Abstract
G protein-coupled receptors (GPCRs) are membrane receptors whose agonist-induced dynamic conformational changes trigger heterotrimeric G protein activation, followed by GRK-mediated phosphorylation and arrestin-mediated desensitization. Cytosolic regions of GPCRs have been studied extensively because they are direct contact sites with G proteins, GRKs, and arrestins. Among various cytosolic regions, the role of helix 8 is least understood, although a few studies have suggested that it is involved in G protein activation, receptor localization, and/or internalization. In the present study, we investigated the role of helix 8 in dopamine receptor signaling focusing on dopamine D1 receptor (D1R) and dopamine D2 receptor (D2R). D1R couples exclusively to Gs, whereas D2R couples exclusively to Gi. Bioinformatic analysis implied that the sequences of helix 8 may affect GPCR-G protein coupling selectivity; therefore, we evaluated if swapping helix 8 between D1R and D2R changed G protein selectivity. Our results suggest that helix 8 is not involved in D1R-Gs or D2R-Gi coupling selectivity. Instead, we observed that D1R with D2R helix 8 or D1R with an increased number of hydrophobic residues in helix 8 relative to wild-type showed diminished β-arrestin-mediated desensitization, resulting in increased Gs signaling.
Keywords: GPCR, G protein, Helix 8, Dopamine receptor, Arrestin
INTRODUCTION
G protein-coupled receptors (GPCRs) are the largest family of membrane proteins, and more than 800 GPCRs in humans are distributed throughout the body (Fredriksson et al., 2003). GPCRs control numerous cellular processes, which makes them attractive drug targets for treatment of many human diseases including diabetes, osteoporosis, obesity, cancer, and neurological and cardiovascular diseases; approximately 40% of all FDA-approved drugs target GPCRs (Santos et al., 2017).
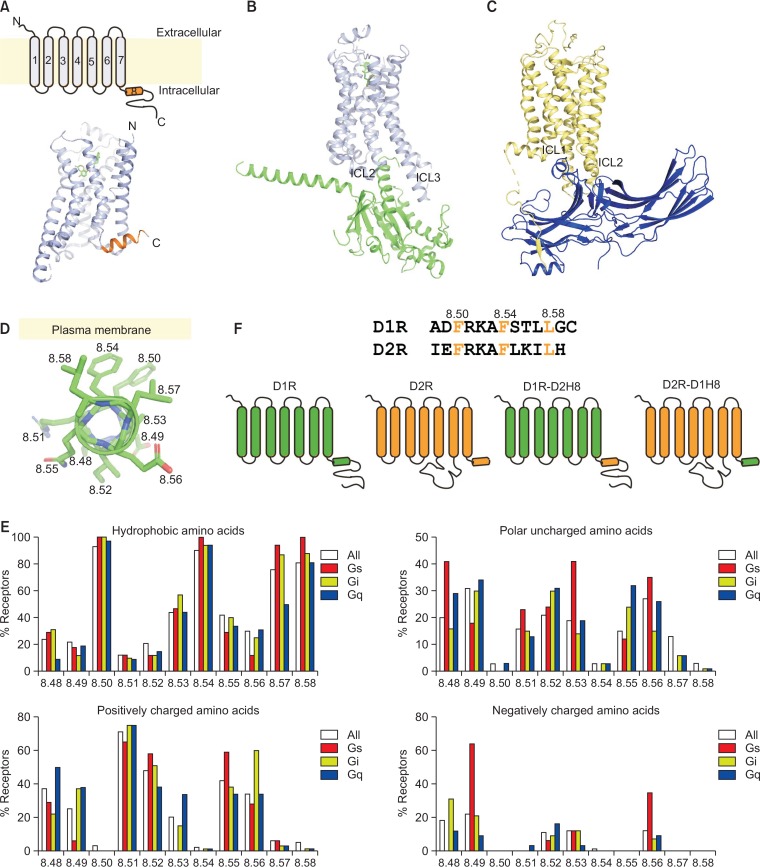
Because of the importance of GPCRs in physiology and pathology, the structural mechanism of GPCR signaling is of great interest. GPCRs have seven α-helical transmembrane domains (TMs 1–7) and an 8th α-helical domain (helix 8) located parallel to the plasma membrane (Fig. 1A). High-resolution structural and biochemical/biophysical studies have provided information about ligand-binding pockets, ligand-induced dynamic conformational changes, and binding interactions with downstream signaling molecules such as G proteins and arrestins (Hilger et al., 2018; Weis and Kobilka, 2018; Edward Zhou et al., 2019). Outward movement of TM6 appears to be the key structural element for coupling to G proteins, while intracellular loops 2 and 3 (ICL2 and 3) and a cytoplasmic pocket formed by TMs directly contact G protein α subunit (Fig. 1B) (Rasmussen et al., 2011). Arrestin interacts with the cytoplasmic pocket, ICL2, ICL3, and phosphorylated C-tail of the receptor (Fig. 1C) (Zhou et al., 2017). Because the arrestin binding sites overlap with the G protein binding sites of GPCRs, arrestin binding may prevent G protein binding, which could be the mechanism of arrestin-mediated desensitization of GPCR-G protein coupling.
Fig. 1.
