Abstract
Severe hypertriglyceridemia (HTG) is a relatively common form of dyslipidemia with a complex pathophysiology and serious health complications. HTG can develop in the presence of rare genetic factors disrupting genes involved in the triglyceride (TG) metabolic pathway, including large-scale copy-number variants (CNVs). Improvements in next-generation sequencing technologies and bioinformatic analyses have better allowed assessment of CNVs as possible causes of or contributors to severe HTG. We screened targeted sequencing data of 632 patients with severe HTG and identified partial deletions of the LPL gene, encoding the central enzyme involved in the metabolism of TG-rich lipoproteins, in four individuals (0.63%). We confirmed the genomic breakpoints in each patient with Sanger sequencing. Three patients carried an identical heterozygous deletion spanning the 5′ untranslated region (UTR) to LPL exon 2, and one patient carried a heterozygous deletion spanning the 5′UTR to LPL exon 1. All four heterozygous CNV carriers were determined to have multifactorial severe HTG. The predicted null nature of our identified LPL deletions may contribute to relatively higher TG levels and a more severe clinical phenotype than other forms of genetic variation associated with the disease, particularly in the polygenic state. The identification of novel CNVs in patients with severe HTG suggests that methods for CNV detection should be included in the diagnostic workup and molecular genetic evaluation of patients with high TG levels.
Keywords: bioinformatic analysis, diagnostic tools, dyslipidemias, genetic testing, human genetics, next-generation sequencing, triglycerides, lipoprotein lipase
Elevations in fasting plasma triglyceride (TG) levels are diagnosed as hypertriglyceridemia (HTG). TG levels ≥10 mmol/l (885 mg/dl) are classified as severe HTG (1) and are seen in ∼1 in 600 individuals (2). As a relatively common form of dyslipidemia with serious health complications that include pancreatitis (2, 3), there is a focus on identifying and understanding factors that can increase susceptibility to or cause severe HTG.
A combination of rare single-nucleotide variants and common SNPs can contribute permissively or causally toward the presentation of this complex disease (3). The monogenic form of severe HTG, also referred to as familial chylomicronemia syndrome (FCS), is caused by bi-allelic variants disrupting canonical genes involved in TG metabolism, such as LPL, APOC2, APOA5, glycosylphosphatidylinositol-anchored HDL-binding protein 1 (GPIHBP1), or lipase maturation factor 1 (LMF1) (4). Conversely, increased susceptibility for the polygenic form of severe HTG is due to a combination of heterozygous rare variants and common TG-raising alleles at certain SNP loci (5–13).
Previously, it has been shown that copy-number variants (CNVs) are an additional type of genetic variation that can markedly contribute to extreme perturbations of TG levels (14–17), as well as other lipid traits and disorders (18–22). Assessment of CNVs is becoming easier due to improvements in sequencing technologies and bioinformatic analysis tools (18, 23). Because of this, it is possible to screen for CNVs in patient samples concurrently with rare single-nucleotide variants and SNPs (19), and assess them as possible causes or contributors toward severe HTG.
A previous study of 563 patients with severe HTG led to the identification of one individual who was likely carrying a heterozygous CNV deletion in LPL (13). From our next-generation sequencing method and data archive (24), we expanded our search for additional LPL CNVs that might be contributing toward the presentation of severe HTG in a larger cohort of patients. We discovered a total of four out of 632 patients with severe HTG who were heterozygous carriers for one of two novel CNV deletions disrupting LPL. We molecularly confirm and characterize each deletion and discuss their likely contribution to severe HTG.
METHODS
Study subjects
Severe HTG patients (defined as TG ≥10 mmol/l or 885 mg/dl on at least one occasion) from the Lipid Genetics Clinic at the London Health Sciences Centre, University Hospital (London, Ontario, Canada), the Genomic Resource in Arteriosclerosis and Metabolic Disease recruited at the Lipid, Diabetes, or Cardiology Clinics (University of California, San Francisco, CA), or patient samples directly from collaborating research centers were screened for CNVs. Patients provided signed consent with approval from the Western University ethics review board (no. 07290E) or from the originating institution. The study of these patients is in compliance with the principles outlined by Declaration of Helsinki.
