. 3E). Both Atf4 and Tead1 motifs were enriched in peaks proximal to genes comprising their associated regulons in control CFs post-MI and in Lats1/2 CKO sham, but not in control sham (Fig. 3E). These data revealed that Tead-Yap motifs were occupied in control CFs post-MI and after deletion of Hippo signaling in Lats1/2 CKO CFs.
Previous studies found that Hippo signaling plays a role in regulating ER stress and UPR in a Yap-dependent fashion (Wu et al. 2015). To more directly validate the SCENIC results, we implemented CUT&RUN for mapping transcription factor binding sites (Skene and Henikoff 2017). Among the several ER stress and UPR associated transcription factors predicted to be activated in MFLs, we chose the Ets transcription factor GABP (hetero-tetramer of Gabpa and Gabpb1) as a suitable candidate for further validation (Fig. 3F; Baumann et al. 2018). CUT&RUN revealed that both GABP components, Gabpa and Gabpb1, preferentially bound proximal to gene promoters (Fig. 3G). Additionally, both exhibited clear ETS motif footprints (Fig. 3H). Out of the total 1834 overlapping Gabpa and Gabpb1 peaks (Supplemental Table S3), 255 intersected with the corresponding regulon target genes (Fig. 3I,J; Supplemental Table S4). The expansion of the GABP regulon in Lats1/2 CKO CFs is consistent with a previous study that identified Lats1 as a direct repressor of GABP transcriptional activity in the liver (Wu et al. 2013) and further supports the conclusion that MFLs are Lats1/2 CKO CFs. These data suggest that Lats1/2 repress injury-induced myofibroblast cell identity acquisition in a cell-autonomous fashion.
To further validate the distinction between aCFs (CF3-5) and MFLs, we interrogated our scRNA-seq data to look for specific markers of each cell state. Our scRNA-seq data revealed specific markers for CF3-5, including Serpina3n (Supplemental Fig. S3B). In situ hybridization for Serpina3n revealed that Lats1/2 CKO sham hearts contained elevated numbers of Serpina3n-expressing cells compared with controls (Supplemental Fig. S3C). Importantly, Serpina3n-positive CFs were located adjacent to GFP-expressing Tcf21iCre lineage cells but were themselves Tcf21iCre lineage negative (Supplemental Fig. S3C). Conversely, in situ hybridization for a pan-MFL marker and direct Yap target gene, Plac8, in control and Lats1/2 CKO hearts revealed that Lats1/2 CKO MFLs that expressed GFP also expressed Plac8, while GFP-negative CF3–5 and control CFs did not (Supplemental Fig. S3D). Our data support the conclusion that CF3–5 clusters are derived from CFs that escaped Lats1/2 deletion in the Lats1/2 CKO hearts and are activated non-cell-autonomously.
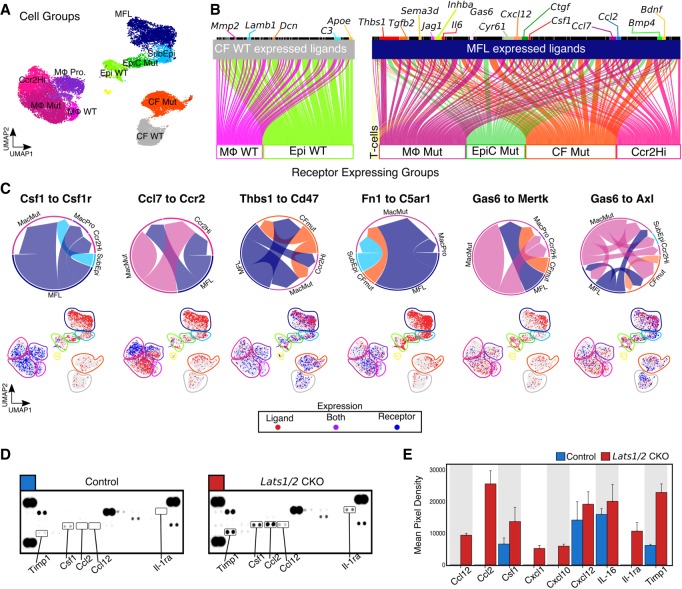
Lats1 and Lats2 deletion results in pro-inflammatory signaling between noncardiomyocytes in the heart
Our data suggested that Lats1/2 prevent the induction of a CF-derived pro-inflammatory cascade that promotes myeloid cell influx, activation, and expansion. To investigate the crosstalk between MFLs and other noncardiomyocytes in Lats1/2 CKO sham hearts, we performed intercellular communication network analysis on groupings of cardiac cell clusters based on the mouse orthologs of the FANTOM5 human ligand-receptor connectome (Fig. 4A,B; Ramilowski et al. 2015). MFLs showed a larger number of significant ligand-receptor pairs (4.96-fold increase) compared with control CFs (Supplemental Table S5; Supplemental Table S6). In controls, CF ligand receptor interactions were between CFs and macrophages and epicardial cells and were primarily a result of extracellular matrix (ECM)-integrin signaling.
Figure 4.
