Abstract
Objectives
To investigate using patient posts in social media as a resource to profile off-label prescriptions of cancer drugs.
Methods
We analyzed patient posts from the Inspire health forums (www.inspire.com) and extracted mentions of cancer drugs from the 14 most active cancer-type specific support groups. To quantify drug-disease associations, we calculated information component scores from the frequency of posts in each cancer-specific group with mentions of a given drug. We evaluated the results against three sources: manual review, Wolters-Kluwer Medi-span, and Truven MarketScan insurance claims.
Results
We identified 279 frequently discussed and therefore highly associated drug-disease pairs from Inspire posts. Of these, 96 are FDA approved, 9 are known off-label uses, and 174 do not have records of known usage (potentially novel off-label uses). We achieved a mean average precision of 74.9% in identifying drug-disease pairs with a true indication association from patient posts and found consistent evidence in medical claims records. We achieved a recall of 69.2% in identifying known off-label drug uses (based on Wolters-Kluwer Medi-span) from patient posts.
Keywords: social media, data mining, off-label drug use, chemotherapy, cancer
INTRODUCTION
Nearly 20% of drugs and 30% of chemotherapeutics are prescribed for non-FDA-approved indications.1,2 This off-label prescribing is both common and costly—at $5 billion annually for just 10 drugs2—yet very poorly characterized.3,4 Therefore, it is necessary to develop innovative ways to detect and aggregate information about off-label uses from existing data sources such as electronic health records3 and patient data in social media.5
In recent years, social health networks have seen significant growth in patient activities in health forums.6,7 User posts in social media contain valuable information about prescription drug uses,8 drug safety,9 and particularly adverse drug reactions.10–12 Social media also contain various other health-related information that augment existing health data sources13,14 and can be utilized for medical research.15,16 While a large number of patients use online health forums to discuss disease and treatment options,12,17 social media is still underutilized as a source of information about off-label drugs. In a recent study, Chancellor et al.18 analyzed user posts on Reddit related to Opioid addiction and found related alternative treatments discussed and promoted by users in online communities. In prior related research, Frost et al.19 analyzed structured information entered by users of PatientsLikeMe, about two drugs with known off-label usage, and suggested that patient reported data can be used as a new source of information about off-label prescriptions. Here, we analyze patient posts in online health forums, using text mining techniques to extract information about cancer drugs prescribed off-label, and their unapproved indications. To our knowledge, this work is among the first to identify off-label drug usage from user posts in social media.
METHODS
Data
We analyzed patient posts from the Inspire health forums (www.inspire.com) from 2005 to 2016. Inspire includes 226 forums with more than 7 million discussion posts. These forums serve as a resource for patients to connect with one another, offering support and sharing their treatment experiences. We extracted data on cancer drug prescriptions by analyzing 2,425,893 patient posts from the 14 most active cancer support groups (lung, ovarian, thyroid, bladder, cervical, breast, prostate, head and neck, colorectal, appendix, kidney, stomach, skin, lymphoma).
Drug-disease candidate pair generation
By preliminary manual review of a subset of posts, we found that the vast majority of posts within a forum were from patients suffering from the disease or their caregivers, allowing posts within each cancer forum to be attributed to that disease. For every cancer support group, we paired the disease with the drugs mentioned in the messages posted in the group.
Because we were interested in including posts about direct patient experiences, rather than news or general information, we used regular expression rules to exclude posts containing hyperlinks. We extracted mentions of drugs from each post using a lexicon developed from RxNorm. We used Lucene20 to index the lexicon. Lucene has been applied as a successful tool for concept extraction in earlier studies.21,22 To identify the drug mentions in user posts, we passed the tokenized and lemmatized sentences as queries to the Lucene index. We used Medi-span to obtain each drug’s current FDA approved and known off-label usages. The extracted drugs, along with other details including the disease name, post ID, anonymized user ID, and drug approval and usage information were stored in a database for analysis. We included only cancer drugs discussed by at least six users in our analyses.
Computing drug-disease associations
To quantify the association of a drug with a disease, as a measure of potential off-label use, we calculated the information component (IC; Equation 1), a metric commonly used for pharmacovigilance signal detection.23,24 Here, we adopted the IC formula to compute an association score between a given drug and a disease (indication), based on the above-mentioned records of drug-disease pairs extracted from user posts. IC (drugx, diseasey) quantifies how frequently drugx and diseasey occur together compared to what we would expect if they were not associated. The resulting drug-disease pairs with no known indication information were considered putative off-label uses. For the purpose of plotting we calculated normalized IC values (NIC) using the normalized pointwise mutual information formula25 (Equation 2). NIC values are in the range of [−1,1], with −1 for pairs with no co-occurrence and 1 for pairs which always co-occur.