Targeted next-generation sequencing
The genomic DNA from each study subject was isolated and prepared for sequencing following our “LipidSeq” panel design. Details on our DNA preparation and sequencing method have been described in detail previously (24, 25).
Bioinformatic processing of sequencing data
Each subject’s sequencing data was imported into CLC Bio Genomics Workbench (version 12.0; CLC Bio, Aarhus, Denmark) for bioinformatic processing. Sequencing data were aligned to human reference genome (build hg19/GRCh37) and subsequently underwent local realignment for increased alignment accuracy. Depth of coverage for each sequencing read was exported as a BAM file for each subject.
Detection of CNVs
Each BAM file was imported into VarSeq® (version 2.1.0; Golden Helix, Inc., Bozeman, MT) to allow for CNV assessment in LPL using the VarSeq-CNV® caller algorithm. The depth-of-coverage information contained within each subject’s BAM file was compared against the average coverage information from a set of samples that have been confirmed to not carry CNVs. Increases and decreases in read-depth indicate a duplication or deletion of genetic material, respectively. The exact criteria used to identify CNVs has been previously described (26).
Validation of partial gene deletions
Breakpoint identification.
To confirm each deletion, we designed primers to flank the regions likely to contain the deletions and used them for PCR amplification (Expand 20 kbplus PCR System; Sigma-Aldrich St. Louis, MO; catalog number 11811002001). The forward (F) and reverse (R) primers used were: F1 5′-TACAAGACGGTGTGTTGTGTTGTGGCACGG-3′ and R1 5′-GTGACTTGATCCACAGCACAGAGCTGGAG-3′ [5′ untranslated region (UTR), exon 1 deletion]; F2 5′-AAGCTAGCTAGCTAGCTGGCTGGCCAG-3′ and R2 5′-GGGTCTCTTGCAGCTAAGTCAGAACTCCAG-3′ (5′UTR, exon 2 deletion). PCR products were run on a gel for visual confirmation of the mutant alleles. Sanger sequencing and primer-walking of the PCR products were performed to identify the deletion breakpoints.
Sanger confirmation.
After identifying deletion breakpoints by primer-walking the PCR products, screening primers spanning the proximal or distal breakpoint were designed for PCR and Sanger sequencing (supplemental Table S1).
RESULTS
Detection of CNV in LPL
We examined next-generation sequencing (NGS) data from a total of 632 individuals with severe HTG. We identified four HTG patients (Table 1) who were carriers for partial deletions in LPL using the VarSeq-CNV® caller algorithm (supplemental Fig. S1).
TABLE 1.
Clinical and demographic characteristics of subjects with LPL CNVs
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | |
| Age | 53 | 48 | 64 | 46 |
| Gender | Male | Male | Male | Female |
| Weight (kg) | 91.7 | 86.4 | 101.0 | 91.9 |
| Height (cm) | 174 | 173 | 180 | 163 |
| Ethnicity | Northern European (Anglo-Saxon); Hispanic | Northern European (Anglo-Saxon) | Northern European | Caucasian |
| Total cholesterol (mmol/l) | 10.1 | 4.98 | 7.54 | 17.4 |
| TG (mmol/l) | 36.1 | 16.7 | 35.9 | 36.4 |
| HDL cholesterol (mmol/l) | 0.59 | 0.76 | 0.45 | 0.39 |
| LDL cholesterol (mmol/l) | 1.76 | Not determined | Not determined | 1.24 |