Yap is a cell-nonautonomous regulator of cardiac cell composition and inflammatory status. (A) UMAP plot of cell groups. MFL, myofibroblast-like cells; SubEpi, sub-epicardial-like cells; EpiC Mut, epicardial cells enriched in Lats1/2 CKO hearts; Epi WT, epicardial cells enriched in control (WT) hearts; CF WT, cardiac fibroblasts enriched in control (WT) hearts; CF Mut, cardiac fibroblasts enriched in Lats1/2 CKO hearts; Mϕ WT, myeloid cells enriched in control (WT) hearts; Mϕ MUT, myeloid cells enriched in Lats1/2 CKO hearts; Mϕ Pro, proliferating myeloid cells; Ccr2Hi, myeloid cell cluster expressing high levels of Ccr2. (B) Ligand-receptor connection analysis. (Left) Control CF ligand-receptor interaction plot. (Right) Lats1/2 CKO-specific MFL-mediated ligand-receptor interaction plot. Lines indicate significant ligand receptor pairs where ligands are expressed in top cell group and receptors are expressed by individual cells in the bottom cell group. Ligands with multiple receptors are largest. (C, top) Circle plots showing the strength of individual ligand-receptor interactions for each indicated group of cells. Size of arrow is proportional to the number of possible cell–cell interactions for each ligand-receptor pair. Base of arrow indicates ligand expressing group, and arrowhead contacts receptor expressing cell group. (Bottom) Feature plots showing the expression of ligand (red), receptor (blue), and cells coexpressing both ligand and receptor (purple). (D) Chemokine protein expression in cardiac lysates determined by cytokine/chemokine protein array. (E) Relative protein expression based on densitometric analysis of Figure 4D. Error bars, SEM.
MFL-expressed ligands consisted of several known components of the wound response (Supplemental Table S7) such as Thbs1, Serpine1, Hbegf, Ereg, and Timp1. There was extensive signaling between MFLs and mutant CFs (CF Mut), including Bmp4 and Tgfb2 containing ligand-receptor pairs. MFLs also expressed well-characterized Yap target genes, such as Ctgf and Cyr6, that signaled to mutant CFs. These results are consistent with the non-cell-autonomous activation of CFs by Lats1/2 CKO MFLs.
Many of the top ligand-receptor pairs also connected MFLs to myeloid cells (Fig. 4B,C). Prominent among the MFL-Mϕ axis is the Csf1–Csf1r pair. Csf1 is important for Mϕ differentiation and survival and links fibroblasts and Mϕs in a stable cell circuit (Hume and MacDonald 2012; Zhou et al. 2018). Furthermore, we detected the mitogen Gas6 connected to its cognate TAM family receptor tyrosine kinases Mertk and Axl. To validate the expression of key signaling molecules, we measured the chemokine/cytokine production of control sham and Lats1/2 CKO sham hearts (Fig. 4D,E). Among the 40 chemokines/cytokines we analyzed, nine were consistently up-regulated in Lats1/2 CKO sham hearts, including TIMP1, CXCL1, CXCL12, and CSF1. Further studies are required to directly dissect the signals dictating myeloid cell recruitment to the myocardium.
Lats1 and Lats2 are required in cardiac fibroblasts for cardiac scar maturation following myocardial infarction
To study CF cell state transitions post-MI in Lats1/2 CKO CFs, we performed MI on Lats1/2 CKO hearts. Interestingly, all Lats1/2 CKO mice died by 3 wk post-MI, indicating that Lats1/2 have an adaptive function post-MI (Supplemental Fig. S4A). In Lats1/2 CKO hearts post-MI, we observed extensive fibrosis both grossly and on histology that was much more severe than the Lats1/2 CKO sham phenotype (Fig. 5A,B). The ischemic area and cardiac lumen of Lats1/2 CKO hearts post-MI were almost completely replaced by fibrotic tissue (Fig. 5B). Genetic reduction of Yap and Taz in Tcf21iCre; Lats1/2 f/f; Yap/Taz f/+ mice revealed that fibrosis was suppressed with improved survival post-MI, indicating that the fibrotic Lats1/2 CKO phenotype post-MI is at least partially Yap/Taz-dependent (Fig. 5A,B; Supplemental Fig. S4A).
Figure 5.
Lats1/2 are required for cardiac scar maturation and compaction following myocardial infarction. (A) Gross heart morphology 3 wk after MI in control, Lats1/2 CKO, and Tcf21-iCre; Lats1/2 f/f; Yap/Taz f/+ mice. Suture used to ligate the left anterior descending coronary artery (LAD) shown with white arrow. Scale bar, 2000 µm. (B) Serial sections treated with Masson's trichrome stain 3 wk after MI in control, Lats1/2 CKO and Tcf21-iCre; Lats1/2 f/f; Yap/Taz f/+ mice. Red color tissue is muscle, and blue color is collagen (fibrotic tissue). Black arrows highlight myocardial muscle tissue. Scale bar, 1000 µm. (C) Representative images of 24 h EdU-labeling of control and Lats1/2 CKO mice after MI. Samples were pulse-chased with EdU (white) and cardiac fibroblasts were labeled with GFP (green). Nuclei stained with DAPI (blue). Scale bar, 25 µm. (D) Quantification of cardiac fibroblast proliferation dynamics after myocardial infarction. Representative image of experiment shown in Figure 5C. Statistical significance was determined by Mann–Whitney U test (P-value <0.1). (E) Stacked bar plot showing percentage of cells within each phase of the cell cycle as determined by flow cytometry analysis. See also Supplemental Figure S4B,C.
In control CFs post-MI, EdU incorporation peaked at three dPMI and tapered off at five dPMI (Fig. 5C,D), consistent with previous reports (Fu et al. 2018). EdU incorporation in Lats1/2 CKO CFs post-MI peaked five dpMI, and finally plateaued by seven dpMI (Fig. 5C,D). FACS analysis at 7 d post-MI revealed that Lats1/2 CKO CFs had an increase in the proportion of cells in S phase and a reduction of cells in G1 phase in contrast to control CFs post-MI (Fig. 5E; Supplemental Fig. S4B,C).