GPCR structures and database analysis of helix 8. (A) Two-dimensional (2D) (upper panel) and 3D (lower panel) structures of the β2-adrenergic receptor (β2AR) (PDB: 3SN6), a representative class A GPCR. Helix 8 is highlighted in orange. (B) High-resolution crystal structure of a β2AR-Gs complex (PDB: 3SN6) with β2AR in light blue and the Gsα Ras-like domain in green. (C) High-resolution crystal structure of a rhodopsin-arrestin complex (PDB: 5W0P), with rhodospin in yellow and arrestin in blue. (D) Representative positions of side chains in helix 8 of β2AR (PDB: 3SN6). (E) Database analysis of helix 8 of all class A GPCRs whose coupling G proteins are known. (F) Schematic illustration of helix 8 chimeric constructs of D1R and D2R.
Although high-resolution structures have provided insight into the signaling mechanisms of GPCRs, these high-resolution structures are mostly energetically stable GPCR-G protein or GPCR-arrestin complexes and do not provide insight into conformational dynamics or the initial interaction modes of the complexes. Although helix 8 has not received much attention in these high-resolution structures, a number of biochemical/biophysical studies has suggested that helix 8 is involved in proper localization of receptors, G protein activation, G protein selectivity, and internalization (Faussner et al., 2005; Ahn et al., 2010; Feierler et al., 2011; Kaye et al., 2011; Kirchberg et al., 2011; Kuramasu et al., 2011; Kawasaki et al., 2015; Zhu et al., 2015). However, some of these studies have reported contradictory results, and the role of helix 8 in GPCR signaling is not clearly understood.
Dopamine is an important neurotransmitter in the central nervous system, regulating locomotor activity, attention, motivation, and memory (Missale et al., 1998). Dopamine acts through binding five class A GPCRs, dopamine D1 through D5 receptors. D1R and D5R are categorized as D1-like dopamine receptors and couple to Gs, while D2R, D3R, and D4R are classified as D2-like dopamine receptors and couple to Gi/o (Missale et al., 1998; Beaulieu and Gainetdinov, 2011). Improved structural understanding of these dopamine receptors would facilitate more effective drug development for diseases involving dopaminergic neurons such as schizophrenia, addiction, ADHD, and Parkinson’s disease (Moritz et al., 2018). In the present study, we investigated the role of helix 8 in D1R and D2R signaling with a focus on Gs- or Gi/o-coupling selectivity and desensitization of D1R-Gs signaling.
MATERIALS AND METHODS
D1R and D2R construct generation
Human D1R and D2R encoded in expression vector pcD-NA3.1 were tagged with the FLAG epitope at the N-terminus. Mutant constructs were created using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, CA, USA) according to the manufacturer’s protocol.
Immunofluorescence to determine the cellular localization of dopamine receptors
HEK293 cells (1×104) were seeded onto glass coverslips coated with poly-L-lysine (SPL, Pocheon, Korea) in 35 mm black dishes. After 24 h, cells were transfected with about 0.5 μg of dopamine receptor construct using Omicsfect (Omics Bio, New Taipei City, Taiwan) in antibiotic-free DMEM for 40–48 h according to the manufacturer’s instructions. After fixing cells with 4% paraformaldehyde for 15 min at room temperature and washed three times for 5 min in PBS. To permeabilize cells, cells were incubated for 5–10 min with 0.1% Triton-X100 in PBS and washed three times with PBS. The non-permeabilized or permieablilized cells were blocked with 5% BSA in PBS for 1 h at room temperature. Cells were labeled with anti-FLAG antibody (Sigma Aldrich, St. Louis, MO, USA) at a 1:500 dilution for 2 h at room temperature. After three washes with PBS, cells were incubated with Rhodamine Red-X- or Alexa488-labeled secondary antibodies (Invitrogen, CA, USA) at a 1:500 dilution for 1 h at room temperature. After a further three washes in PBS, cells were viewed under a laser scanning confocal microscope (LSM 710, Carl Zeiss, Jena, Germany). To visualize nucleus of the permeabilized cells, the cells were stained with DAPI (1 μg/mL, Sigma Aldrich) for 10 min.
Analysis of cell surface expression of D1R
15,000 cells were seeded on the 96 well plate, and on the next day the cells are transiently transfected with different amounts of WT D1R-containing plasmid as described above. Approximately 44–48 h after transfection, cells were washed with ice-cold PBS and then fixed in 4% formaldehyde, followed by three washes with PBS for 5 min. Cells were blocked with 5% BSA in PBS, washed once with PBS, and then labeled with anti-FLAG antibody (Sigma Aldrich) at a 1:500 dilution for 2 h at room temperature. After three washes with PBS, cells were incubated with anti-mouse-HRP at a 1:10,000 dilution (Bethyl, TX, USA) for 1 h, followed by an extensive wash. We added 100 μl ECL solution (GE Healthcare, IL, USA), and luminescence was measured for 1 s using a microplate reader (Tristar2 LB942, Berthold Technologies, Bad Wildbad, Germany).