| apo B (g/l) | 1.28 | 0.69 | 0.84 | 4.44 |
| Fasting glucose (mmol/l) | 10.0 | 6.3 | 10.0 | 11.0 |
| HbA1c, normal <6.0% | 6.8% | 6.1% | 8.0% | 5.6% |
| ALT (U/l), normal <45 U/l | 35 | 53 | 18 | 83 |
| TSH (U/l), normal <4 U/l | 1.75 | 2.18 | 3.4 | 1.34 |
| Co-morbidities and other conditions | Acute pancreatitis ×3; pancreatic pseudocyst; type 2 diabetes; carotid and aortoiliac plaque; hepatic steatosis; gout; historically highest TG was 102 mmol/l (9,000 mg/dl) | Herpes zoster; impaired glucose tolerance; hepatosteatosis | Acute pancreatitis ×3; type 2 diabetes; CABG; MI ×2; gout | Acute pancreatitis; gallstones (cholecystectomy) |
| Medications | Insulin; canagliflozin; pioglitazone; intermittent fasting; gemfibrozil; testosterone supplement | Rosuvastatin; fenofibrate | Gemfibrozil; rosuvastatin; canagliflozin; insulin; metformin; metoprolol; ramipril; amlodipine; acetylsalicylic acid; allopurinol | Gemfibrozil |
| Identified genetic determinants | LPL exon 1 deletion (heterozygous); common LPL p.D36N variant (heterozygous); normal polygenic risk score (<64th percentile) | LPL exon 1-2 deletion (heterozygous); normal polygenic risk score (<43rd percentile) | LPL exon 1-2 deletion (heterozygous); normal polygenic risk score (<77th percentile) | LPL exon 1-2 deletion (heterozygous); normal polygenic risk score (<31st percentile) |
Values provided are from first presentation to specialist lipid clinic or date first obtained. ALT, alanine transaminase; CABG, coronary artery bypass graft; HbA1c, hemoglobin A1C; MI, myocardial infarction; TSH, thyroid stimulating hormone.
Subject 1 was detected as carrying a heterozygous deletion of the 5′UTR to exon 1. From our LipidSeq panel, the CNV was detected to cover a single probe, and had an average ratio of 0.504 and average Z score of −13.030.
Subjects 2, 3, and 4 were all detected as carrying a heterozygous deletion of the 5′UTR to exon 2; the observation of subject 4’s CNV was first reported by our group earlier this year (13). From our LipidSeq panel, the CNV was detected to cover two probes. Subject 2 had an average ratio of 0.566 and average Z score of −7.058. Subject 3 had an average ratio of 0.542 and average Z score of −9.713. Subject 4 had an average ratio of 0.546 and average Z score of −7.248.
Validation of copy-number variants in LPL
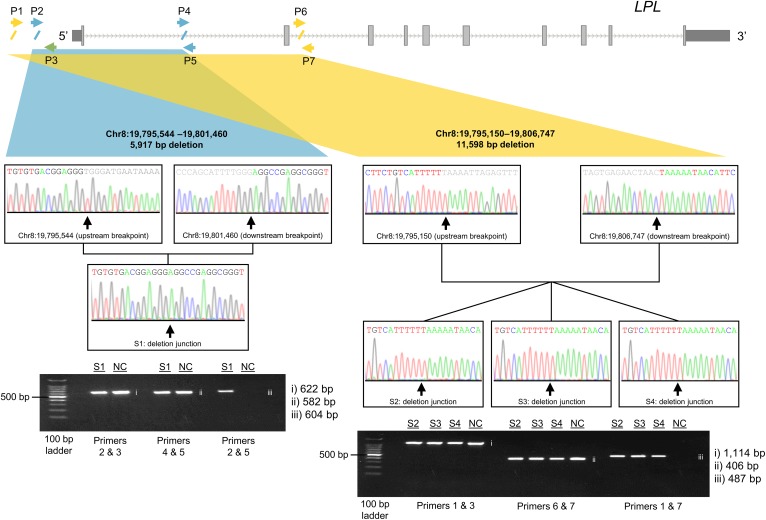
A combination of PCR primer-walking upstream and downstream of the putative CNVs and gel electrophoresis validated the deletions and allowed for their characterization (Table 2). The deletion in subject 1 was found to be 5,917 bp in size. This deletion began 1,038 bp upstream of LPL, covered the 5′UTR and exon 1, and ended 4,420 bp downstream of the splice donor site in intron 1 (Fig. 1). Subjects 2, 3, and 4 were found to have the exact same deletion, which was 11,598 bp in size. This deletion began 1,432 bp upstream of LPL, covering the 5′UTR, exon 1, and exon 2, and ended 895 bp downstream of the splice donor in intron 2 (Fig. 1). We currently do not have any information suggesting that these three individuals are related.
