Abstract
Purpose of Review:
To examine the consequences of metabolism compartmentalized at the sub-cellular level, provide prototypical examples of compartmentalized metabolism, and describe methods to examine compartmentalized metabolism.
Recent findings:
Progress in metabolomics and isotope tracing has underscored the importance of sub-cellular compartments of metabolism. The discovery of biological effects of metabolites as bioenergetic intermediates, anabolic building blocks, signaling mediators, and effectors in post-translation modifications of proteins and nucleic acids have highlighted the role of compartmentalization in determining metabolic fate. Recent advances in both direct and indirect methods to quantify compartmentalized metabolism have improved upon historical approaches. Genetically encoded metabolite sensors, chemical probes, immunoaffinity purification, and compartment resolved metabolic modeling have all been recently applied to study compartmentalization.
Summary:
Accurate measurement of metabolites in distinct sub-cellular compartments is important for understanding and pharmacologically targeting metabolic pathways in diverse disease contexts, including cancer, diabetes, heart failure, obesity, and regulation of the immune system. Direct and indirect approaches to quantify compartmentalized metabolism are advancing rapidly. Yet, major challenges remain in the generalizability, rigor, and interpretation of data from the available methods to quantify compartmentalized metabolism.
Keywords: Compartmentalization, metabolism, mitochondria
Introduction
Compartmentalization is a general principle of metabolism in complex organisms. At both the organism level and cellular level, compartmentalization of processes, pathways, enzymes and metabolites act as a spatial mechanism of control for metabolite function. Within cells, this compartmentalization can derive from physical separation by membrane bound organelles, channeling of metabolites within large enzyme superstructures, or co-localization of enzymes(1). This compartmentalization allows individual metabolites to participate in multiple distinct processes, based on the compartment where the metabolite is generated, transported, and used.
Acetyl-CoA is a prototypical compartmentalized metabolic intermediate
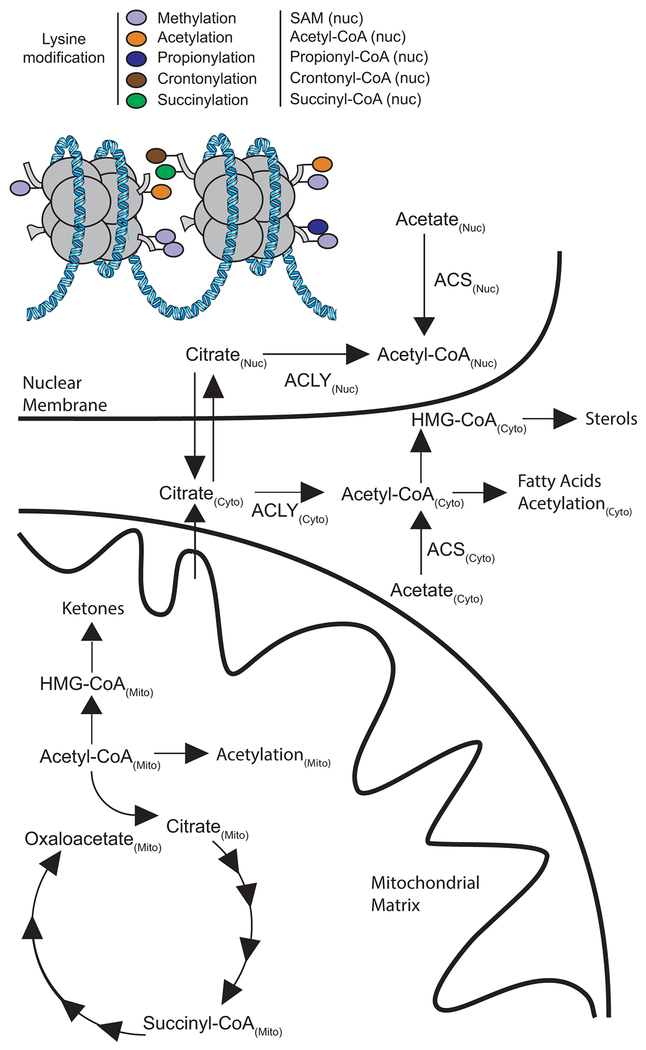
One of the clearest examples of compartmentalized metabolism is the central metabolic intermediate acetyl-Coenyzme A (acetyl-CoA) (Fig. 1). Since acetyl-CoA is the two-carbon carrier for de novo fatty acid and sterol synthesis, as well as the acyl-donor for protein and metabolite acetylation, changing concentrations of acetyl-CoA can have a range of biological consequences. Evidence in both yeast and mammalian systems has clearly shown that the mitochondrial pool of acetyl-CoA is physically and functionally distinct from the nuclear-cytoplasmic pools (2) and recent biological evidence indicates that the nuclear pool may be somewhat distinct as well (3). Both the CoA backbone and the acyl-group of the acyl-CoA are controlled by compartmentalization. Since CoA and acyl-CoAs are too polar to cross lipid membranes and high capacity transport systems for acyl-CoAs are not known to exist on the mitochondrial membrane, a shuttling system of acylcarnitines and organic anion transporters are required for short chain (C2-C6) and long chain (C10+) acyl-moieties (4).