Cardiac fibroblast and myofibroblast cell state transitions after myocardial infarction
We performed scRNA-seq on control and Lats1/2 CKO hearts 1 wk post-MI. We merged CFs from the post-MI Drop-seq data with sham CFs detailed previously (n = 9200 cells) and identified thirteen transcriptionally distinct CF clusters, including four resting cardiac fibroblast clusters (CF1–4), four injury activated fibroblast clusters (aCF1–4), and five MFL clusters (MFL1–5) (Fig. 6A,B; Supplemental Fig. S5A,B; Supplemental Table S8). We used the Monocle2 algorithm, which orders cells along an unsupervised and unbiased differentiation axis (Qiu et al. 2017) and found resting CFs were localized at one extreme, aCFs localized to an intermediate location, and MFLs occupied the distal portion of the trajectory (Supplemental Fig. S5C). The Monocle2 results were consistent with the graph-based clustering results (Supplemental Fig. S5D). We determined the density of all cells from each experiment across pseudotime (Fig. 6C). This analysis demonstrated that cells derived from Lats1/2 CKO shams more closely resembled control CFs post-MI. We next performed Fast-ATAC on sorted GFP positive Lats1/2 CKO CFs post-MI to compare to our other Fast-ATAC data sets. Importantly, Tead motifs proximal to Yap target genes, like Ankrd1, were highly accessible in Lats1/2 CKO post-MI samples (Fig. 6D). Comparative PCA revealed distinct global chromatin accessibility signatures for control sham and Lats1/2 CKO post-MI, however, control post-MI and Lats1/2 CKO sham samples clustered together (Fig. 6E). Thus, Lats1/2 repress the acquisition of an injured myofibroblast cell identity at both the transcriptional and epigenetic level.
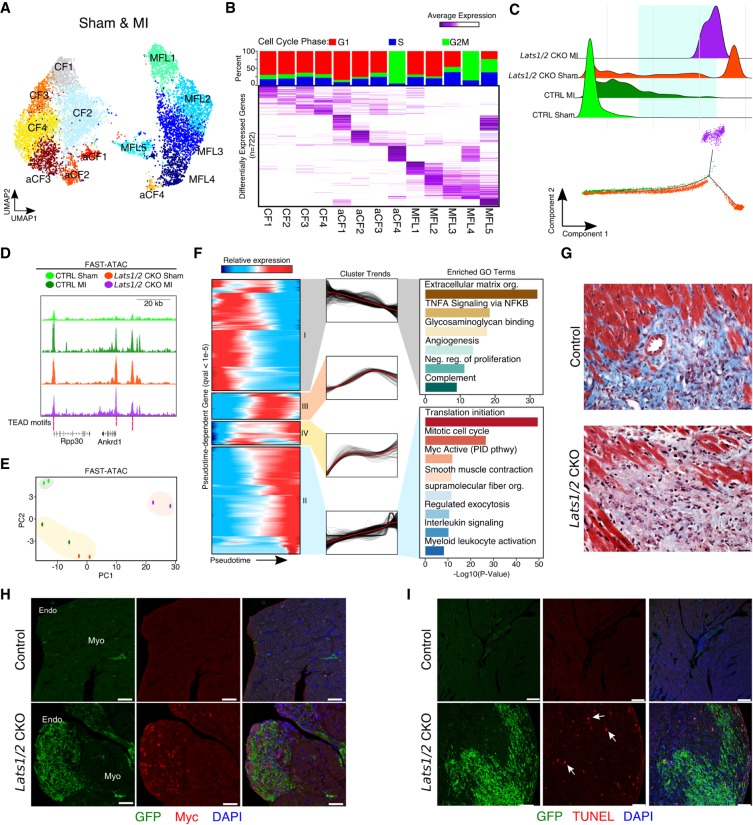
Figure 6.
Lats1/2 inhibit Myc expression and limit homeostatic cell replacement to maintain the proper cellular composition of the heart. (A) UMAP plot of cardiac fibroblast clusters. MFL, myofibroblast-like cells; CF, resting cardiac fibroblasts; aCF, activated cardiac fibroblasts. (B) Differential expression analysis and cell cycle phase of cardiac fibroblasts. (Top) Cell cycle phase analysis stacked bar graph of each cluster. Percentage of single-cell transcriptomes within each cluster scored for S-phase (blue), G1 (red), and G2-to-M phase transition (green) is shown. (Bottom) Average expression heatmap of the top differentially expressed cardiac fibroblast marker genes (n = 722). (C, top) Density plot of experimental compositions across pseudotime. (Bottom) Cardiac fibroblast differentiation trajectory. Green square highlights pseudotemporal overlap of control MI and Lats1/2 CKO CFs. (D) Genome browser tracks for Fast-ATAC. All libraries scaled equally. (E) Principal component analysis (PCA) of Fast-ATAC signals from biological duplicates of FACS sorted GFP+ cardiac fibroblasts from control and Lats1/2 CKO hearts with and without myocardial infarction. (F) Dynamic cardiac fibroblast expression patterns across differentiation following myocardial infarction. (Left) Hierarchically clustered heatmap of gene expression dynamics (q-value <1 × 10−5). (Middle) Global cluster gene expression trends across pseudotime. (Right) Gene ontology analysis for cluster I (top) and cluster II (bottom). (G) High-magnification view of Masson's trichrome stained histological sections derived from control and Lats1/2 CKO hearts 3 wk post-MI. Scale bar, 25 µm. (H) Expression of Myc (red) control and Lats1/2 CKO cardiac fibroblasts (green). Nuclei stained with DAPI (blue). Endo, endocardium; Myo, myocardium. Scale bar, 25 µm. (I) Representative images of TUNEL stained control and Lats1/2 CKO hearts without injury. Cardiac fibroblasts (green), apoptotic cells (red), nuclei (blue). See also Supplemental Figure S5F. Scale bar, 100 µm.