TABLE 2.
Genomic coordinates and breakpoints of LPL CNVs
| CNV | Zygosity State | Breakpoint | ||
| Genomic Coordinates | Length (bp) | HGVS Notation | ||
| 5′UTR, exon 1 | Heterozygous | Chr8: 19,795,544–19,801,460 | 5,917 | g.19795544-19801460del |
| c.1_88del | ||||
| p.Met1? | ||||
| 5′UTR, exon 2 | Heterozygous | Chr8: 19,795,150–19,806,747 | 11,598 | g.19795150-19806747del |
| c.1_249del | ||||
| p.Met1? | ||||
The sequences are in the forward-strand orientation. Chr, chromosome; HGVS, Human Genome Variation Society.
Fig. 1.
Validation of deletions disrupting LPL in patients with severe HTG. The LPL gene transcript with the approximate breakpoints of the smaller CNV deletion encompassing exon 1 (left) and the larger CNV deletion encompassing exons 1 and 2 (right) are indicated in blue and yellow, respectively. The diagonal slashes along the transcript indicate sequence breakpoints, while the arrows demonstrate the position and orientation of primers used in breakpoint identification and Sanger sequencing. Gel electrophoresis of PCR products across upstream and downstream breakpoints and deletion junctions for each deletion are shown for subjects 1–4. The primer pairs used for each PCR are indicated underneath the corresponding gel lanes. Chr, chromosome; NC, normal control; P, primer; S, subject.
DISCUSSION
Out of 632 patients with severe HTG, four (0.63%) were identified as carriers of one of two unique partial gene deletions in LPL. CNVs involving LPL, both deletions and duplications, have previously been identified using older methods (14–17); but to our knowledge, this is one of the first few reports identifying and characterizing LPL CNVs using an NGS-based bioinformatic method, with confirmation of the genomic breakpoints. A recent study identified a LPL CNV deletion in an individual with severe HTG last year using different NGS-based methods (27).
LPL is the primary enzyme responsible for the hydrolysis of TG-rich lipoproteins, such as chylomicrons and VLDLs (28, 29). After being chaperoned by LMF1 from parenchymal cells to endothelial cells, LPL is anchored to the vascular lumen by GPIHBP1 (29). From there, LPL binds to the apoC-II component of circulating TG-rich lipoproteins to initiate the catabolism of their TG-rich cores (29). Molecular disruptions that impair LPL mobilization or activity lead to an overall decrease in the hydrolysis of TG. With fewer TG-rich lipoproteins being catabolized, there is a resultant increase in the circulating concentration of TG, which is the defining feature of HTG.
Considering that the two identified CNVs spanning the 5′UTR to exon 1 and the 5′UTR to exon 2 both delete the initiator codon, it is almost certain that these CNVs are null mutations (30). However, the exact molecular consequences of these partial gene deletions cannot be confirmed without functional data related to mRNA expression, protein expression, or protein function. Because only heterozygous deletions were found, each patient must have multifactorial HTG (3, 12, 13) with additional factors, either genetic or environmental or both, contributing to their clinical phenotype (31). When considering polygenic and environmental factors together, this can be referred to as “multifactorial HTG” or “multifactorial chylomicronemia syndrome”. Interestingly, none of these patients have a high polygenic risk score or any other rare variants in canonical TG metabolism genes. Although the sizes of the CNVs are quite large and the reported TG levels are extremely high, these patients are not considered to have FCS, which refers specifically to a highly penetrant autosomal recessive disease. Only individuals with bi-allelic variants disrupting one of the canonical TG metabolism genes can be diagnosed with FCS.