EQUATION 1
EQUATION 2
Evaluating extracted drug-disease associations
We evaluated the resulting drug-disease associations using three sources: manual review, Wolters-Kluwer Medi-span, and Truven MarketScan insurance claims.
Evaluating ranked list of extracted associations
We calculated mean average precision (MAP)26 to evaluate the ranked list of drug-disease pairs. Average precision is a score that combines precision and recall and is often used to evaluate ranked retrieval results.27,28 To calculate MAP, we selected three diseases (head and neck, thyroid, and prostate cancer) with number of extracted drugs around average among other diseases in the study, and calculated the mean of the average precision scores for the three extracted lists of drug-disease pairs. An extracted pair was considered as true positive if it had records of a known indication association (based on Wolters-Kluwer Medi-span), or if it represented an off-label association based on manual review of the related user posts. For each pair with a previously unknown association, we selected 10 random posts reported by unique users, and considered it as true positive if the majority of the users (50% or more of the randomly selected ones) reported the actual usage of the drug. We manually reviewed a total of 214 posts related to 23 potential off-label drug-disease pairs.
Measuring recall using known off-label drug usage
Next, we used the curated information in Medi-span, which captures FDA approved indications and off-label uses of drugs, to validate the usage information of the drugs frequently discussed in patient disease forums, and to compare the distribution of IC values of putative off-label uses extracted from Inspire with known off-label usages and FDA approved usages.
Last, to estimate the coverage of off-label drug usage in user posts, we generated a set of known off-label cancer drug-disease pairs (13 pairs) using the Wolters-Kluwer Medi-span drug database as the ground truth. For every disease in the study with more than 200 active users (as of June 2016), we compared the set of known off-label drugs with the extracted drugs. The drug-disease pairs in the truth set with no match in the extracted pairs by our system are considered as false negatives.
Validating extracted associations using medical insurance claim records
Finally, we validated extracted off-label uses using Truven Health MarketScan Research Databases. The Truven data capture person-specific insurance claim records across inpatient, outpatient, prescription drug, and carve-out services, and comprise diagnosis, procedure, and medication prescription records for more than 120 million individuals. For any extracted drug-disease pair, we counted patients who started taking the drug during the course of the disease. We defined the course of the disease as the time period starting with the first mention of the disease and ending 6 months after the last mention of the disease in each patient’s claims data. To compare these counts with a “negative control” set, we generated a random set of 44 drug-disease pairs that were not discussed in Inspire in the context of usage, are not known treatments, and do not have a clinical trial underway.
RESULTS
We analyzed patient posts in 14 different cancer support groups from Inspire. Table 1 shows support groups included in the study and lists information about number of users and the posts in each group. We identified 279 frequently discussed drug-disease pairs from Inspire posts. Of these, 96 are FDA approved drugs for the corresponding disease, 9 are known off-label uses, and 174 do not have records of known usage (and are therefore potentially novel, off-label uses). Figure 1 shows the distribution of Inspire NIC scores, calculated based on extracted records from posts in Inspire, for these groups. The distributions of NIC calculated based on insurance claims data are illustrated in Figure 2. The negative control drug-disease pairs often (79.3%) did not co-occur in claims data (mean co-occurrence count of 1.69; NIC value of −1). We did not illustrate the distribution of Inspire NIC for the negative control group (Figure 1), since negative controls were selected from drug-disease pairs that were not mentioned in patient posts; therefore, they all have an Inspire NIC value of −1.
Table 1.
Included cancer support groups from inspire, number of users (with at least one post), number of posts and average length of posts (average word count)
| Inspire support group | No. of users | No. of posts | Average post length | Inspire support group | No. of users | No. of posts | Average post length |
|---|---|---|---|---|---|---|---|
| Lung cancer | 19 907 | 7 94 766 | 115.1 | Head and neck cancer | 1597 | 27 821 | 131.2 |
| Ovarian cancer | 12 371 | 5 81 615 | 114.2 | Colorectal cancer | 1147 | 10 081 | 127.5 |
| Thyroid cancer | 10 375 | 2 58 587 | 115.4 | Pseudomyxoma peritonei (appendix cancer) | 325 | 2585 | 113.9 |
| Bladder cancer | 8201 | 3 08 202 | 113.3 | Kidney cancer | 314 | 2099 | 137.3 |
| Cervical cancer and HPV | 9192 | 1 34 422 | 134.8 | Stomach cancer | 132 | 696 | 127.2 |
| Advanced breast cancer | 6407 | 2 21 288 | 120.9 | Skin cancer | 153 | 432 | 109.3 |
| Prostate cancer | 3816 | 82 906 | 137.2 | Lymphoma | 122 | 393 | 116.0 |
Figure 1.