Figure 1.
Compartmentalization of acetyl-CoA metabolism and function across the mitochondria, cytoplasm, and nucleus. Mitochondrial acetyl-CoA can contribute to the TCA cycle, mitochondrial acetylation, and ketone synthesis (via mitochondrial HMG-CoA). Cytoplasmic acetyl-CoA can be used for de novo lipid synthesis, cytoplasmic protein acetylation, and synthesis of sterols (via cytoplasmic HMG-CoA). Nuclear acetyl-CoA can be used for histone acetylation. A diversity of other histone modifications come from similarly compartmentalized metabolites.
In addition to control by substrate transport systems, mitochondrial and nucleo/cytoplasmic pools of acyl-CoAs are generated by distinct enzymatic systems. Within mitochondria, the pyruvate dehydrogenase complex (PDC) converts pyruvate (largely derived from glucose metabolism) into acetyl-CoA (5). Other important mitochondrial sources of acetyl-CoA include the pathways of fatty acid β-oxidation and branched chain amino acid catabolism. This mitochondrial acetyl-CoA is important to contribute 2 units of carbon to the TCA cycle, and can also be used to produce ketone bodies via mitochondrial 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA). In comparison, ATP citrate lyase (ACLY) is a primary enzyme responsible for cytoplasmic/ nuclear acetyl-CoA generation by ATP-dependent cleavage of citrate into oxaloacetate while transferring an acetyl group to CoASH. Additionally, the acetyl-CoA synthetases (ACSS1 and ACSS2), catalyze the ATP-dependent synthesis of acetyl-CoA from acetate and CoASH in the mitochondrial matrix (ACSS1) or cytoplasm (ACSS2). Acetyl-CoA in the cytoplasm then contributes to fatty acid synthesis, as well as other reactions including cholesterol synthesis via cytoplasmic HMG-CoA. Cytosolic acetyl-CoA production is now a validated therapeutic target, with a positive phase III trial for lowering LDL-cholesterol by the ACLY inhibitor bempedoic acid (6). Notably, bempedoic acid is the prodrug for the active acyl-CoA form which would require cytosolic compartmentalization for the pharmacologic mechanism of action. Additionally, ACSS inhibitors are under development for other uses including targeting cancer metabolism (7).
Although acetyl-CoA can likely traverse the nuclear pores making nuclear and cytoplasmic pools continuous, ACLY and ACSS2 are present in both compartments (8), suggesting that local production is important for regulation of specific processes. Interestingly, exogenous acetate appears to be inefficiently used for nuclear histone acetylation despite readily contributing to cytosolic acetyl-CoA pools (9), and nuclear ACSS2 has been shown to be important for the recycling of acetate produced by histone deacetylases reactions (10). Two recent studies have demonstrated that the nuclear localization and chromatin recruitment of ACSS2 are important for gene regulation in neurons and in cancer cells, suggesting the possibility that acetyl-CoA is produced in close proximity to nucleosomes to be acetylated (11, 12). Moreover, nuclear ACLY has been demonstrated to participate in the regulation of histone acetylation during DNA double strand break repair, promoting repair by homologous recombination (13). Further reinforcing this distinction, another study documented that the PDC is also present and functional within the nucleus (14). These findings align with isolated nuclei experiments with addition of exogenous acyl-CoAs correlating tightly with quantification of histone acylation (15). Thus, it remains possible that the nuclear/cytosolic compartment is in fact two functionally separate compartments, especially in the case of changing nutrient environments where an equilibrium between locally produced nuclear versus cytosolic pools of acetyl-CoA has not been reached.