To our knowledge, these particular LPL CNV deletions have never been reported. Overall, publications on LPL CNVs have been infrequent. In 1989, Langlois et al. (14) identified several LPL-deficient individuals with either a 2 kb insertion or a 6 kb deletion in LPL using Southern blotting. The next year, Devlin et al. (15) further characterized the insertion and showed that it was a 2 kb tandem duplication event disrupting exon 6 of LPL. Some years later, the first report of a homozygous CNV deletion in LPL was reported by Benlian et al. (16) who used a PCR-based approach to define a 2.1 kb deletion encompassing exon 9 and flanking intronic sequence in a patient with LPL deficiency. The next report on a LPL CNV was published more than a decade later, when Okubo et al. (17) described a complex deletion-insertion event. By using both Southern blot analysis and PCR, they found their LPL-deficient proband was a homozygous carrier for a 2.3 kb deletion across exon 2 and 150 bp insertion at the break junction (17). When considering more modern detection methods, a recent study by Marmontel et al. (27) identified a heterozygous LPL deletion of exons 3 to 7 in a young patient with severe HTG (TG = 87 mmol/l); this individual also carried a heterozygous single-nucleotide variant in LPL (c.642A>C) and was classified as having bi-allelic mutations and thus FCS.
Given the rarity of LPL CNVs, it was interesting to find the same deletion in three of our patients, subjects 2–4, who have no known relationship between them. Because their deletion breakpoints are identical by sequencing, it is possible that these individuals have a distant common ancestor who carried the CNV. Although these patients presently live in different geographical locations, they all self-report similar ancestry. An alternative, albeit far less likely, explanation is that the exact same CNV event occurred independently in each separate patient lineage. For this deletion, there is sequence homology and repeated sequence around the breakpoint junction, which increases the likelihood of slippage, replication errors, and CNV events (32). Despite having features that promote CNV events, the rarity of this LPL deletion in the literature and public databases suggests that this CNV is more likely shared by a common ancestor, rather than a reoccurring independent deletion.
When considering TG levels in these patients, we noted that subject 1 who had the smallest CNV also had the highest measured TGs at 102 mmol/l (9,000 mg/dl), while subjects 2 to 4 who shared the larger CNV had somewhat lower TG measurements ranging between 16.7 mmol/l (1,480 mg/dl) and 36.4 mmol/l (3,224 mg/dl). It is unclear as to whether CNV size corresponds to magnitude in TG elevation, or if it gives any indication for the function of the resultant protein product. Overall, the patients ranged from 48 to 53 years old and presented with a variety of comorbidities. Interestingly, subjects 1, 3, and 4 had reported past instances of acute pancreatitis requiring hospitalization; subjects 1 and 3 each actually had three reported episodes. Acute pancreatitis has heretofore been a more frequent manifestation among individuals with the monogenic form of severe HTG (33). We speculate that these predicted null mutations may have predisposed them to relatively higher TG levels than other types of genetic variation. Without functional studies and larger cohorts, it is difficult to isolate the CNV-specific effects. Disparities in genotype-phenotype relationships have previously been observed with CNVs underlying depressed HDL cholesterol levels, in which the same genetic variants were found in individuals with variable lipid profiles (20, 34).
The reported nongenetic factors, including comorbidities such as diabetes (Table 1), are likely contributing toward the overall severity of these patients’ HTG phenotypes and, in turn, may help to explain the frequency of acute pancreatitis episodes in these individuals. As stated previously, a single heterozygous variant is not enough to cause HTG; given that the patients did not have any additional related genetic factors identified as contributing toward their phenotypes, these nongenetic factors must be considered as likely contributory factors.
Future studies are required to characterize the functional impact of our identified CNVs on LPL activity and TG clearance pathways. Moving forward, it is also important to screen for CNVs in the other canonical TG metabolism genes, such as GPIHBP1 and APOC2, as CNVs in these genes have been previously identified in individuals with HTG (35–37).