To detail the temporal transcriptional shifts occurring during MI induced cardiac fibroblast differentiation, we determined the gene expression dynamics that change as a function of progress (q-value <1 × 10−5) through the cardiac fibroblast differentiation axis (Fig. 6F; Supplemental Table S9). GO analysis revealed that as CFs differentiate they shift their ECM expression patterns (Fig. 6F). To highlight these shifts, we constructed a collagen and ECM-related gene list. We collapsed all CF clusters into three representative groups (CFs, aCFs, and MFLs) and determined their ECM signatures (Supplemental Fig. S5E). We observed differential expression of several matrix metalloproteases (MMPs), as well as Tissue Inhibitors of Metalloproteases (TIMPs), and many other ECM modifiers. Masson's Trichrome staining on Lats1/2 CKO hearts 3 wk post MI revealed that Lats1/2 CKO hearts post-MI exhibited differences in collagen content compared with controls post-MI, consistent with our transcriptomics data (Fig. 6G). These results detail the ECM shifts associated with Lats 1/2 inactivation in CFs. Further work is essential to clarify these findings.
GO analysis of up-regulated genes identified increased Myc activity as a hallmark of MI-induced CF differentiation (Fig. 6F, bottom). Lats1/2 CKO sham CFs expressed high levels of nuclear Myc (Fig. 6H), which is a critical regulator of tissue composition and homeostasis during development (de la Cova et al. 2004). Myc up-regulation can induce non-cell-autonomous apoptotic death. We scored apoptosis by TUNEL in Lats1/2 CKO hearts and found increased apoptotic cells that were located peripheral to GFP+ cells, suggesting that high levels of Myc contributed to the expansion of Lats1/2 CKO CFs (Fig. 6I; Supplemental Fig. S5F). These results suggest that Lats1/2 inhibit Myc expression to limit CF expansion and maintain physiologic cardiac cell composition.
Genome-wide myofibroblast Yap chromatin occupancy mapping
To gain deeper insight into the direct transcriptional targets of Yap, we analyzed genome-wide Yap chromatin binding using CUT&RUN in NIH3T3 fibroblasts, a cell line that has high Yap activity and models myofibroblasts (Ota and Sasaki 2008). We profiled the epigenetic landscape of NIH3T3 myofibroblasts using ATAC-seq along with H3K27ac, H3K4me3, and CTCF CUT&RUN (Fig. 7A). After peak calling and filtering, we identified 5941 Yap binding sites (Supplemental Table S10). The median distance of Yap to the transcriptional start site (TSS) was 29 kb (Fig. 7B), consistent with previous YAP ChIP-seq data (Galli et al. 2015).Yap binding was predominantly localized to intergenic and intronic regions (84% combined), and only 13% of peaks were found in promoters (Fig. 7C). To estimate the resolution of CUT&RUN in NIH3T3 fibroblasts, we plotted the sequenced fragment ends centered around CTCF motifs and TEAD motifs. For CTCF, we observed a large, ∼20-bp footprint, characteristic of CTCF occupancy (Supplemental Fig. S6A). The TEAD footprint at Yap-bound sites was half the size of the CTCF footprint (Fig. 7D), commensurate with the length of the Tead DNA binding motif, indicating that CUT&RUN maps Yap occupancy at TEAD motif containing regulatory elements with high precision.
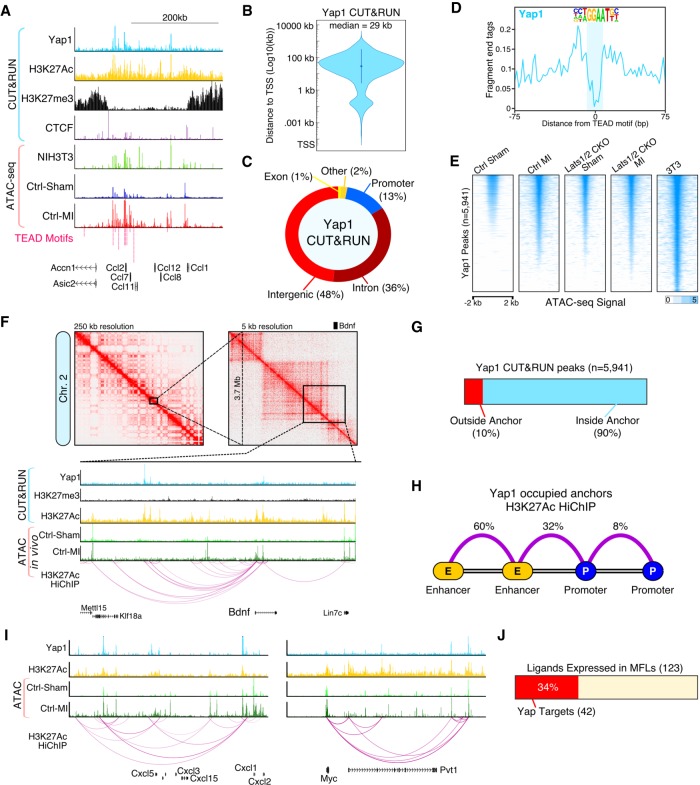
Figure 7.