Levels of other acyl-CoAs are important rheostats with regulatory functions. For example, cytoplasmic malonyl-CoA inhibits carnitine palmitoyl transferase 1 to prevent futile catabolic/anabolic cycling (4). This mechanism of inhibition leverages the compartmentalization of fatty acid synthesis in the cytosol and β-oxidation in the mitochondria. Downstream of acetyl-CoA as noted above, mitochondrial HMG-CoA is an intermediate in ketone body synthesis, whereas cytoplasmic HMG-CoA is an intermediate in the mevalonate pathway used for cholesterol synthesis. Moreover, compartment specific roles of other acyl-CoAs, including succinyl-, crotonoyl-, propionyl-, (iso)butyryl-, malonyl-, palmitoyl-, paltmitoleoyl-, glutaryl-CoA and even CoA itself in modifying histones and other proteins at lysine and potentially cysteine are also emerging (16).
Likewise, the localized pools of TCA cycle intermediates, the one carbon donor S-adenosyl-methionine (SAM), and the major carriers of reducing equivalents NAD(P)/NAD(P)H have disparate generation and functions depending on sub-cellular localization (1). Resolution of subcellular levels of these metabolites would contribute significantly to understanding the effects of changing each unique pool on cell function as well as growth and survival advantages provided by modulating each pool.
Considering sub-cellular compartmentalization in studies of metabolism
If a metabolite fills a distinct role or is indicative of unique processes within different compartments, measurement of a metabolite within a specific compartment provides more information on the biology of that metabolite. Thus, compartmentalization of metabolism is recognized as important but most studies of cellular metabolism do not quantitatively differentiate the metabolism within different compartments within a cell (1). The vast majority of bioanalytical techniques rely on analyzing entire populations of cells or complex fluids that may or may not contain cellular components.
Despite this predominance of the whole cell approach, some attempts to measure sub-cellular compartmentalization have been published for over 40 years. For categorization purposes, we can separate the different approaches as direct or indirect measurements of compartments. Direct approaches take the compartmentalized metabolite as the unit of analysis (i.e. mitochondrial acetyl-CoA is measured). Indirect approaches are based on a unit of analysis that is not the compartmentalized metabolite (i.e. acetylcarnitine is measured assuming an equilibrium between acetyl-CoA and acetylcarnitine). As we will illustrate, the major challenges in compartmentalized metabolomics overlap with wider metabolomics concerns including coverage of the metabolome, specificity, sensitivity, analyte stability throughout extraction, and artifacts of analysis . Additional challenges introduced by compartmentalization include the purity and specificity of the compartment being measured as well as the disruptive nature of the additional processing steps used to separate compartments.
Direct methods to quantify compartmentalized metabolites
The most commonly directly measured compartmentalization of metabolites is the distinction between extra- vs intra-cellular metabolites. Metabolites excreted from cells have obviously different functions, with the capability to act in a paracrine fashion as exhibited by succinate, arachidonic acid metabolites, and itaconate (17). Extracellular metabolites are generally more limited in undergoing further metabolism. Sampling media, interstitial fluid, or plasma/serum from effluent blood vessels all capture extra-cellular metabolites and thus measure one degree of compartmentalization.
In comparison, examination of sub-cellular compartmentalization within the cell is relatively sparse. Direct measurements of sub-cellular compartments span back to fundamental work in the 1960s-1970s with a large literature focused on the process of mitochondrial β-oxidation using isolated fractions (18). For the purposes of this review, we draw a distinction between assays using isolated compartments and studies attempting to provide a measure of a compartmentalized metabolism within a cell. The former is beyond the scope and purpose of this review. For the latter, the majority of studies on compartmentalized metabolism have used differential centrifugation, selective permeabilization, and at least one study using non-aqueous fractionation. A classic study by Idell-Wenger, Grotyohann, and Neely of total CoA and carnitine in the heart remains instructive as to the rationale, assumptions, and calculations that go into this measurement (18). Specifically, this study examined free carnitine and CoA, distinguishing between acid soluble and fat-soluble acylated forms, compared unfractionated levels with the fractionated levels, and then used a combination of experimental perturbations of the metabolism under study to examine the robustness of findings in regards to perfused ischemic or non-ischemic rat heart ventricular muscle. They found that ischemia altered the proportion of acyl derivatives in mitochondria but the cofactors (carnitine and CoA) remained constant or greater for carnitine. They were unable to measure cytoplasmic acyl-CoA. Concomitant with advances in analytical platforms, recent studies have examined systems level sub-cellular metabolomics by either gas chromatography-mass spectrometry (GC-MS) or liquid chromatography-mass spectrometry (LC-MS) based assays. Fractionation of S. cerevisiae coupled with metabolic profiling by GC-MS to probe specific mitochondrial mutants revealed pathway level perturbations resulting from two ETC mutants with deletions in complex II and complex V (19). An untargeted metabolomics pipeline using two modes of LC coupled to MS captured over 2,000 metabolomic features from isolated liver mitochondria (20). Select analytes from this analysis could differentiate sex of the animal and multivariate analysis could differentiate the wild type versus thioredoxin-2 transgenic mice used in the study.