In summary, although they are relatively infrequent, LPL CNVs are an important type of genetic variation that should be screened for when establishing the genetic basis of HTG, given their disruptive nature. With developments and improvements to next-generation sequencing techniques and more accessible CNV detection methods, CNV assessment can be easily incorporated into routine screens of rare single-nucleotide variants and polygenic risk score calculations (18, 19). Efforts must be taken to carefully characterize different determinants, including CNVs, single-nucleotide variants, and the accumulation of SNPs. By assessing a larger spectrum of genetic factors, we can achieve a more comprehensive understanding of the genetic etiology underlying severe HTG.
Supplementary Material
Acknowledgments
The authors would like to acknowledge and thank the patients involved in this study.
Footnotes
Abbreviations:
- CNV
- copy-number variant
- FCS
- familial chylomicronemia syndrome
- GPIHBP1
- glycosylphosphatidylinositol-anchored HDL-binding protein 1
- HTG
- hypertriglyceridemia
- LMF1
- lipase maturation factor 1
- NGS
- next-generation sequencing
- TG
- triglyceride
- UTR
- untranslated region
J.S.D. is supported by the Canadian Institutes of Health Research (Doctoral Research Award). R.A.H. is supported by the Jacob J. Wolfe Distinguished Medical Research Chair, the Edith Schulich Vinet Canada Research Chair in Human Genetics, the Martha G. Blackburn Chair in Cardiovascular Research, and operating grants from the Canadian Institutes of Health Research (Foundation Grant) and the Heart and Stroke Foundation of Ontario (G-18-0022147). The work at University of California San Francisco was supported by the Joseph Drown Foundation, the Campini Foundation, and gifts from Peter Read, Harold Dittmer, Susan Boeing, and Donald Yellon. R.A.H. reports consulting fees from Acasti, Aegerion, Akcea/Ionis, Amgen, HLS Therapeutics, and Sanofi. P.B.D. reports consulting fees and/or institutional research grants from Akcea, Astra Zeneca, Esperion, Regeneron, RegenXBio, and Retrophin. The other authors have no disclosures.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Hegele R. A., Ginsberg H. N., Chapman M. J., Nordestgaard B. G., Kuivenhoven J. A., Averna M., Boren J., Bruckert E., Catapano A. L., Descamps O. S., et al. 2014. The polygenic nature of hypertriglyceridaemia: implications for definition, diagnosis, and management. Lancet Diabetes Endocrinol. 2: 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dron J. S., and Hegele R. A.. 2017. Genetics of triglycerides and the risk of atherosclerosis. Curr. Atheroscler. Rep. 19: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahm A. J., and Hegele R. A.. 2015. Chylomicronaemia–current diagnosis and future therapies. Nat. Rev. Endocrinol. 11: 352–362. [DOI] [PubMed] [Google Scholar]
- 4.Johansen C. T., Kathiresan S., and Hegele R. A.. 2011. Genetic determinants of plasma triglycerides. J. Lipid Res. 52: 189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansen C. T., Wang J., Lanktree M. B., McIntyre A. D., Ban M. R., Martins R. A., Kennedy B. A., Hassell R. G., Visser M. E., Schwartz S. M., et al. 2011. An increased burden of common and rare lipid-associated risk alleles contributes to the phenotypic spectrum of hypertriglyceridemia. Arterioscler. Thromb. Vasc. Biol. 31: 1916–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansen C. T., Wang J., McIntyre A. D., Martins R. A., Ban M. R., Lanktree M. B., Huff M. W., Peterfy M., Mehrabian M., Lusis A. J., et al. 2012. Excess of rare variants in non-genome-wide association study candidate genes in patients with hypertriglyceridemia. Circ Cardiovasc Genet. 