Generation of HEK293 stably expressing 22F (HEK293-p22F)
HEK293 cells were cultivated in DMEM medium supplemented with 1% penicillin-streptomycin solution (Wellgene, Gyeongsan, Korea) and FBS (10% v/v) at 37°C in a humidified atmosphere with 5% CO2. Early passage HEK293 cells were seeded in a 6-well plate, and cells were transfected with 1 μg of 22F plasmid (Promega, WI, USA). Positive clones were selected using hygromycin B (200 μg/mL), and stable expression of 22F was determined using the Glo-SensorTM cAMP assay kit (Promega) according to the manufacturer’s instructions.
Glo-SensorTM cAMP assay to measure Gs signaling
HEK293-p22F cells were transiently transfected with dopamine receptor constructs as described above. After 40–48 h, intracellular cAMP accumulation was measured using the Glo-SensorTM cAMP assay (Promega) according to the manufacturer’s instructions. Briefly, on the day of assay, culture media was removed, and cells were pre-equilibrated for 120 min with 2% v/v of GloSensor cAMP reagent in reaction medium (90% CO2-independent medium, 10% FBS) at 37°C and 5% CO2. Cells were then treated with various concentrations of dopamine for 5 min at room temperature, and luminescence was measured for 20 min using a microplate reader. Forskolin (10 μM) was used as a positive control. All experiments were performed at least three times independently.
Luciferase reporter gene assay for Gs and Gq signaling
CHO cells were plated in a 96-well white plate (Corning, NY, USA) in DMEM (Hyclone, GE Healthcare Life Sciences, UT, USA) containing 10% FBS and 1% penicillin at a concentration of 1.5×104 cells/well. After a 24-h incubation, SRE plasmid (pGL4.33[luc2P/SRE /Hygro]) (Promega) or CRE plasmid (pGL4.29[luc2P/CRE /Hygro]) (Promega) was co-transfected with D1R WT or D1R-K8.52A plasmid using Lipofectamine 2000 (Invitrogen). After 3 h of incubation, transfected CHO cells were treated with various concentrations of dopamine. Control cells were treated with 1% DMSO. After a 24-h incubation, firefly luciferase activity was measured using the Bright-GloTM assay system (Promega) following the manufacturer’s instructions.
Data analysis
Experimental data were analyzed and graphically processed using GraphPad Prism 5 (GraphPad Prism Software Inc., CA, USA). Data are expressed as mean ± SEM.
RESULTS
Effects of helix 8 on coupling selectivity of D1R-Gs and D2R-Gi
A few studies suggested that helix 8 is involved in G protein signaling; site-directed mutagenesis revealed that helix 8 is important for agonist-induced activation of histamine H3 receptor (Kuramasu et al., 2011); helix 8 is important for odorant receptor-mediated G protein signaling (Kawasaki et al., 2015); Alanine substitution mutations showed that helix 8 of chemokine receptors CCR2a and CCR2b mediates Gq coupling (Markx et al., 2019); Scanning mutagenesis of M1 muscarinic acetylcholine receptor helix 8 delineated G protein recognition sites (Kaye et al., 2011); Moreover, an NMR study with μ-opioid receptor suggested that helix 8 might be the initial interface between a GPCR and G protein implying that the initial interface may determine receptor-G protein selectivity (Sounier et al., 2015). However, there has been no systemic bioinformatics analysis of the relationship between helix 8 and GPCR-G protein selectivity.
To gain insight into the role of helix 8 in GPCR-G protein pair selectivity, we analyzed and compared the sequence of helix 8 of all class A GPCRs with known coupling G proteins according to the database GPCRdb (http://gpcrdb.org/) (Fig. 1E) (Pandy-Szekeres et al., 2018). We adopted the modified Ballesteros-Weinstein numbering scheme to identify specific residues on receptors (Isberg et al., 2015). Within helix 8, conserved residue 8.50 is mostly a bulky hydrophobic amino acid such as Phe, Leu, Val, or Tyr because this residue interacts with the plasma membrane (Fig. 1D, 1E). Similarly, 8.54 and 8.58, which are residues facing the plasma membrane, are composed of hydrophobic amino acids (Fig. 1D, 1E). In contrast, residues facing the cytosol tend to be hydrophilic amino acids, and residues 8.51 and 8.52 are generally positively charged (approximately 75% and 50% of analyzed GPCRs, respectively) (Fig. 1D, 1E). When we analyzed and grouped residues on helix 8 depending on the coupling G protein, we found that residue 8.49 is more frequently negatively charged in Gs-coupling receptors (approximately 60% of Gs-coupling receptors), while residue 8.56 is more frequently positively charged in Gi-coupling receptors (approximately 60% of Gi-coupling receptors) (Fig. 1E). Therefore, we hypothesized that helix 8 may be involved in GPCR-Gs or GPCR-Gi coupling selectivity.
To test this hypothesis, we generated chimeric constructs in which helix 8 of D1R and D2R was swapped (Fig. 1F). Based on GPCRdb, although many receptors can couple to more than one type of G protein, D1R and D2R couple exclusively to Gs and Gi, respectively. Therefore, these receptors are not only important drug targets but also good model systems to study the Gs- or Gi-coupling mechanism. Although both D1R and D2R have negatively charged amino acids at position 8.49 (i.e., Asp for D1R and Glu for D2R), residue 8.56 is positively charged in D2R (i.e., Thr for D1R and Lys for D2R) (Fig. 1F). We reasoned that, if helix 8 has a crucial role in D1R-Gs and D2R-Gi coupling selectivity, we would observe reversed coupling selectivity when we swapped helix 8 between D1R and D2R.
