Abstract
This study sought to examine the human immunodeficiency virus type 1 (HIV-1) genetic diversity on drug resistance among men who have sex with men (MSM) with virologic failure in antiretroviral therapy (ART), and investigate linking-associated factors for genetic transmission networks.
Seven hundred and thirty-four HIV-positive MSM with virologic failure in ART were recruited into our study from 2011 to 2017. HIV-1 pol gene sequences were used for phylogenetic and genotypic drug resistance analyses. The drug resistance mutations were determined using the Stanford University HIV Drug Resistance Database. The genetic transmission networks were analyzed for CRF01_AE and CRF07_BC sequences by the genetic distance-based method.
Of 734 subjects, 372 (50.68%) showed drug resistance, in which CRF01_AE and CRF07_BC were the predominating subtypes. Drug resistance more frequently occurred in non-nucleoside reverse transcriptase inhibitors (NNRTIs) treatment (48.64%), and followed by nucleoside reverse transcriptase inhibitors (NRTIs) (36.51%) and PIs (4.03%). The most common drug resistance-associated mutations in protease inhibitors (PIs), NRTIs and NNRTIs were K20I/R, M184V/I and K103N/KN, respectively. For 283CRF01_AE sequences, 64 (22.61%) fell into clusters at a genetic distance of 0.011, resulting in 17 clusters ranging in size from 2 to 16 individuals. For 230 CRF07_BC sequences, 66 (28.69%) were connected to at least one other sequence with 0.005 genetic distances, resulting in 8 clusters ranging in size from 2 to 52 individuals. Individuals who showed drug resistance to ART were less likely to fall into clusters than those who did not. The genetic linkage was robust by the exclusion of sites associated with drug resistance.
CRF01_AE and CRF07_BC were the main strains among MSM with virologic failure in ART, and the drug resistance more frequently occurred in NNRTIs, followed by NRTIs and PIs. Genetic transmission networks revealed a complexity of transmission pattern, suggesting early-diagnosis and in-time intervention among MSM.
Keywords: drug resistance, genetic transmission networks, HIV-1, MSM
1. Introduction
The number of HIV-infected individuals in Sichuan province is increasing dramatically. By the end of 2017, a total of 110,872 human immunodeficiency virus infection/acquired immune deficiency syndrome (HIV/AIDS) cases had been reported, making it the highest number of HIV/AIDS in China.[1] Homosexual transmission has gradually become the fastest growing transmission way in Sichuan province,[2] and HIV prevalence in MSM in this province has significantly increased in the last few years (from 9.5% in 2011 to 12.8% in 2015),[3] both of which made Sichuan the most impacted province in China.[4]
ART is an effective approach for treating HIV-infected patients, which can regulate the HIV life cycle, suppress HIV replication and preserve normal immune function.[5] The scale-up of ART has led to dramatic reductions in morbidity and mortality among HIV-infected patients.[5] HIV-infected people on suppressive ART have a low risk of HIV transmission.[6] However, increasing genotypic HIV drug resistance inevitably occurs after a period of ART, and the number of drug resistance has risen in the last 10 years in China. For MSM, drug resistance of MSM receiving ART increases more rapidly than other populations, which is due in part to frequent high-risk behaviors of MSM and a low HIV detection rate in these patients.[7–9] The high incidence of drug resistance and low adherence to prescribed treatment are associated with drug resistance-related mutations that can reduce the efficacy of ART and raise the risk of treatment failure.[10] The genetic mutations of HIV strains could influence the therapy response to different medication and the occurrence of treatment failure. Therefore, it is necessary to identify the drug resistance-related mutations of HIV genotypes.
Interestingly, the high prevalence of virologic failure in ART across the world is observed in regions with well-established use of ART, such as in Europe and United States of American.[11,12] Treatment failure in HIV infected patients could lead to new challenges, such as increasing HIV transmission among individuals with risk behaviors and transmitted drug resistance.[13] The prevalence (about 10–17.2%) of transmitted drug resistance in ART-naïve individuals remains high and stable in most developed countries.[11,14] Identification of transmission network in treatment failure in HIV-infected individuals provides an estimate of unobserved transmission network and potential routes of viral spread, and understands the transmitted drug resistance.[15,16] Techniques for reconstructing genetic transmission network can indicate a set of sequences from potential transmission partners and show the relationships between infected individuals.