The Yap enhancer connectome engages the immune system. (A) Genome browser tracks for Yap CUT&RUN and Fast-ATAC. (B) Violin plot showing the absolute distance of Yap peaks to the nearest transcription start site (TSS). (C) Fraction of Yap CUT&RUN peaks annotated with each category. (D) CUT&RUN footprint analysis for Yap at TEAD motifs. (E) Heatmap showing Fast-ATAC signal (read depth) across all myofibroblast Yap binding sites (n = 5941, P-value <1 × 10−5) for indicated experimental conditions. (F) H3K27Ac HiChIP interaction maps for in vitro myofibroblasts (NIH3T3 cells). (Top) Knight-Ruiz matrix-balanced HiChIP interaction maps plotted using Juicebox (Durand et al. 2016) at 250-and 5-kb resolutions. (Bottom) Browser tracks showing CUT&RUN, Fast-ATAC, and 3D HiChIP interaction signals. (G) Fraction of Yap CUT&RUN peaks located within H3K27Ac loop anchors. (H) H3K27Ac HiChIP loop interaction summary for Yap occupied loops. (I) Genome browser tracks for Cxcl5 and Myc. (J) Fraction of MFL expressed ligands (Fig. 4B) that are direct Yap target genes.
We used our in vivo ATAC-seq data, from CFs, to further validate the high confidence Yap binding sites. While the highest chromatin accessibility of Yap binding sites was found in NIH3T3 cells, the accessibility of Yap binding sites in CFs increased post-MI to levels similar to that observed in Lats1/2 CKO sham CFs (Fig. 7E). Overall, 1512 of the significant TEAD motif-containing ATAC peaks identified in CFs post-MI were also high confidence Yap binding sites (Supplemental Fig. S6B). Yap peaks had an active chromatin status, they displayed H3K27ac enrichment and H3K27me3 depletion consistent with previous findings (Supplemental Fig. S6C; Galli et al. 2015). Taken together, these data support the notion that Yap activity is increased in CFs post-MI.
Active distal enhancers form topological loops with target promoters. The precise annotation of Yap target genes requires a 3D contact map of active enhancer–target looping. To more firmly identify Yap target genes, we characterized the Yap-associated enhancer loop connectome in the NIH3T3 myofibroblasts using H3K27ac HiChIP (Mumbach et al. 2017). We found that Yap occupied distal enhancers that interacted with the promoters of key myofibroblast genes, like Bdnf (Fig. 7F). Importantly, 90% of Yap peaks were positioned on H3K27ac loop anchors (Fig. 7G). In addition, we found that Yap predominantly occupies enhancer–enhancer loops (60%), and enhancer–promoter loops (32%) (Fig. 7H). For further validation, we investigated the accessibility of Yap-occupied enhancer–promoter loops using our ATAC-seq data from control and Lats1/2 CKO CFs. Promoters isolated from Yap occupied loops increased in chromatin accessibility in control CFs post-MI to similar levels observed in Lats1/2 CKO CFs with or without MI (Supplemental Fig. S6D).
We next ranked and filtered the enhancer–promoter loops based on promoter gene expression from CFs post-MI and performed GO analysis (Supplemental Fig. S6E). Enriched gene categories included inflammatory signaling pathways, myeloid leukocyte migration, and TNFA signaling via NFkß. To illustrate this finding, we looked at the Cxcl5 locus and found prolific looping from Yap binding sites across this chemokine-rich locus (Fig. 7I). In total, 34% of the ligands identified as MFL-specific from our scRNA-seq analysis (Fig. 4B; Supplemental Table S11) could be annotated as direct Yap targets (Fig. 7J). Moreover, we found that many of the Yap target genes have been previously classified as Myc targets including the Myc gene itself (Fig. 7I). Indeed, we found that in NIH3T3 fibroblasts Myc was a direct Yap target, with a complex 3D enhancer “clique” (Fig. 7I). Consistent with the above findings, knockdown of Lats1/2 with siRNAs increased Myc protein levels in NIH3T3 fibroblasts (Supplemental Fig. S6F). Our results indicate that Yap occupies active enhancers in CFs and directly promotes the transcription of genes involved in the cardiac injury response and immunostimulation.
Discussion
CFs respond to injury by transitioning through cell states that provide an acute response to injury and the formation of an adaptive, long-term scar. Our data reveal that in the uninjured heart Hippo signaling maintains the resting CF cell state and prevents the spontaneous acquisition of the MFL cell state. MFLs had characteristics of an increased stress response, including increased ER stress and UPR activity. Moreover, MFLs signal to other CFs, with intact Lats1/2 activity, and myeloid cells to nonautonomously induce an inflammatory phenotype revealing a mechanism promoting progressive fibrosis and inflammation (Supplemental Fig. S7). In the post-MI context, Lats1/2 function to limit Yap activity and promote scar maturation.
Hippo signaling autonomously inhibits fibroblast proliferation and the myofibroblast cell state transition
We found that Lats1/2 CKO CFs differentiated into MFLs without injury revealing an essential role for Lats1/2 in preventing the activation of the cardiac wound response. Previously, we applied scRNA-seq to characterize the role of epicardial Hippo signaling during cardiac development and found that the Hippo pathway functions to promote proper epicardial-to-CF differentiation (Xiao et al. 2018). Thus, Hippo signaling plays essential roles in the achievement of the resting CF cell identity during embryonic development and subsequently regulates essential CF fate transitions during postnatal homeostasis and post-MI.