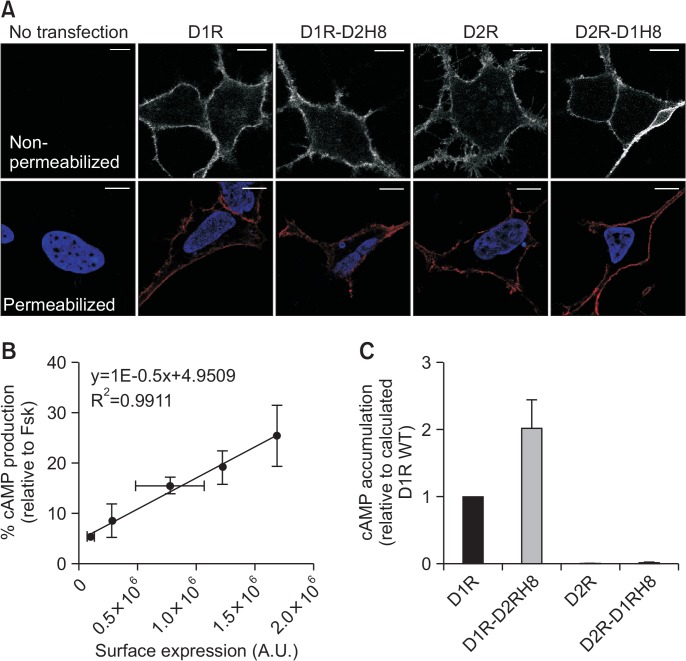
Wild type (WT) and chimeric constructs localized correctly at the plasma membrane (Fig. 2A), but expression levels differed despite transfecting the same amount of DNA (data not shown). Therefore, we adjusted expression levels prior to measuring G protein activation efficiency. We generated a standard curve with surface-expression level of D1R on the x-axis and dopamine-induced cAMP generation plotted on the y-axis (Fig. 2B). After measuring surface expression level of each construct (i.e., D1R-D2H8, D2R, D2R-D1H), we calculated the expected cAMP accumulation level using the equation in Fig. 2B. This calculated level (i.e., cAMP accumulation level assuming that the surface-expressed receptor is WT D1R) was compared to experimentally derived levels (Fig. 2C). This strategy has been successfully used in a previous study (Schonegge et al., 2017).
Fig. 2.
Functional analysis of helix 8 chimeras of D1R and D2R. (A) Subcellular localization of WT and helix 8 chimeras of D1R and D2R was visualized by immunofluorescence using confocal microscopy. Upper panels show non-permeabilized cells, and lower panels show permeabilized cells with blue presenting nucleus (DAPI staining) and red presenting receptors. Scale bars represent 10 μm. (B) Standard curve of cAMP generation according to surface-expression of WT D1R. Surface-expression level is plotted on the x-axis, and the y-axis is a percentage of maximal cAMP accumulation induced by 100 nM dopamine treatment relative to 10 μM forskolin (Fsk). (C) Effect of chimeric mutation on cAMP accumulation induced by 100 nM dopamine. Error bars represent STDEV.
As expected, dopamine did not induce cAMP accumulation when WT D2R was activated because D2R couples to Gi (Fig. 2C). If helix 8 determines D1R-Gs or D2R-Gi coupling selectivity, cAMP accumulation level of the chimeric construct D1R-D2H8 or D2R-D1H8 was expected to decrease or increase relative to the WT receptor, respectively. However, contrary to our expectations, cAMP accumulation was enhanced for D1R-D2H8 relative to WT D1R, and there was no difference in cAMP accumulation between D2R-D1H8 and WT D2R (Fig. 2C). These results suggest that helix 8 of D1R and D2R does not determine Gs or Gi coupling selectivity, but rather that D2R helix 8 improves the coupling efficiency of D1R to Gs.