Different from other high risk population of HIV, Chinese MSM are more likely to be in close social and sexual networks because of stigma and social discrimination, and HIV strains from linked individuals are more similar than those from unassociated ones.[17,18] For MSM with virologic failure in ART, identifying potential transmission partners by genetic transmission network could help selection of more effective therapeutic regimens,[19] prevention of transmitted drug resistance,[20,21] and understanding of linking-associated factors of transmission networks among MSM.[22] However, no previous studies have described HIV drug resistance and genetic transmission networks among MSM with virologic failure in ART in China. Therefore, this study aimed to examine the HIV-1 genetic diversity among MSM with virologic failure in ART in Sichuan province between 2011 and 2017, and use a genetic distance-based method to investigate the genetic transmission networks.
2. Materials and methods
2.1. Ethics
All subjects voluntarily participated in our study and signed informed consent forms before enrollment. The study protocol was approved by the Ethics Committee of the Sichuan Center for Disease Control and Prevention (CDC), and the study was carried out following the Helsinki Declaration of 1964.
2.2. Study participants
Participants who met the inclusion criteria were included into our study:
-
(1)
MSM living in Sichuan province,
-
(2)
being confirmed with positive HIV-1,
-
(3)
receiving the ART for at least 6 months,
-
(4)
being followed up from Jan 1st 2011 to Dec 31th 2017, and
-
(5)
being diagnosed as virologic failure in ART.
These sources that are available in Sichuan province included:
-
(1)
Provider-initiated HIV testing & counseling (PITC) service for HIV,
-
(2)
Voluntary counseling and testing (VCT) service through fixed VCT sites and non-government organizations (NGOs),
-
(3)
HIV sentinel surveillance (HSS) for MSM, and
-
(4)
scientific studies conducted in Sichuan Province to find the HIV infections among MSM.
PITC has been routinely recommended by health care providers to provide HIV testing services to patients who seek medical treatment at health care facilities.
Subjects who were infected through the sexual transmission of homosexuality and had self-reported oral or anal sex at least once with another male were eligible for the study.
A total of 734 HIV-1infected MSM individuals with virologic failure in ART met the inclusion criteria. Each participant provided 5 mL of venous blood for the viral load of HIV-1. Plasma was isolated and sent under the cold chain to the Sichuan CDC where the HIV viral load was measured. Virologic failure was defined as an HIV RNA level >1000 copies/ml. Samples with a viral load of ≥1000 copies/mL were performed HIV drug resistance genotyping in Sichuan CDC by using an in-house PCR.[23]
2.3. Data collection
Participants were asked about their socio-demographic information, including age, occupation, marital status, education level attained. The occupation was categorized as student, farmer, employed (workers, administers or managers, professional technicians, sales or service workers, private owners, etc) and unemployed. The demographic characteristics of participants were collected by the staff at the local Center for Disease Prevention and Control (CDCs) and community health center during the follow-up period.
2.4. Nucleic acid extraction, amplification, and sequencing
The viral nucleic acid was extracted from 200-μl plasma of subjects with virologic failure in ART (according to WHO guideline, we defined viral load ≥1000 copies/mL after 6 months of ART treatment as virologic failure) by using an automatic extraction machine (MagNA Pure LC 2.0 system, Roche, Branchburg, NJ). The RT-PCR (Reverse Transcription-Polymerase Chain Reaction) was conducted to amplify the full-length protease gene in Pol region and the first 300 codons of the reverse transcriptase gene. The PCR products were dealt with electrophoresis with 1% agarose gel. The amplified products were sent to Beijing Genomics Research Center Ltd. for purification and gene sequencing (Table 1). Detailed amplification and sequencing methodology has been published previously.[24]
Table 1.
The sequencing primers.