5: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansen C. T., Wang J., Lanktree M. B., Cao H., McIntyre A. D., Ban M. R., Martins R. A., Kennedy B. A., Hassell R. G., Visser M. E., et al. 2010. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 42: 684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surendran R. P., Visser M. E., Heemelaar S., Wang J., Peter J., Defesche J. C., Kuivenhoven J. A., Hosseini M., Peterfy M., Kastelein J. J., et al. 2012. Mutations in LPL, APOC2, APOA5, GPIHBP1 and LMF1 in patients with severe hypertriglyceridaemia. J. Intern. Med. 272: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., Mora S., et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Ban M. R., Zou G. Y., Cao H., Lin T., Kennedy B. A., Anand S., Yusuf S., Huff M. W., Pollex R. L., et al. 2008. Polygenic determinants of severe hypertriglyceridemia. Hum. Mol. Genet. 17: 2894–2899. [DOI] [PubMed] [Google Scholar]
- 13.Dron J. S., Wang J., Cao H., McIntyre A. D., Iacocca M. A., Menard J. R., Movsesyan I., Malloy M. J., Pullinger C. R., Kane J. P., et al. 2019. Severe hypertriglyceridemia is primarily polygenic. J. Clin. Lipidol. 13: 80–88. [DOI] [PubMed] [Google Scholar]
- 14.Langlois S., Deeb S., Brunzell J. D., Kastelein J. J., and Hayden M. R.. 1989. A major insertion accounts for a significant proportion of mutations underlying human lipoprotein lipase deficiency. Proc. Natl. Acad. Sci. USA. 86: 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devlin R. H., Deeb S., Brunzell J., and Hayden M. R.. 1990. Partial gene duplication involving exon-Alu interchange results in lipoprotein lipase deficiency. Am. J. Hum. Genet. 46: 112–119. [PMC free article] [PubMed] [Google Scholar]
- 16.Benlian P., Etienne J., de Gennes J. L., Noe L., Brault D., Raisonnier A., Arnault F., Hamelin J., Foubert L., Chuat J. C., et al. 1995. Homozygous deletion of exon 9 causes lipoprotein lipase deficiency: possible intron-Alu recombination. J. Lipid Res. 36: 356–366. [PubMed] [Google Scholar]
- 17.Okubo M., Horinishi A., Saito M., Ebara T., Endo Y., Kaku K., Murase T., and Eto M.. 2007. A novel complex deletion-insertion mutation mediated by Alu repetitive elements leads to lipoprotein lipase deficiency. Mol. Genet. Metab. 92: 229–233. [DOI] [PubMed] [Google Scholar]
- 18.Iacocca M. A., and Hegele R. A.. 2018. Role of DNA copy number variation in dyslipidemias. Curr. Opin. Lipidol. 29: 125–132. [DOI] [PubMed] [Google Scholar]
- 19.Iacocca M. A., Dron J. S., and Hegele R. A.. 2019. Progress in finding pathogenic DNA copy number variations in dyslipidemia. Curr. Opin. Lipidol. 30: 63–70. [DOI] [PubMed] [Google Scholar]
- 20.Dron J. S., Wang J., Berberich A. J., Iacocca M. A., Cao H., Yang P., Knoll J., Tremblay K., Brisson D., Netzer C., et al. 2018. Large-scale deletions of the ABCA1 gene in patients with hypoalphalipoproteinemia. J. Lipid Res. 59: 1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iacocca M. A., Wang J., Sarkar S., Dron J. S., Lagace T., McIntyre A. D., Lau P., Robinson J. F., Yang P., Knoll J. H., et al. 2018. Whole-gene duplication of PCSK9 as a novel genetic mechanism for severe familial hypercholesterolemia. Can. J. Cardiol. 34: 1316–1324. [DOI] [PubMed] [Google Scholar]