Effect of increasing the number of hydrophobic residues in helix 8 on D1R signaling
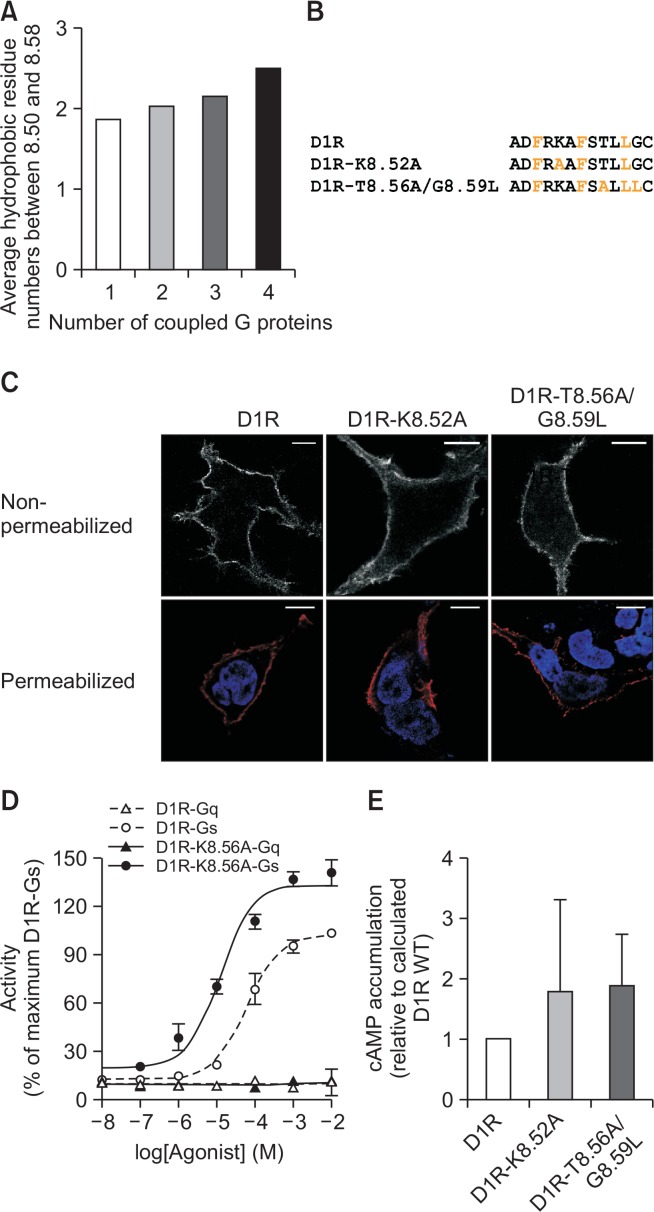
In addition to residues 8.50, 8.54, and 8.58, other residues may also be hydrophobic (Fig. 1E). Approximately 50% and 80% of class A GPCRs contain hydrophobic amino acids at residues 8.53 and 8.57 (Fig. 1E); D1R and D2R also contain hydrophobic amino acids at these positions (Fig. 1F). Residues 8.52, 8.55, and 8.56 are hydrophobic in approximately 20, 40, and 50% of class A GPCRs, respectively (Fig. 1E). Interestingly, the number of hydrophobic residues within helix 8 increases as the number of G proteins that a receptor can couple increases (Fig. 3A). Therefore, we hypothesized that the greater the number of hydrophobic residues in helix 8, the more promiscuous the receptor coupling with G proteins.
Fig. 3.
Effects of increasing the number of hydrophobic residues in helix 8 of D1R. (A) Database analysis of the number of hydrophobic residues. Hydrophobic residues between 8.50 and 8.58 were counted for the class A GPCRs D1R and D2R. Conserved hydrophobic residues at positions 8.50, 8.54, and 8.58 were not counted. (B) Mutant constructs in which D1R residues 8.52 and 8.56/8.59 were changed to hydrophobic residues. (C) Subcellular localization of WT and helix 8 mutant constructs of D1R were visualized by immunofluorescence using confocal microscopy. Upper panels show non-permeabilized cells, and lower panels show permeabilized cells with blue presenting nucleus (DAPI staining) and red presenting receptors. Scale bars represent 10 μm. (D) Dopamine-induced Gq and Gs signaling in WT D1R and D1R-K8.56A measured by luciferase reporter gene assay. (E) Effect of mutation on cAMP accumulation induced by 100 nM dopamine measured as described in Fig. 2. Error bars represent STDEV.
To test this hypothesis, we generated a mutant construct in which Lys at 8.52 was switched to Ala (Fig. 3B, D1R-K8.52A) and measured Gq activation by using luciferase reporter gene assay (Fig. 3D). D1R-K8.52A localized at the plasma membrane (Fig. 3C) but failed to induce Gq activation (Fig. 3D). Unexpectedly, D1R-K8.52A showed higher Gs signaling than WT D1R with higher potency (LogEC50 of −1.184 ± 0.06294 vs. −1.883 ± 0.1178) and efficacy (100 vs. 140.51 ± 5.9) (Fig. 3D); therefore, we performed cAMP accumulation analysis as described in Fig. 2 and confirmed that D1R-K8.52A showed enhanced Gs signaling relative to WT D1R (Fig. 3E). To test if replacement of non-hydrophobic with hydrophobic residues in the C-terminal part of helix 8 would have a similar effect (i.e., enhanced Gs signaling), we generated another mutant construct in which residues Thr8.56 and Gly8.59 were switched to Ala and Leu, respectively (Fig. 3B). This mutant localized at the plasma membrane (Fig. 3C) and induced more Gs signaling than WT D1R (Fig. 3E). Together with Fig. 2, these results suggest that helix 8 of D1R does not regulate Gs coupling selectivity.