2.5. Sequencing analysis of HIV-1 pol region and drug-resistance analyses
The obtained sequences were edited and aligned using ChromasPro l.33, and the sequence alignments were performed with reference sequences retrieved from the Los Alamos Sequence Database (http://www.hiv.lanl.gov/) by BioEdit Sequence Alignment Editor software (Ibis Bisciences, Carlsbad, CA). FastTree 2.1.8 (http://meta.microbesonline.org/fasttree/) was used to estimate an approximately-maximum likelihood phylogenetic tree for pol sequences. More than 90% of neighbor-joining trees had bootstrap value above 70, which showed the high reliability of phylogenetic tree. The phylogenetic tree was visualized by FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Drug resistance mutations were analyzed using the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu/). All experimental protocols were following the manufacturer's instructions.
2.6. Analysis of HIV-1 genetic transmission network
Genetic distance-based methods ascribe a putative transmission link to any pair of viral sequences within a predetermined genetic distance threshold. It is expected that viral genetic diversity between transmission partners should approximate the diversity of the source partners, and allow for some degree of onward evolution in the recipient partners.[25] The flowchart of genetic transmission networks was performed in 4 steps, including comparing sequence, constructing phylogenetic tree, calculating pairwise distance, and visualizing the network.
After comparing the sequences and constructing phylogenetic tree described above, calculations of pairwise genetic distances of all sequences within the available clusters were performed using Run HyPhy 2.2.4 (http://www.hyphy.org). Among the genetic distances within 0.001 to 0.015, the gene distance with the largest number of clusters was selected as the linkages within a cluster. Finally, the network was visualized and analyzed by using Cytoscape 3.5 software (https://cytoscape.org).
2.7. Statistical analysis
The database was established by excel, and the statistical analyses were performed using SPSS 21.0 (SPSS Inc. Chicago, IL). Categorical variables were presented as frequencies and percentages, and were compared using fisher exact tests. A 2-sided P value <.05 was considered statistically significant.
3. Results
3.1. Characteristics of drug resistance among MSM with virologic failure in ART
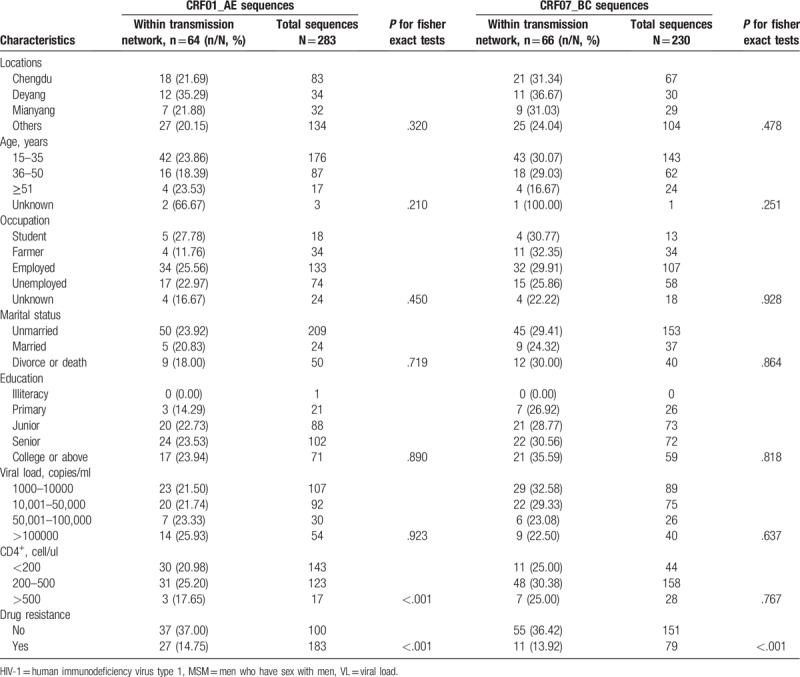
Among 734 MSM with virologic failure in ART, 50.68% (372/734 showed drug resistance between 2011 and 2017 (Table 2), and 63.89% (469/734) of the participants were 15 to 35 years old. Over half attained senior or above education level (58.45%, 429/734), 38.56% (283/734) were employed, and 71.11% (522/734) were unmarried. The phylogenetic analyses based on the pol regions showed that CRF01_AE and CRF07_BC were the most common strains in Sichuan province, and the CRF01_AE had the highest drug resistant rate (67.40%, 246/365) (Fig. 1). We observed significant differences in drug resistant rates in terms of different subtypes of HIV-1 (P < .05).