Lats1/2 promote resolution of the cardiac wound response after myocardial infarction
Our data indicate that Hippo signaling in CFs prevents precocious myofibroblast fate acquisition. Cardiac myofibroblasts are highly proliferative and resistant to apoptosis; however, the cardiac myofibroblast cell state is transitory and lost by 10 d post-infarct (Fu et al. 2018). Consistent with the proliferative potential of myofibroblasts, removal of Lats1/2 in CFs resulted in increased cell proliferation. Interestingly, in Lats1/2 CKO sham hearts, we observed large homogenous aggregates of Lats1/2 CKO CFs. Previous work found that myofibroblasts join stress fibers at cadherin-type intercellular adherens junctions (Hinz et al. 2004). The expression of cadherins, like the direct Yap target N-cadherin (Cdh2), play a role in homotypic segregation of myofibroblasts (Hinz et al. 2004). Thus, cell–cell adhesion and/or cell repulsion may segregate Lats1/2 CKO CFs and prevent intermingling within the myocardium. Notably, Lats1/2 CKO post-MI hearts displayed a large noncompacted scar in the post-MI period that was more severe than Lats1/2 CKO sham hearts. Our data are consistent with Hippo-independent mechanisms regulating Yap in CFs post-MI. Mechanical cues regulate Yap activity (Aragona et al. 2013) and cardiac tissue matrix stiffness progressively increases following MI (Fomovsky and Holmes 2010). Indeed, mechanotransduction induced by a stiff ECM has also been implicated in driving the myofibroblast phenotypic transition in pancreatic stellate cells (Lachowski et al. 2017). Moreover, YAP and TAZ have fibrogenic properties in pathologically stiff human lung tissue (Liu et al. 2015). Further experiments are required to determine the pathophysiologic inputs that enhance Yap activity post-MI.
Hippo signaling inhibits myeloid cell recruitment during cardiac homeostasis
The role of Hippo-Yap in inducing the immune response is still poorly understood. While some labs have found evidence for Yap-driven recruitment of M2 macrophages in the context of cancer (Guo et al. 2017) overexpression of YAP5SA in the liver failed to induce myeloid invasion. Moreover, emerging data in a cancer model suggest that LATS1/2-null cells release vesicles containing nucleic acids that engage circulating immune cells via type I Interferons and Toll-like receptors (Moroishi et al. 2016). Incidentally, IFNICs, or Interferon inducible cells, are considered post-phagocytic cardiac Mϕs with high levels of self-DNA-activated interferon regulatory factor 3 (Irf3) transcriptional activity that were originally identified as being specific to the ischemic myocardium (King et al. 2017). We speculate that the increased cell death induced by Lats1/2 CKO cells and/or the release of pro-inflammatory vesicles may partly explain the injury-like myeloid cell phenotypic expansion and the appearance of IFNICs (Mϕ6) during homeostasis.
Cell competition may contribute to maintenance of cardiac cellular composition
Cell competition is an evolutionarily conserved mechanism to maintain tissue homeostasis whereby “fit” cells (“winners”) with higher anabolic capacity eliminate “unfit” cells (“losers”) with lower anabolic capacity. In Lats1/2 CKO CFs, we observed pronounced Myc up-regulation, while nonmutant cells adjacent to CFs in Lats1/2 CKO sham hearts displayed elevated levels of apoptosis, which agrees with a cell-competition phenotype. This suggests a role for Hippo-induced cell competition in CFs during homeostasis which is extendable to the post-infarct tissue environment. One limitation of this study is that we did not investigate the cellular identity of the apoptotic cells in proximity to Lats1/2 CKO CFs. An intriguing possibility is that myofibroblast-mediated cell competition, a mechanism typically associated with the preservation of tissue fitness, promotes not only the removal of damaged cells post-MI, but also helps to generate an adaptive scar. However, further work is essential to clarify the fate of both CMs and other non-CMs in Lats1/2 CKO mice. Consistent with the nonautonomous activity of myofibroblasts, cell competition in Drosophila has been found to take place via Toll related receptors (TRRs) that are activated by cytokines to promote apoptosis in an NFKβ-dependent manner (Meyer et al. 2014).
Targeting the Hippo signaling pathway in CMs may be a viable treatment option for heart failure. In this study, we found that Hippo pathway inactivation in CFs promotes cardiac fibrosis and adversely effects cardiac function. We also found that a reduction in Yap activity suppressed the fibrotic phenotype observed in Lats1/2 CKO post-MI hearts. Importantly, these results emphasize the need for highly cell type-specific therapeutic targeting of the Hippo pathway for safe and effective heart failure therapy.
Materials and methods
Detailed methods are in the Supplemental Material (Supplemental Methods).
Mice
Tcf21iCre, Lats1/2flox/flox, Yap/Tazflox/flox (Xin et al. 2011), and Rosa26mTmG alleles have been described previously. Mice were on a mixed genetic background of C57BL/6 and 129SV. A total of 3 mg tamoxifen was administrated to 6–8-wk-old mice by intraperitoneal injection for 6 d. Surgery was performed after the six doses of tamoxifen.
Echocardiography
Cardiac function was analyzed by echocardiography every week post-surgery. Imaging was performed on VisualSonics Vevo 2100 system with 550-s probe. B-mode images and M-mode images were captured on short-axis projection. Ejection fraction, fraction shortening, and cardiac output were calculated using cardiac measurement package installed in Vevo2100 system.
Histology and immunofluorescence
For histology and immunofluorescence staining, hearts were fixed in 4% PFA overnight at 4°C and dehydrated in a serial ethanol, xylene, and embedded in paraffin. TUNEL assay was performed according to manufacturer's instruction (Progema, G3250). Immunofluorescence images were captured on a Leica TCS SP5 confocal microscope.
EdU incorporation assay
Mice post-MI were injected with EdU (0.5 mg) 24 h before collecting heart tissue. Hearts were processed as described above. EdU incorporation was assayed by Click-it EdU imaging kit (Life Technologies C10340).