- 22.Iacocca M. A., Chora J. R., Carrie A., Freiberger T., Leigh S. E., Defesche J. C., Kurtz C. L., DiStefano M. T., Santos R. D., Humphries S. E., et al. ; ClinGen FH Variant Curation Expert Panel . 2018. Clinvar database of global familial hypercholesterolemia-associated DNA variants. Hum. Mutat. 39: 1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valsesia A., Mace A., Jacquemont S., Beckmann J. S., and Kutalik Z.. 2013. The growing importance of CNVs: new insights for detection and clinical interpretation. Front. Genet. 4: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen C. T., Dube J. B., Loyzer M. N., MacDonald A., Carter D. E., McIntyre A. D., Cao H., Wang J., Robinson J. F., and Hegele R. A.. 2014. LipidSeq: a next-generation clinical resequencing panel for monogenic dyslipidemias. J. Lipid Res. 55: 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dron J. S., Wang J., Low-Kam C., Khetarpal S. A., Robinson J. F., McIntyre A. D., Ban M. R., Cao H., Rhainds D., Dube M. P., et al. 2017. Polygenic determinants in extremes of high-density lipoprotein cholesterol. J. Lipid Res. 58: 2162–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iacocca M. A., Wang J., Dron J. S., Robinson J. F., McIntyre A. D., Cao H., and Hegele R. A.. 2017. Use of next-generation sequencing to detect LDLR gene copy number variation in familial hypercholesterolemia. J. Lipid Res. 58: 2202–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marmontel O., Charriere S., Simonet T., Bonnet V., Dumont S., Mahl M., Jacobs C., Nony S., Chabane K., Bozon D., et al. 2018. Single, short in-del, and copy number variations detection in monogenic dyslipidemia using a next-generation sequencing strategy. Clin. Genet. 94: 132–140. [DOI] [PubMed] [Google Scholar]
- 28.Olivecrona G. 2016. Role of lipoprotein lipase in lipid metabolism. Curr. Opin. Lipidol. 27: 233–241. [DOI] [PubMed] [Google Scholar]
- 29.Young S. G., and Zechner R.. 2013. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 27: 459–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter M. C., Witt T. N., Weigel B. S., Reilich P., Richard P., Pongratz D., Bonne G., Wehnert M. S., and Lochmuller H.. 2005. Deletion of the LMNA initiator codon leading to a neurogenic variant of autosomal dominant Emery-Dreifuss muscular dystrophy. Neuromuscul. Disord. 15: 40–44. [DOI] [PubMed] [Google Scholar]
- 31.Cole C. B., Nikpay M., and McPherson R.. 2015. Gene-environment interaction in dyslipidemia. Curr. Opin. Lipidol. 26: 133–138. [DOI] [PubMed] [Google Scholar]
- 32.Hastings P. J., Lupski J. R., Rosenberg S. M., and Ira G.. 2009. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10: 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paquette M., Bernard S., Hegele R. A., and Baass A.. 2019. Chylomicronemia: differences between familial chylomicronemia syndrome and multifactorial chylomicronemia. Atherosclerosis. 283: 137–142. [DOI] [PubMed] [Google Scholar]
- 34.Brunham L. R., Singaraja R. R., and Hayden M. R.. 2006. Variations on a gene: rare and common variants in ABCA1 and their impact on HDL cholesterol levels and atherosclerosis. Annu. Rev. Nutr. 26: 105–129. [DOI] [PubMed] [Google Scholar]
- 35.Patni N., Brothers J., Xing C., and Garg A.. 2016. Type 1 hyperlipoproteinemia in a child with large homozygous deletion encompassing GPIHBP1. J. Clin. Lipidol. 10: 1035–1039.e2. [DOI] [PubMed] [Google Scholar]
- 36.Hegele R. A., Berberich A. J., Ban M. R., Wang J., Digenio A., Alexander V. J., D’Erasmo L., Arca M., Jones A., Bruckert E., et al. 2018. Clinical and biochemical features of different molecular etiologies of familial chylomicronemia. J. Clin. Lipidol. 12: 920–927.e4. [DOI] [PubMed] [Google Scholar]
- 37.Rios J. J., Shastry S., Jasso J., Hauser N., Garg A., Bensadoun A., Cohen J. C., and Hobbs H. H.. 2012. Deletion of GPIHBP1 causing severe chylomicronemia. J. Inherit. Metab. Dis. 35: 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.