Involvement of helix 8 in arrestin-mediated desensitization of D1R-Gs signaling
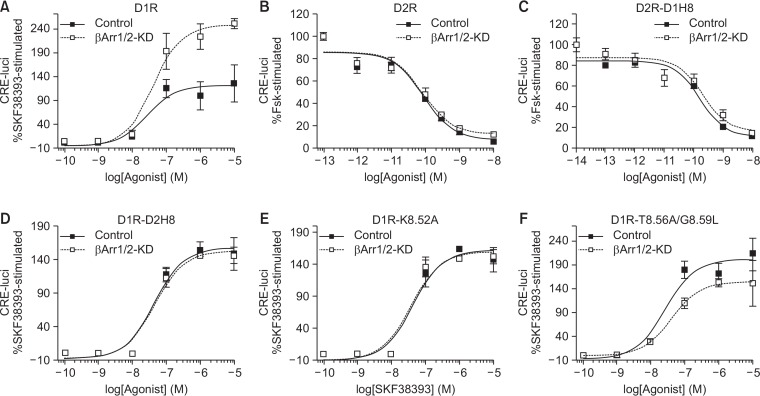
The increased Gs signaling in D1R-D2H8, D1R-K5.82A, and D1R-T5.86A/G5.89L (Fig. 2C, 3E) could be due to increased Gs coupling or decreased desensitization. To test if increased Gs signaling was due to decreased sensitization, we used β-arrestin-1/2 knockdown HEK293 cells. When D1R was activated in control and β-arrestin-1/2 knockdown cells, the efficacy of Gs signaling was higher in the β-arrestin-1/2 knockdown cells compared to the control cells (126 ± 38 vs. 251 ± 11) although the potency was similar (LogEC50 of −7.587 ± 0.322 vs. −7.383 ± 0.176) (Fig. 4A), showing that D1R-mediated Gs signaling is desensitized by a β-arrestin-1/2-mediated mechanism in control cells. In contrast, there was no difference in Gi signaling between control and β-arrestin-1/2 knockdown cells when D2R was activated (Fig. 4B), suggesting that D2R-mediated Gi signaling is not desensitized by a β-arrestin-1/2-mediated mechanism, consistent with previous reports (Westrich and Kuzhikandathil, 2007; Namkung et al., 2009; Min et al., 2013).
Fig. 4.
Involvement of helix 8 in β-arrestin-1/2-mediated desensitization of D1R-Gs signaling. HEK293 cells were stably transfected with the shRNAs for human β-arrestin1 and β-arrestin2 to generate β-arrestin-1/2 knockdown (βArr1/2-KD) cells (Zhang et al., 2008). About 80–90% cellular β-arrestin1 and β-arrestin2 were knocked down in β-arrestin KD cells (Min et al., 2013). Control and βArr1/2-KD cells were transfected with indicated receptor constructs along with the plasmids required for the determination of cellular cAMP. Receptor expression levels were maintained between 2.1–2.7 pmol/mg protein. (A) Control and βArr1/2-KD cells expressing D1R were treated with increasing concentrations of SKF38393. Forskolin (10 μM)-induced increase in CRE-luci was calculated as 100%, and SKF38393-induced increase in CRE-luci was normalized to it. The same protocols were applied for (D) (D1R-D2H8), (E) (D1R-K8.52A), and (F) (D1R-T8.56A/G8.59L). (B) Control and βArr1/2-KD cells expressing D2R were treated with 2 μM forskolin and increasing concentrations of quinpirole. The same protocols were applied for (C) (D2R-D1H8). Error bars represent STDEV.
When D1R-D2H8 was activated, we did not observe enhanced Gs signaling in β-arrestin-1/2 knockdown cells compared to the control cells (Fig. 4D). β-Arrestin-1/2-mediated desensitization was also abolished in D1R-K8.52A and D1R-T8.56A/G8.59L (Fig. 4E, 4F), suggesting that the enhanced Gs signaling observed for these mutant constructs compared to WT D1R is potentially due to lack of β-arrestin-1/2-mediated desensitization. On the other hands, there were no differences in Gi signaling between control and β-arrestin-1/2 knockdown cells upon D2R-D1H8 activation (Fig. 4C) suggesting that helix 8 of D1R alone would not trigger the β-arrestin-1/2-mediated desensitization but needs other regions of the receptor as discuss below.
DISCUSSION
The present study investigated the role of helix 8 in dopamine receptor signaling by swapping helix 8 between D1R (a Gs-coupling receptor) and D2R (a Gi-coupling receptor). The results suggest that helix 8 does not determine the Gs or Gi selectivity of the dopamine receptors but rather affects the intensity of D1R-mediated Gs signaling by regulating the arrestin-dependent desensitization of D1R signaling.
Previously, a few studies reported that helix 8 is involved in G protein coupling selectivity. For example, helix 8 of the thyrotropin receptor regulates G protein coupling selectivity (Kleinau et al., 2010). However, another study reported that helix 8 of β2AR is not involved in G protein coupling selectivity (Liggett et al., 1991). Therefore, it is possible that the involvement of helix 8 in the GPCR-G protein coupling selectivity is dependent on receptor type. We showed that, in D1R and D2R, helix 8 dose not regulate Gs or Gi-coupling selectivity.
Only a few studies have proposed a potential role for helix 8 in GPCR-arrestin interactions or GPCR-mediated G protein signaling desensitization (Okuno et al., 2003; Feierler et al., 2011; Kirchberg et al., 2011). A previous study with rhodopsin suggested that the dynamic mobility and bulkiness of helix 8 affect its interaction with visual arrestin (Kirchberg et al., 2011). In the present study, we showed for the first time that helix 8 of D1R has a crucial role in β-arrestin-1/2-mediated desensitization of the receptor by domain swapping and introducing hydrophobic residues in the helix 8.