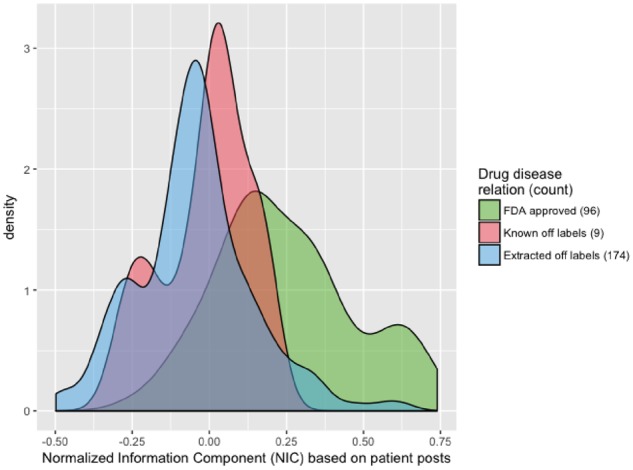
Distribution of the normalized information component (NIC) score for the extracted drug-disease pairs from Inspire posts.
Figure 2.
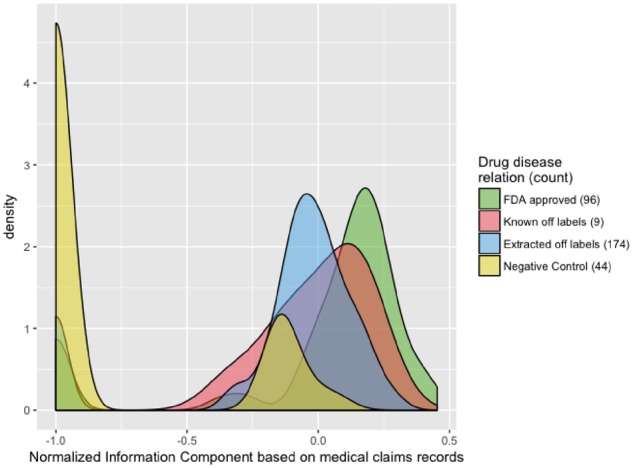
Distribution of the normalized information component (NIC) score for the extracted drug-disease pairs from insurance claims records.
We measured the average precision of the extracted ranked list results for the three selected diseases and achieved a high MAP score of 0.749.
To evaluate recall, we used known off-label pairs from Wolters-Kluwer Medi-span as the ground truth set. From a total of 13 known off-label cancer uses, 9 pairs were identified by our method, corresponding to a recall of 0.692.
Table 2 shows a list of selected off-label pairs with the number of Inspire users mentioning the pair, Inspire IC and NIC, insurance claims patient counts and claims IC and NIC.
Table 2.
DRUG off-label uses ranked by inspire information component (IC) that were discussed more than average (inspire count) in the related disease support group; claims count is the number of patients in Truven data with a record of the drug starting within the course of the corresponding disease. Claims IC is the calculated information component based on claims records. NIC is normalized information component
| Drug | Disease | Inspire count | Inspire IC (NIC) | Claims count | Claims IC (NIC) |
|---|---|---|---|---|---|
| Temodar | Skin cancer | 10 | 6.03 (0.56) | 1062 | −2.60 (−0.20) |
| Oxaliplatin | Stomach cancer | 12 | 4.88 (0.38) | 49 | 2.98 (0.17) |
| Pazopanib | Thyroid cancer | 21 | 4.36 (0.36) | 120 | 0.24 (0.01) |
| Mutamycin | Bladder cancer | 653 | 4.22 (0.63) | 169 | 3.49 (0.23) |
| Cisplatin | Head and neck cancer | 264 | 2.33 (0.29) | 34 | 2.66 (0.15) |
| Platinol | Cervical cancer | 399 | 2.31 (0.31) | 28 | 2.70 (0.15) |
| Pazopanib | Ovarian cancer | 83 | 0.39 (0.04) | 125 | 1.02 (0.06) |
| Erbitux | Lung cancer | 271 | 0.35 (0.04) | 73 | 2.49 (0.15) |
| Carboplatin | Prostate cancer | 49 | −0.14 (−0.02) | 42 | −1.49 (−0.08) |
| Avastin | Breast cancer | 562 | −0.82 (−0.12) | 173 | −0.34 (−0.02) |
| Paclitaxel | Colorectal cancer | 9 | −3.69 (−0.29) | 32 | −0.35 (−0.02) |
DISCUSSION
Our methods demonstrate that it is possible to extract information about off-label usage of drugs from patient posts to social health networks. We observed a consistent pattern in the distribution of NIC when comparing the NICs calculated from Inspire posts and insurance claims records related to different drug-disease groups (Figure 1). The NIC values for the FDA approved pairs were generally larger compared to the values associated with off-label usage. NIC values reflect how commonly the drug and the disease were observed together with negative values indicating less frequent events, which is expected in the case of off-label usage of drugs. As illustrated in Figure 2, the negative control group often had no co-occurrence in insurance claim records. There are only four drug-disease pairs in the negative control set that had co-occurrence counts of more than 5 in the claims records. These pairs may indicate off-label usage, and can be further investigated in future studies.