Cytokine arrays
Tissue lysate was prepared according to the manufacturer's instruction. Tcf21 iCre and Tcf21 iCre; Lats1/2 fl/fl hearts were collected at 10-d post tamoxifen injection. Tissue were further homogenized with Dounce homogenizer in PBS supplemented with protease inhibitors. Ten percent Trion X-100 was added to make final concentration of 1%. Samples were frozen at −80°C for 2 h, thawed, and centrifuged at 10,000g for 5 min. Supernatant was collected for protein array. Protein loading was normalized by GAPDH. Approximately 1000–2000 µg total protein was used for each sample.
FACS analysis for cell cycle scoring
Cardiac fibroblasts were isolated from hearts at 1 wk post-MI by langendorff perfusion. GFP-positive cells were gated for analysis and DAPI were used for analyzing DNA content. FACS were performed on BD Biosciences SORP Aria I and BD Biosciences LSRII and cell cycle modeling were processed with FlowJo software.
RNAscope in situ hybridization
Formaldehyde-fixed paraffin-embedded heart sections were processed for RNA in situ detection using the RNAscope2.5 Assay (Advanced Cell Diagnostics, Inc.) according to the manufacturer's instructions.
Fast-ATAC
Approximately 10,000 FACS-sorted GFP+ cells were used as input for Fast-ATAC. Fast-ATAC was performed according to Corces et al. (2016).
Drop-seq
Adult hearts from indicated genotypes were dissected, cannulated, and then dissociated into a single-cell suspension via collagenase digestion (Collagenase A, Roche) on a custom built Langendorff apparatus. Dissociated cells were diluted to a concentration of 200 cells per microliter in PBS with 0.01% BSA. Drop-seq was then performed according to Macosko et al. (2015). Re-mapping of the data to quantify mEgfp expression was performed with zUMIs (Parekh et al. 2018) using a custom genome and gtf file that included the mEgfp sequence (including the bovine growth hormone polyA signal).
CUT&RUN
CUT&RUN experiments were carried out as described in Skene et al. (2018). Protein A-MNase (batch 6) and Yeast spike-in DNA were kindly provided by Dr. Steve Henikoff.
H3K27ac HiChIP
H3K27ac HiChIP was performed according to Mumbach et al. (2017) with only minor modifications. All libraries were sequenced on a NextSeq 500 platform.
Data availability
Data are available in the Gene Expression Omnibus (GEO) under accession number GSE135296.
Competing interest statement
J.F.M. is a founder and owns shares in Yap Therapeutics.
Supplementary Material
Acknowledgments
We thank Dr. Todd Heallen for help with densitometry. This work was supported by grants from the National Institutes of Health (DE023177, HL127717, HL130804, HL118761 [J.F.M.]; F31HL136065 [M.C.H.]; R56HL142704, R03DE025873, K01DE026561 [J.W.]), American Heart Association 14SDG19840000 (J.W.), Vivian L. Smith Foundation (J.F.M.), State of Texas funding (J.F.M.), and Fondation LeDucq Transatlantic Networks of Excellence in Cardiovascular Research (14CVD01) “Defining the genomic topology of atrial fibrillation” (J.F.M.). This work was also supported by Intellectual and Developmental Disabilities Research Center grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development and the Mouse Phenotyping Core at Baylor College of Medicine (U54 HG006348).
Author contributions: Y.X., M.C.H., and J.F.M. conceived the project; Y.X. and M.C.H. were responsible for methodology; Y.X., M.C.H., L.L., and V.D. performed the investigation; M.C.H. and Y.X. wrote the original draft; M.C.H., Y.X., and J.F.M. reviewed and edited the manuscript; J.F.M., J.W., and M.C.H. acquired funding; J.F.M. and J.W. obtained resources; J.F.M. supervised the project; M.C.H. and Y.X. visualized the project; M.C.H. and T.J.M. curated the data.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.329763.119.
References
- Acharya A, Baek ST, Banfi S, Eskiocak B, Tallquist MD. 2011. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis 49: 870–877. 10.1002/dvg.20750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VAA, Imrichova H, Hulselmans G, Rambow F, Marine J-CC, Geurts P, Aerts J, et al. 2017. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14: 1083–1086. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154: 1047–1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Baumann J, Ignashkova T, Chirasani S, Ramírez-Peinado S, Alborzinia H, Gendarme M, Kuhnigk K, Kramer V, Lindemann R, Reiling J. 2018. Golgi stress–induced transcriptional changes mediated by MAPK signaling and three ETS transcription factors regulate MCL1 splicing. Mol Biol Cell 29: 42–52. 10.1091/mbc.E17-06-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitsch CM, Kanisicak O, van Berlo JH, Molkentin JD, Yutzey KE. 2013. Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. J Mol Cell Cardiol 65: 108–119. 10.1016/j.yjmcc.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. 2016. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48: 1193–1203. 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. 2004. Drosophila myc regulates organ size by inducing cell competition. Cell 117: 107–116. 10.1016/S0092-8674(04)00214-4 [DOI] [PubMed] [Google Scholar]
- Del Re DP, Matsuda T, Zhai P, Gao S, Clark GJ, Van Der Weyden L, Sadoshima J. 2010. Proapoptotic Rassf1A/Mst1 signaling in cardiac fibroblasts is protective against pressure overload in mice. J Clin Invest 120: 3555–3567. 10.1172/JCI43569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3: 99–101. 10.1016/j.cels.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomovsky GM, Holmes JW. 2010. Evolution of scar structure, mechanics, and ventricular function after myocardial infarction in the rat. Am J Physiol Heart Circ Physiol 298: H221–H228. 10.1152/ajpheart.00495.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC, et al. 2018. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest 128: 2127–2143. 10.1172/JCI98215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan W-CC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, et al. 2015. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol Cell 60: 328–337. 10.1016/j.molcel.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhao Y, Yan H, Yang Y, Shen S, Dai X, Ji X, Ji F, Gong X-GG, Li L, et al. 2017. Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev 31: 247–259. 10.1101/gad.294348.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL. 2011. Hippo signaling: growth control and beyond. Development 138: 9–22. 10.1242/dev.045500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B, Pittet P, Smith-Clerc J, Chaponnier C, Meister J-JJ. 2004. Myofibroblast development is characterized by specific cell-cell adherens junctions. Mol Biol Cell 15: 4310–4320. 10.1091/mbc.e04-05-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA, MacDonald KP. 2012. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood 119: 1810–1820. 10.1182/blood-2011-09-379214 [DOI] [PubMed] [Google Scholar]