Immunoaffinity enrichment of mitochondrial (21) and lysosomal (22) sub-cellular compartments was recently reported. This approach requires an affinity tag expressed on the target compartments, and for coupling to LC-MS required the optimization of an immunoprecipitation compatible buffer that was not suppressive in downstream analysis. Reported processing time was around 10 minutes. For the mitochondria, a systems level approach was used to quantify a large panel of metabolites in HeLa cells with 132 major mitochondrial metabolites identified and quantified. Recently, the generation of a mouse model expressing the mito specific tag significantly extended the applications of this approach enabling analysis of murine tissues ex vivo (23).
A major caveat of these direct approaches remains in how to interpret and compare the results. A fundamental question remains as to how much the measurements resulting from these techniques reflects the state before fractionation or the result of the fractionation. Although fractionation as an approach to measure exactly what is in a fraction has high face validity, little systemic effort has been conducted to address the bias that specific steps in a fractionation might create that can lead to disparate interpretations of the same observation across studies. For example, Idell-Wenger reported that they could not detect mitochondrial leakage of CoAs, but could detect active oxidation of acyl-CoAs during mitochondrial isolation that could be inhibited by pre-addition of cyanide or rotenone (18). This result was paralleled by, but interpreted differently, by Chen et al. that observed acetyl-CoA in the immuno-purified mitochondrial fraction going from an undetectable level to an observable concentration only with complex I inhibition (21).
A second critical consideration is that the timescale of these fractionations and the processing steps used to prepare cells for fractionation remains long in the context of metabolism. “Rapid” protocols on the order of 10 minutes are still slower than the reported kinetics of enzymatic reactions and likely longer than the half-lives of many of the metabolites under study. This was well illustrated by a study incorporating stable isotope tracing with a rapid digitonin based selective permeabilization (24). The rapidity of the digitonin based permeabilization used by Lee et al., allowed the authors to examine the fractional change in pool sizes over processing time, with about half of the metabolites undergoing a 50% decrease in metabolite pool size with the mitochondrial fraction kept in PBS for 10 minutes. Even the rapid-digitonin and the immunocapture methods still relied on cell lifting (for plated cells), followed by homogenization of the tissue before incubation with the capture antibodies. Other metabolomics studies have demonstrated large changes in metabolism from washing, lifting cells and/or trypsinization. Even less data is available on the effect of selective permeabilization used in most nuclear isolations. Thus, similar to the argument that studies on isolated fractions include non-physiologic conditions, fractionation (to a lesser extent) currently introduces non-physiological buffers that do not faithfully recapitulate the unfractionated environment of a cell.
Third, the interpretation of the recovered fractions should be seen as an enrichment of a given compartment. Mitochondria form intimate contacts with other compartments including endoplasmic reticulum, and existing methods enrich for these associated subcellular structures. This was acknowledged by Chen and Lee, with the details of the “contaminating” compartments specific to the fractionation used (21, 24).
Finally, it is also unclear if there is a systemic bias within or across different metabolite assays based on the compartment studied. The enzymatic and flourometric assays used in some studies have known (and suspected) cross-reactivity with other similar analytes- e.g. succinyl-CoA and malonyl-CoA with common acetyl-CoA assays. Any difference in suppression and matrix effects for LC-MS and GC-MS across different compartments has not been investigated although suppression across different isolation buffers for LC-MS was examined across two common immunoprecipitation buffers (21). Furthermore, the range of analyte concentrations measured by Chen, et al., incorporated non-linear (quadratic log-log) calibrators, an analyte response relationship not typically observed for chromatographic methods. Robustness of coupling fractionations with higher throughput metabolomics has also not been investigated and reported fractionations contain high concentrations of non-volatile phosphate buffers or supra-physiologic sucrose concentrations that may impact assay performance in larger experiments.