We observed abolished β-arrestin-1/2-mediated desensitization in D1R-D2H8, and therefore it is tempting to suggest that a specific sequence of helix 8 is necessary for β-arrestin-1/2-mediated desensitization of D1R. However, helix 8 alone was not sufficient to trigger β-arrestin-1/2-mediated desensitization, as we did not observe β-arrestin-1/2-mediated desensitization in D2R-D1H8. Interestingly, D1R has a C-tail that can be phosphorylated by GRK and interact with arrestins, whereas D2R does not (Fig. 1F); instead, D2R has a large ICL3 that has been reported to interact with β-arrestin-1/2, but the role of this interaction in receptor desensitization is unknown (Macey et al., 2004; Lan et al., 2009; Namkung et al., 2009). Thus, helix 8 of D1R regulates β-arrestin-1/2-mediated desensitization in aid of other regions of D1R (e.g. phosphorylated C-terminal tail), and it is possible that D2R helix 8 (Fig. 1F) or mutant helix 8 (Fig. 3B) has different conformational dynamics than WT D1R helix 8, resulting in defective arrestin interactions even though the mutants has intact C-terminal tail. As a next step, a precise molecular mechanism of helix 8 involvement in D1R desensitization needs to be further investigated by using purified receptor systems; for example, we need to analyze the conformational dynamics of D1R helix 8 in WT, D1R-D2R, and D1R-K8.52A or D1R-T8.56A/G8.59L and also analyze the conformation of D1R-arrestin complexes to seek the involvement of D1R helix 8 in D1R-arrestin complex formation.
In conclusion, we evaluated the role of helix 8 in D1R and D2R signaling and verified that helix 8 does not determine D1R-Gs or D2R-Gi coupling selectivity but rather regulates β-arrestin-1/2-mediated desensitization. We did not evaluate if this is due to interaction of D1R helix 8 with GRK or β-arrestin-1/2. Nevertheless, helix 8 could be a good drug target site for biased ligand development targeting D1R. In addition to desensitization or internalization of GPCRs, receptor-activated arrestin triggers signaling cascades different from those triggered by G protein-mediated signaling, which can result in different physiological and pathological outcomes (Rankovic et al., 2016; Smith et al., 2018). For this reason, selective regulation of G protein- or arrestin-mediated signaling through biased ligands has been suggested as a new stratagem for more effective and/or safer drug development. The findings of the present study suggest that D1R helix 8 could be a potential drug target site to selectively inhibit arrestin-mediated signaling without affecting Gs coupling.
Acknowledgments
This work was supported by grants from the National Research Foundation of Korea funded by the Korean government (NRF-2017K1A3A1A12072316 to K.Y.C., NRF-2018R1D1A1 B07051124 to H.-J. P., and KRF-2017R1A2A2A05001227 to K.M.K.).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Ahn KH, Nishiyama A, Mierke DF, Kendall DA. Hydrophobic residues in helix 8 of cannabinoid receptor 1 are critical for structural and functional properties. Biochemistry. 2010;49:502–511. doi: 10.1021/bi901619r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Gainetdinov RR. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev. 2011;63:182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- Edward Zhou X, Melcher K, Eric Xu H. Structural biology of G protein-coupled receptor signaling complexes. Protein Sci. 2019;28:487–501. doi: 10.1002/pro.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faussner A, Bauer A, Kalatskaya I, Schussler S, Seidl C, Proud D, Jochum M. The role of helix 8 and of the cytosolic C-termini in the internalization and signal transduction of B(1) and B(2) bradykinin receptors. FEBS J. 2005;272:129–140. doi: 10.1111/j.1432-1033.2004.04390.x. [DOI] [PubMed] [Google Scholar]
- Feierler J, Wirth M, Welte B, Schussler S, Jochum M, Faussner A. Helix 8 plays a crucial role in bradykinin B(2) receptor trafficking and signaling. J Biol Chem. 2011;286:43282–43293. doi: 10.1074/jbc.M111.256909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Hilger D, Masureel M, Kobilka BK. Structure and dynamics of GPCR signaling complexes. Nat Struct Mol Biol. 2018;25:4–12. doi: 10.1038/s41594-017-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, Mordalski S, Pin JP, Stevens RC, Vriend G, Gloriam DE. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Saka T, Mine S, Mizohata E, Inoue T, Matsumura H, Sato T. The N-terminal acidic residue of the cytosolic helix 8 of an odorant receptor is responsible for different response dynamics via G-protein. FEBS Lett. 2015;589:1136–1142. doi: 10.1016/j.febslet.2015.03.025. [DOI] [PubMed] [Google Scholar]