We performed an analysis to identify sources of errors in false positive extracted pairs. We found that the system extracts and assigns high scores to pairs related to popular clinical trials that are often actively discussed by patients in the forums. Interestingly, out of 7 false positive pairs, 6 (85.7%) belong to this category and the majority of the related posts were about ongoing or future clinical trials and contained information about the new drugs’ effectiveness, and associated adverse reactions, for example. We found that 4 out of 6 pairs in the clinical trial category were related to immune checkpoint inhibitors (Pembrolizumab, Nivolumab, Ipilimumab), newer drugs with relatively recent FDA approval dates considering the time of the discussions. Future studies may apply natural language processing methods to classify and exclude posts related to clinical trials to obtain a more refined set of drug-disease pairs with true indication associations.
Health-related information extraction from social media can be challenging since user postings are often informal and may contain noise. For instance, some posts only contain information about news or scientific findings in literature and are not first-hand patient experiences. Future studies may explore using machine learning text classification methods as a preprocessing step for both noise detection and selecting posts with first-hand experiences.
CONCLUSION
We demonstrate the proof-of-principle detection of off-label drug use from patient-generated content in social media using text mining methods. We identified off-label uses by analyzing medical insurance claims data and a database of known off-label uses.
Further characterization of off-label drug use in social media may aid in understanding why such use occurs, and also offer insight into temporal usage patterns. For example, Avastin appeared as the top ranked off-label breast cancer prescription, with continued mentions after market withdrawal in 2011. Additionally, while most chemotherapeutics are approved for organ-specific malignancies, prescriber knowledge of a targetable, shared mutation in a cancer of another organ may lead to off-label use such as the use of Enasidenib for cancers with IDH-2 mutations. Finally, complexities of health insurance coverage may lead to the selection of off-label alternatives. Mining social health network data offers a view of off-label prescribing practices from the patient’s perspective and augments existing data to better profile this phenomenon. User posts in social media also contain information about off-label drug efficacy, safety and overall patient satisfaction about the drug; future research may explore natural language processing and sentiment analysis techniques to mine such valuable information.
FUNDING STATEMENT
This work was supported by the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) grant number 5R01GM101430-05.
CONTRIBUTORSHIP STATEMENT
A.N., J.D.R. and N.H.S. drafted the manuscript and subsequent revisions. A.N. carried out all data analyses. J.D.R., A.C. and V.P. contributed to data collection and analysis. J.D.R., A.C., and N.H.S. edited and provided feedback on all drafts and responses to reviewers.
COMPETING INTERESTS
The authors have no competing interests to declare.
ACKNOWLEDGEMENTS
We thank the Inspire team for making this dataset available to us for analysis, including: Erik Jones, Brian Loew, Peter Hartzler, Jeff Terkowitz, and Kathryn Ticknor.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Radley DC, Finkelstein SN, Stafford RS.. Off-label prescribing among office-based physicians. Arch Intern Med 2006; 1669: 1021–6. [DOI] [PubMed] [Google Scholar]
- 2. Conti RM, Bernstein AC, Villaflor VM, et al. Prevalence of off-label use and spending in 2010 among patent-protected chemotherapies in a population-based cohort of medical oncologists. J Clin Oncol 2013; 319: 1134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jung K, LePendu P, Chen WS, et al. Automated detection of off-label drug use. PLoS One 2014; 9. doi: 10.1371/journal.pone.0089324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dang T-T, Ouankhamchan P, Ho T-B. Detection of new drug indications from electronic medical records. In: Computing & Communication Technologies, Research, Innovation, and Vision for the Future (RIVF), 2016 IEEE RIVF International Conference on 2016: 223–8. Hanoi, Vietnam.