Thus, major technical questions in direct measurements include the purity, efficiency, reproducibility, variability, and recovery of the extractions. These questions remain difficult to answer since no study to date has compared multiple fractionations. Thus, establishing a simple generalizable fractionation technique to compare the rapidly evolving landscape of potential fractionation methods against would be of benefit. Likewise, approaches to test potential artifacts created by each step of a fractionation method would be useful.
Indirect methods to quantify compartmentalized metabolism
Indirect fractionation via analysis of perturbation on specific biochemical pathways offers another route to quantifying compartmentalized metabolism. This approach relies on understanding of the metabolic pathways being examined. The most common indirect method is the quantification of the failure of compartmentalized metabolism that can be observed in a number of genetic diseases including peroxisomal and mitochondrial disorders, mutations in transporters, as well as in the effects of toxic compounds including complex I inhibitors and fluoroacetate. In these cases, compartmentalized metabolism is inferred by knowledge about the specific pathological biochemistry. This knowledge has been effectively scaled from studying mitochondrial pathway dysfunction in cells to a population health level with newborn screening. For example, in propionic acidemia caused by mutations in the gene encoding propionyl carboxylase, increases in propionyl-carnitine reflect the mitochondrial failure to convert propionyl-CoA into methylmalonyl-CoA for entry into the TCA cycle as succinyl-CoA. Thus, external propionylcarnitine measurement has content validity to measure one aspect of mitochondrial metabolism, but may in fact be confounded by other issues (including carnitine availability, other fates of propionate, transporters, enzyme specificity, and other errors of metabolism) (25). Similarly, measurement of organic acids in urine can also provide insight into the sub-cellular function of the cells within the larger organism but the signal is complicated by other pathways, other compartments, organ differences, exchange with microbiota, and contributions from diet.
Genetic models can also be employed to query specific subcellular processes. For example, disruption of ACLY provides an indirect fractionation by uncoupling glucose derived citrate from the cytoplasmic acetyl-CoA pool(9). Coupling this genetic model with isotope tracing allows inferences to be made about the mitochondrial and cytosolic distribution of acetyl-CoA pools, as well as the interrogation of compartment-specific outputs of acetyl-CoA on histone acetylation and de novo fatty acid synthesis.
Use of specific isotopic tracers provides one way to measure compartment specific metabolism. Dissection of NAD(P)/NAD(P)H compartmentalization has seen especially clever approaches. Through deuterium (2H) labeled glucose, cytosolic and mitochondrial NADPH pools can be dissected. 3-2H-glucose produces hydride labeled NADPH via the pentose phosphate pathway, resulting in labeling of saturated fatty acids and cholesterol(26). 4-2H-glucose produces a more complicated result due to some incompletely explained incorporation of the isotopic label in fatty acids via NADPH, but can be used to trace NADH pool derived from glycolysis. 2,3,3-2H-labeled serine can also be used to dissect cytosolic versus mitochondrial metabolism by taking advantage of the compartment specific serine hydroxymethyltransferase (SHMT1(cytosolic)/SHMT2 (mitochondrial)) that converts serine to glycine as part of the 1 carbon folate cycle. This compartment specificity produces a unique label on thymidine, maintaining 2 deuteriums from cytosolic metabolism (before any exchange), and only 1 deuterium from mitochondrial serine metabolism (27). Targeting disruption of the compartment specific SHMTs also provides an indirect fractionation and demonstrates a differential nutrient dependent reliance on cytoplasmic versus mitochondrial folate metabolism.