- Kaye RG, Saldanha JW, Lu ZL, Hulme EC. Helix 8 of the M1 muscarinic acetylcholine receptor: scanning mutagenesis delineates a G protein recognition site. Mol Pharmacol. 2011;79:701–709. doi: 10.1124/mol.110.070177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberg K, Kim TY, Moller M, Skegro D, Dasara Raju G, Granzin J, Buldt G, Schlesinger R, Alexiev U. Conformational dynamics of helix 8 in the GPCR rhodopsin controls arrestin activation in the desensitization process. Proc Natl Acad Sci USA. 2011;108:18690–18695. doi: 10.1073/pnas.1015461108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinau G, Jaeschke H, Worth CL, Mueller S, Gonzalez J, Paschke R, Krause G. Principles and determinants of G-protein coupling by the rhodopsin-like thyrotropin receptor. PLoS ONE. 2010;5:e9745. doi: 10.1371/journal.pone.0009745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramasu A, Sukegawa J, Sato T, Sakurai E, Watanabe T, Yanagisawa T, Yanai K. The hydrophobic amino acids in putative helix 8 in carboxy-terminus of histamine H(3) receptor are involved in receptor-G-protein coupling. Cell Signal. 2011;23:1843–1849. doi: 10.1016/j.cellsig.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Lan H, Liu Y, Bell MI, Gurevich VV, Neve KA. A dopamine D2 receptor mutant capable of G protein-mediated signaling but deficient in arrestin binding. Mol Pharmacol. 2009;75:113–123. doi: 10.1124/mol.108.050534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett SB, Caron MG, Lefkowitz RJ, Hnatowich M. Coupling of a mutated form of the human beta 2-adrenergic receptor to Gi and Gs. Requirement for multiple cytoplasmic domains in the coupling process. J Biol Chem. 1991;266:4816–4821. [PubMed] [Google Scholar]
- Macey TA, Gurevich VV, Neve KA. Preferential interaction between the dopamine D2 receptor and Arrestin2 in neo-striatal neurons. Mol Pharmacol. 2004;66:1635–1642. doi: 10.1124/mol.104.001495. [DOI] [PubMed] [Google Scholar]
- Markx D, Schuhholz J, Abadier M, Beier S, Lang M, Moepps B. Arginine 313 of the putative 8th helix mediates Galphaq/14 coupling of human CC chemokine receptors CCR2a and CCR2b. Cell Signal. 2019;53:170–183. doi: 10.1016/j.cellsig.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Min C, Zheng M, Zhang X, Caron MG, Kim KM. Novel roles for beta-arrestins in the regulation of pharmacological sequestration to predict agonist-induced desensitization of dopamine D3 receptors. Br J Pharmacol. 2013;170:1112–1129. doi: 10.1111/bph.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: from structure to function. Physiol Rev. 1998;78:189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- Moritz AE, Free RB, Sibley DR. Advances and challenges in the search for D2 and D3 dopamine receptor-selective compounds. Cell Signal. 2018;41:75–81. doi: 10.1016/j.cellsig.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkung Y, Dipace C, Javitch JA, Sibley DR. G protein-coupled receptor kinase-mediated phosphorylation regulates post-endocytic trafficking of the D2 dopamine receptor. J Biol Chem. 2009;284:15038–15051. doi: 10.1074/jbc.M900388200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno T, Ago H, Terawaki K, Miyano M, Shimizu T, Yokomizo T. Helix 8 of the leukotriene B4 receptor is required for the conformational change to the low affinity state after G-protein activation. J Biol Chem. 2003;278:41500–41509. doi: 10.1074/jbc.M307335200. [DOI] [PubMed] [Google Scholar]
- Pandy-Szekeres G, Munk C, Tsonkov TM, Mordalski S, Harpsoe K, Hauser AS, Bojarski AJ, Gloriam DE. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankovic Z, Brust TF, Bohn LM. Biased agonism: an emerging paradigm in GPCR drug discovery. Bioorg Med Chem Lett. 2016;26:241–250. doi: 10.1016/j.bmcl.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonegge AM, Gallion J, Picard LP, Wilkins AD, Le Gouill C, Audet M, Stallaert W, Lohse MJ, Kimmel M, Lichtarge O, Bouvier M. Evolutionary action and structural basis of the allosteric switch controlling beta2AR functional selectivity. Nat Commun. 2017;8:2169. doi: 10.1038/s41467-017-02257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Lefkowitz RJ, Rajagopal S. Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov. 2018;17:243–260. doi: 10.1038/nrd.2017.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sounier R, Mas C, Steyaert J, Laeremans T, Manglik A, Huang W, Kobilka BK, Demene H, Granier S. Propagation of conformational changes during mu-opioid receptor activation. Nature. 2015;524:375–378. doi: 10.1038/nature14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis WI, Kobilka BK. The molecular basis of g protein-coupled receptor activation. Annu Rev Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich L, Kuzhikandathil EV. The tolerance property of human D3 dopamine receptor is determined by specific amino acid residues in the second cytoplasmic loop. Biochim Biophys Acta. 2007;1773:1747–1758. doi: 10.1016/j.bbamcr.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang F, Chen X, Chen Y, Ma L. Post-endocytic fates of delta-opioid receptor are regulated by GRK2-mediated receptor phosphorylation and distinct beta-arrestin isoforms. J Neurochem. 2008;106:781–792. doi: 10.1111/j.1471-4159.2008.05431.x. [DOI] [PubMed] [Google Scholar]
- Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, Barty A, Latorraca NR, Chapman HN, Hubbell WL, Dror RO, Stevens RC, Cherezov V, Gurevich VV, Griffin PR, Ernst OP, Melcher K, Xu HE. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell. 2017;170:457–469.e13. doi: 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhang M, Davis JE, Wu WH, Surrao K, Wang H, Wu G. A single mutation in helix 8 enhances the angiotensin II type 1a receptor transport and signaling. Cell Signal. 2015;27:2371–2379. doi: 10.1016/j.cellsig.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]