- 5. Frost JH. Expert review of pharmacoeconomics & outcomes research. Expert Rev Pharmacoecon Outcomes Res 2011; 11: 371–3. [DOI] [PubMed] [Google Scholar]
- 6. Swan M. Emerging patient-driven health care models: an examination of health social networks, consumer personalized medicine and quantified self-tracking. Int J Environ Res Public Health 2009; 62: 492–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wicks P, Massagli M, Frost J, et al. Sharing health data for better outcomes on PatientsLikeme. J Med Internet Res 2010; 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvaro N, Conway M, Doan S, et al. Crowdsourcing Twitter annotations to identify first-hand experiences of prescription drug use. J Biomed Inform 2015; 58: 280–7. [DOI] [PubMed] [Google Scholar]
- 9. Sarker A, Smith K, O’Connor K, et al. Social media mining for toxicovigilance: automatic monitoring of prescription medication abuse from Twitter. Drug Saf 2016; 393: 231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leaman R, Wojtulewicz L, Sullivan R, et al. Towards internet-age pharmacovigilance: extracting adverse drug reactions from user posts to health-related social networks. In: Proceedings of the 2010 workshop on biomedical natural language processing, 2010: 117–25. http://acl.eldoc.ub.rug.nl/mirror/W/W10/W10-19.pdf#page=131 Accessed November 11, 2012.
- 11. Nikfarjam A, Sarker A, O’Connor K, et al. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc 2015; 22 (3): 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sarker A, Ginn R, Nikfarjam A, et al. Utilizing social media data for pharmacovigilance: A review. J Biomed Inform 2015; 54: 202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harpaz R, DuMouchel W, Shah NH, et al. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Ther 2012; 916: 1010–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harpaz R, DuMouchel W, Schuemie M, et al. Toward multimodal signal detection of adverse drug reactions. J Biomed Inform 2017; 76: 41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mossanen M, Chu A, Smith AB, et al. Inferring bladder cancer research prioritization from patient-generated online content. World J Urol 2018;37: 1–6. [DOI] [PubMed] [Google Scholar]
- 16. Sarker A, Chandrashekar P, Magge A, et al. Discovering cohorts of pregnant women from social media for safety surveillance and analysis. J Med Internet Res 2017; 1910: e361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou WS, Hunt YM, Beckjord EB, et al. Social media use in the united states : implications for health communication. J Med Internet Res 2009; 114: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chancellor S, Nitzburg G, Hu A, et al. Discovering Alternative Treatments for Opioid Use Recovery Using Social Media. In: CHI Conference on Human Factors in Computing Systems. Glasgow, Scotland; 2019.
- 19. Frost J, Okun S, Vaughan T, et al. Patient-reported outcomes as a source of evidence in off-label prescribing: analysis of data from PatientsLikeMe. J Med Internet Res 2011; 131: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Apache Lucene 2018. https://lucene.apache.org/
- 21. Leaman R, Dogan RI, Lu Z.. DNorm: Disease name normalization with pairwise learning to rank. Bioinformatics 2013; 2922: 2909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nikfarjam A, Sarker A, O’Connor K, et al. Pharmacovigilance from social media: Mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Informatics Assoc 2015; 22: 671–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bate A, Lindquist M, Edwards IR, et al. A data mining approach for signal detection and analysis. Drug Saf 2002; 256: 393–7. [DOI] [PubMed] [Google Scholar]
- 24.Madigan D, Ryan P, Simpson S, Zorych I. Bayesian methods in pharmacovigilance. Bayesian Statistics. 2010; 9: 421–38.
- 25. Bouma G. Normalized (Pointwise) mutual information in collocation extraction. In: Proceedings of GSCL; 2009: 31–40. Potsdam, Germany.
- 26. Manning C, Raghavan P, Hinrich S. Introduction to Information Retrieval. Natural Language Engineering. 2010; 16 (1): 100–103.
- 27. Harpaz R, Vilar S, DuMouchel W, et al. Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Am Med Informatics Assoc 2013; 203: 413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zorych I, David M, Patrick R, et al. Disproportionality methods for pharmacovigilance in longitudinal observational databases. Stat Methods Med Res 2013; 221: 39–56. [DOI] [PubMed] [Google Scholar]