Reporter metabolites and genetically encoded sensors of specific metabolites can also be used effectively to gauge sub-cellular metabolism. For example, Ryu and colleagues recently used such a biosensor to elucidate differential regulation of NAD pools in the nucleus versus the cytosol (28). In differentiating adipocytes, PARP1-dependent poly ADP-ribosylation (PARylation) constrains adipocyte differentiation and is rapidly suppressed upon induction of differentiation. Ryu et al., rationalized that the distribution of different isoforms of a NAD synthesizing enzyme across the nucleus and cytoplasm indicated the potential for distinct metabolite pools between these compartments. Using a genetically encoded fluorescent NAD sensor, microscopy, and flow cytometry, they demonstrated that a localized decrease in nuclear NAD, along with a commensurate increase in cytosolic NAD, occurred rapidly in differentiation reflecting both temporal and sub-cellular compartmentalization. The sum of their experimental data suggested that the critical functions of each compartmentalized pool was distinct during differentiation, with nuclear NAD production limiting nuclear PARP1 activity, thereby reducing PARylation to allow differentiation to proceed. Limitations of this approach include that flow cytometry can induce changes in metabolism (29) and the required ability to express the construct in the experimental system. Such biosensors are extremely valuable in that they allow analysis of subcellular metabolite pools in intact living cells without disruptive fractionation. However, biosensors are currently available for analysis of only a limited number of metabolites and are limited in multiplexing (multiple analyte) coverage. Further technological innovation to develop and validate sensitive biosensors for additional metabolites with distinct subcellular functions has potential for substantial impact in elucidating novel aspects of metabolic regulation.
Measurement of localized protein modifications also provides some insight into compartmentalized metabolite pools. Histone acylation is controlled by a mix of enzymatic and non-enzymatic processes, and enzymes dependent on acyl-CoAs, S-adenosyl-methionine (SAM), NAD/NADH, and alpha-ketoglutarate play roles in adding and removing histone modifications (30). DNA methylation is similarly dependent on one-carbon metabolism and cofactors for enzymes that catalyze the addition and removal of methylation (31). Likewise, mitochondrial protein post-translational modifications have been proposed to reflect similar processes within mitochondria. For example, acetylation and succinylation of mitochondrial proteins is thought to represent some degree of acetyl-CoA and succinyl-CoA metabolism (32). The limitation of this approach is that it reflects only analytes that can create a stable modification and may be influenced by processes that remove or alter the modifications being assayed.
Finally, one overarching issue in measuring metabolites within a given compartment is that many metabolites are channeled via enzymatic complexes. Channeled metabolites are a class of compartmentalized intermediates where measurement does not necessarily reflect every process dependent on those metabolites equally (33). Quantifying the fate of these channeled metabolites is more difficult, and traditionally established by isotope tracing where a labeled precursor can label the ultimate substrate, but introduction of a labeled but channeled intermediate is inefficient in labeling the product. For example, as Coleman points out, incorporation of fatty acyl-groups into complex lipids is a channeled process, thus, measurement of compartmentalized fatty acyl-CoAs would not completely measure the substrate pool for complex lipid synthesis (33). Therefore, for these metabolites, even a compartment specific measure would not provide a complete description of the substrate/product relationships. This highlights the potential for isotope tracing within compartments to complement and extend the information provided by sub-cellular metabolite quantification.
Future Directions
Examining of sub-cellular compartmentalization of metabolism is likely to become increasingly common as the tools to do so become more available. Fields that have an established history integrating metabolomics, including metabolism of the heart and liver as well as cancer metabolism are early adopters. Fundamental biochemical questions including the distinction between nuclear and cytosolic pools of metabolites remain incompletely addressed. Major challenges in interpretation, rigor, generalizability, sensitivity, and experimental feasibility still exist in moving the resolution of metabolite measurements down to the level of sub-cellular compartments.
Key points:
Metabolites in distinct subcellular compartments (i.e. mitochondria, cytosol, nucleus, etc.) have different biochemical functions.
Measurement of metabolites within subcellular compartments can be accomplished directly by measuring the fractionated pools or indirectly by using inference from other metabolites or via designed systems.
Recent advances in analytical technology have enhanced the capability of subcellular metabolite measurement, but unique challenges remain.
Comparisons of different techniques for sub-cellular metabolite measurements are needed for interpretation of results, to widen the use of compartmentalized measurement, and to aid in cross-study comparisons.
Acknowledgements:
We would like to thank Drs. Sophie Trefely, Andrew Worth, and Sanhka “Bobby” Basu for conversations contributing to this review.
Financial support and sponsorship:
KEW is supported by NIH grants R01DK116005 and R01CA228339. NWS is supported by NIH grant R03HD092630.
Footnotes
Conflicts of interest
The authors have no relevant conflicts of interest.