- King KR, Aguirre AD, Ye Y-XX, Sun Y, Roh JD, Ng RP, Kohler RH, Arlauckas SP, Iwamoto Y, Savol A, et al. 2017. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat Med 23: 1481–1487. 10.1038/nm.4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowski D, Cortes E, Pink D, Chronopoulos A, Karim SA, Morton JP, Del Río Hernández AE. 2017. Substrate rigidity controls activation and durotaxis in pancreatic stellate cells. Sci Rep 7: 2506 10.1038/s41598-017-02689-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach JP, Heallen T, Zhang M, Rahmani M, Morikawa Y, Hill MC, Segura A, Willerson JT, Martin JF. 2017. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550: 260–264. 10.1038/nature24045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lagares D, Choi KM, Stopfer L, Marinković A, Vrbanac V, Probst CK, Hiemer SE, Sisson TH, Horowitz JC, et al. 2015. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol 308: L344–L357. 10.1152/ajplung.00300.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Reginensi A. 2017. Lats1/2 Regulate Yap/Taz to control nephron progenitor epithelialization and inhibit myofibroblast formation. J Am Soc Nephrol 28: 852–861. 10.1681/ASN.2016060611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SN, Amoyel M, Bergantiños C, de la Cova C, Schertel C, Basler K, Johnston LA. 2014. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science 346: 1258236 10.1126/science.1258236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Hayashi T, Pan W-WW, Fujita Y, Holt MV, Qin J, Carson DA, Guan K-LL. 2016. The hippo pathway kinases LATS1/2 suppress cancer immunity. Cell 167: 1525–1539.e17. 10.1016/j.cell.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Satpathy AT, Boyle EA, Dai C, Gowen BG, Cho SW, Nguyen ML, Rubin AJ, Granja JM, Kazane KR, et al. 2017. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet 49: 1602–1612. 10.1038/ng.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. 2007. A global double-fluorescent Cre reporter mouse. Genesis 45: 593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Ota M, Sasaki H. 2008. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135: 4059–4069. 10.1242/dev.027151 [DOI] [PubMed] [Google Scholar]
- Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I. 2018. zUMIs - a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7 10.1093/gigascience/giy059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14: 979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski J, Goldberg T, Harshbarger J, Kloppman E, Lizio M, Satagopam V, Itoh M, Kawaji H, Carninci P, Rost B, et al. 2015. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 6: 7866 10.1038/ncomms8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook BA, Wasko RR, Rivera-Gonzalez GC, Salazar-Gatzimas E, López-Giráldez F, Dash BC, Muñoz-Rojas ARR, Aultman KD, Zwick RK, Lei V, et al. 2018. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362: eaar2971 10.1126/science.aar2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6: e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, Henikoff S. 2018. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13: 1006–1019. 10.1038/nprot.2018.015 [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Molkentin JD. 2017. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol 14: 484–491. 10.1038/nrcardio.2017.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. 2018. Recovering gene interactions from single-cell data using data diffusion. Cell 174: 716–729.e27. 10.1016/j.cell.2018.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, Theis FJ. 2019. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 20: 59 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xiao Y, Zhang S, Ji S, Wei L, Fan F, Geng J, Tian J, Sun X, Qin F, et al. 2013. The Ets transcription factor GABP is a component of the hippo pathway essential for growth and antioxidant defense. Cell Rep 3: 1663–1677. 10.1016/j.celrep.2013.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wei L, Fan F, Ji S, Zhang S, Geng J, Hong L, Fan X, Chen Q, Tian J, et al. 2015. Integration of Hippo signalling and the unfolded protein response to restrain liver overgrowth and tumorigenesis. Nat Commun 6: 6239 10.1038/ncomms7239 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Hill MC, Zhang M, Martin TJ, Morikawa Y, Wang S, Moise AR, Wythe JD, Martin JF. 2018. Hippo signaling plays an essential role in cell state transitions during cardiac fibroblast development. Dev Cell 45: 153–169.e6. 10.1016/j.devcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. 2011. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal 4: ra70 10.1126/scisignal.2002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Franklin RA, Adler M, Jacox JB, Bailis W, Shyer JA, Flavell RA, Mayo A, Alon U, Medzhitov R. 2018. Circuit design features of a stable two-cell system. Cell 172: 744–757.e17. 10.1016/j.cell.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi M, Maqsood A, Prehar S, Mohamed TM, Abou-Leisa R, Robertson A, Cartwright EJ, Ray SG, Oh S, Lim D-SS, et al. 2014. The mammalian Ste20-like kinase 2 (Mst2) modulates stress-induced cardiac hypertrophy. J Biol Chem 289: 24275–24288. 10.1074/jbc.M114.562405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the Gene Expression Omnibus (GEO) under accession number GSE135296.