References
- 1.Zecchin A, Stapor PC, Goveia J, Carmeliet P. Metabolic pathway compartmentalization: an underappreciated opportunity? Current opinion in biotechnology. 2015;34:73–81. Epub 2014/12/17. doi: 10.1016/j.copbio.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Molecular cell. 2006;23(2):207–17. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 3.Sivanand S, Viney I, Wellen KE. Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem Sci. 2018;43(1):61–74. Epub 2017/1½8. doi: 10.1016/j.tibs.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerner J, Hoppel C. Fatty acid import into mitochondria. Biochim Biophys Acta. 2000;1486(1):1–17. Epub 2000/06/17. [DOI] [PubMed] [Google Scholar]
- 5.Carrer A, Wellen KE. Metabolism and epigenetics: a link cancer cells exploit. Current opinion in biotechnology. 2014;34C:23–9. doi: 10.1016/j.copbio.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray KK, Bays HE, Catapano AL, Lalwani ND, Bloedon LT, Sterling LR, Robinson PL, Ballantyne CM, Trial CH. Safety and Efficacy of Bempedoic Acid to Reduce LDL Cholesterol. N Engl J Med. 2019;380(11):1022–32. Epub 2019/03/14. doi: 10.1056/NEJMoa1803917. [DOI] [PubMed] [Google Scholar]
- 7.Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, Horton JD, Hammer RE, McKnight SL, Tu BP. Acetate dependence of tumors. Cell. 2014;159(7):1591–602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao S, Torres A, Henry RA, Trefely S, Wallace M, Lee JV, Carrer A, Sengupta A, Campbell SL, Kuo YM, Frey AJ, Meurs N, Viola JM, Blair IA, Weljie AM, Metallo CM, Snyder NW, Andrews AJ, Wellen KE. ATP-Citrate Lyase Controls a Glucose-to-Acetate Metabolic Switch. Cell Rep. 2016;17(4):1037–52. Epub 2016/10/21. doi: 10.1016/j.celrep.2016.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Genetic model that allows “pseudo” fractionation of acetyl-CoA between mitochondrial and nucleo/cytoplasmic compartments.
- 10.Bulusu V, Tumanov S, Michalopoulou E, van den Broek NJ, MacKay G, Nixon C, Dhayade S, Schug ZT, Vande Voorde J, Blyth K, Gottlieb E, Vazquez A, Kamphorst JJ. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 2017;18(3):647–58. Epub 2017/01/19. doi: 10.1016/j.celrep.2016.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mews P, Donahue G, Drake AM, Luczak V, Abel T, Berger SL. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature. 2017;546(7658):381–6. doi: 10.1038/nature22405. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Demonstrated a role for acetate as a precursor for nuclear acetyl-CoA and histone acetylation.
- 12.Li X, Yu W, Qian X, Xia Y, Zheng Y, Lee JH, Li W, Lyu J, Rao G, Zhang X, Qian CN, Rozen SG, Jiang T, Lu Z. Nucleus-Translocated ACSS2 Promotes Gene Transcription for Lysosomal Biogenesis and Autophagy. Molecular cell. 2017;66(5):684–97 e9 Epub 2017/05/30. doi: 10.1016/j.molcel.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivanand S, Rhoades S, Jiang Q, Lee JV, Benci J, Zhang J, Yuan S, Viney I, Zhao S, Carrer A, Bennett MJ, Minn AJ, Weljie AM, Greenberg RA, Wellen KE. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Molecular cell. 2017;67(2):252–65 e6 Epub 2017/07/12. doi: 10.1016/j.molcel.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158(1):84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]; ** Demonstrated a functional metabolic complex previously thought to be mitochondrial as also nuclear with a critical functional role in histone acetylation.
- 15.Simithy J, Sidoli S, Yuan ZF, Coradin M, Bhanu NV, Marchione DM, Klein BJ, Bazilevsky GA, McCullough CE, Magin RS, Kutateladze TG, Snyder NW, Marmorstein R, Garcia BA. Characterization of histone acylations links chromatin modifications with metabolism. Nat Commun. 2017;8(1):1141 Epub 2017/10/27. doi: 10.1038/s41467-017-01384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nature reviews Molecular cell biology. 2017;18(2):90–101. Epub 2016/12/08. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy MP, O’Neill LAJ. Krebs Cycle Reimagined: The Emerging Roles of Succinate and Itaconate as Signal Transducers. Cell. 2018;174(4):780–4. Epub 2018/08/11. doi: 10.1016/j.cell.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 18.Idell-Wenger JA, Grotyohann LW, Neely JR. Coenzyme A and carnitine distribution in normal and ischemic hearts. J Biol Chem. 1978;253(12):4310–8. Epub 1978/06/25. [PubMed] [Google Scholar]; ** One of the original studies on sub-cellular fractionation for measurement of comparmentalized metabolites.
- 19.Pan D, Lindau C, Lagies S, Wiedemann N, Kammerer B. Metabolic profiling of isolated mitochondria and cytoplasm reveals compartment-specific metabolic responses. Metabolomics. 2018;14(5):59 Epub 2018/04/10. doi: 10.1007/s11306-018-1352-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Fractionation of yeast using multiple genetic models and gas chromatography-mass spectrometry demonstrating compartment specific metabolic changes.
- 20.Roede JR, Park Y, Li S, Strobel FH, Jones DP. Detailed mitochondrial phenotyping by high resolution metabolomics. PLoS One. 2012;7(3):e33020 Epub 2012/03/14. doi: 10.1371/journal.pone.0033020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell. 2016;166(5):1324–37 e11 Epub 2016/08/28. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Use of an affinity tag for purification of mitochondria coupled to highly multiplexed liquid chromatography-mass spectrometry measurement of metabolites.
- 22.Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, Freinkman E, Sabatini DM. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358(6364):807–13. Epub 2017/10/28. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Extension of the affinity tag approach to sub-cellular fractionation to isolate lysosomes.
- 23.Bayraktar EC, Baudrier L, Ozerdem C, Lewis CA, Chan SH, Kunchok T, Abu-Remaileh M, Cangelosi AL, Sabatini DM, Birsoy K, Chen WW. MITO-Tag Mice enable rapid isolation and multimodal profiling of mitochondria from specific cell types in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(1):303–12. Epub 2018/12/14. doi: 10.1073/pnas.1816656115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee WD, Mukha D, Aizenshtein E, Shlomi T. Spatial-fluxomics provides a subcellular-compartmentalized view of reductive glutamine metabolism in cancer cells. Nat Commun. 2019;10(1):1351 Epub 2019/03/25. doi: 10.1038/s41467-019-09352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Rapid digitonin based fractionation coupled to isotope tracing and metabolite quantification.
- 25.Wang Y, Christopher BA, Wilson KA, Muoio D, McGarrah RW, Brunengraber H, Zhang GF. Propionate-induced changes in cardiac metabolism, notably CoA trapping, are not altered by l-carnitine. Am J Physiol Endocrinol Metab. 2018;315(4):E622–E33. Epub 2018/07/18. doi: 10.1152/ajpendo.00081.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Molecular cell. 2014;55(2):253–63. Epub 2014/06/03. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell metabolism. 2016;23(6):1140–53. Epub 2016/05/24. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, Kraus WL. Metabolic regulation of transcription through compartmentalized NAD(+) biosynthesis. Science. 2018;360(6389). Epub 2018/05/12. doi: 10.1126/science.aan5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llufrio EM, Wang L, Naser FJ, Patti GJ. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 2018;16:381–7. Epub 2018/04/09. doi: 10.1016/j.redox.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni RA, Montgomery DC, Meier JL. Epigenetic regulation by endogenous metabolite pharmacology. Curr Opin Chem Biol. 2019;51:30–9. Epub 2019/03/19. doi: 10.1016/j.cbpa.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao X, Reid MA, Kong M, Locasale JW. Metabolic interactions with cancer epigenetics. Mol Aspects Med. 2017;54:50–7. Epub 2016/09/14. doi: 10.1016/j.mam.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer JG, Softic S, Basisty N, Rardin MJ, Verdin E, Gibson BW, Ilkayeva O, Newgard CB, Kahn CR, Schilling B. Temporal dynamics of liver mitochondrial protein acetylation and succinylation and metabolites due to high fat diet and/or excess glucose or fructose. PLoS One. 2018;13(12):e0208973 Epub 2018/12/27. doi: 10.1371/journal.pone.0208973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coleman RA. It takes a village: channeling fatty acid metabolism and triacylglycerol formation via protein interactomes. J Lipid Res. 2019;60(3):490–7. Epub 2019/0½7. doi: 10.1194/jlr.S091843. [DOI] [PMC free article] [PubMed] [Google Scholar]