Table 2.
The demographic characteristics of drug resistance in MSM with virologic failure in ART.

Figure 1.
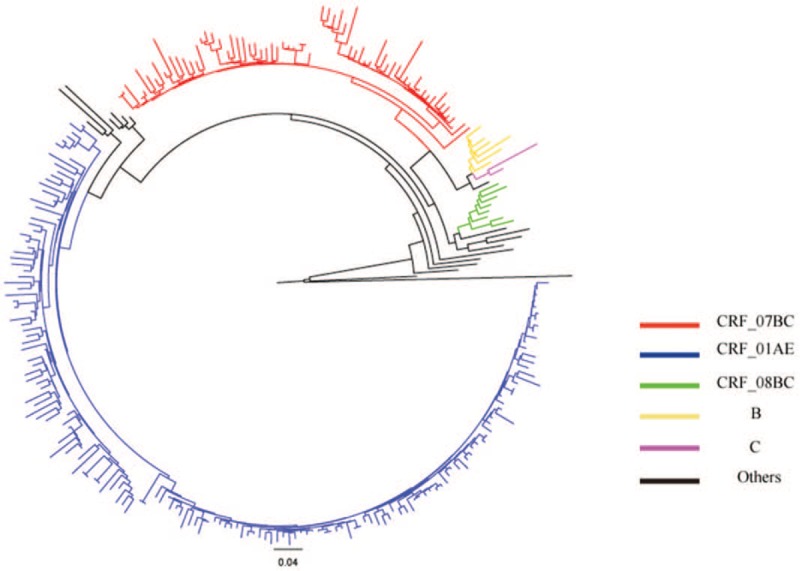
Phylogenetic tree analyses of the HIV-I pol sequences in men who have sex with men with virologic failure in antiretroviral therapy.
Drug resistance was most likely to occur in MSM treated with nucleoside reverse transcriptase inhibitors (NRTIs) (drug resistant rate: 59.68%, 37/62, Table 2). MSM treated with non-nucleoside reverse transcriptase inhibitors (NNRTIs) also showed high drug resistant rates (AZT+3TC+EFV/NVP: 48.59%; D4T+3TC+EFV/NVPNNRTIs: 52.88; TDF+3TC+EFV/NVPNNRTIs: 52.03%).
3.2. Drug resistance-associated mutations
The most common PI-related mutations were K20I/R (18/372, 4.84%) and V82I (13/372, 3.49%) (Table 3). M184V/I (236/372, 63.44%), K65KR/R (78/372, 20.97%), D67DN/N (77/372, 20.70%), and K70E/R/KR (72/372, 19.35%) were most common NRTI-related mutations. K103N/KN (155/372, 41.67%), V106M/MA/MV (107/372, 28.76%), G190A/AG/S (103/372, 27.69%), and Y181C/CY (89/372, 23.92%) were the major NNRTI-related mutations.
Table 3.
Drug resistance associated mutations among MSM with virologic failure in ART.

3.3. The transmission networks of MSM with virologic failure in ART
A total of 283 MSM with CRF01_AE and 230 with CRF07_BC pol sequences were identified and used for genetic transmission network analysis between 2011 and 2017. Of 283 CRF01_AE pol sequences, 64 (22.61%) fell into clusters at a genetic distance of 0.011, resulting in 17 clusters ranging in size from 2 to 16 individuals (Fig. 2A). Of 230 CRF07_BC pol sequences, 66 (28.69%) were connected to at least one other sequence with 0.005 genetic distances, resulting in 8 clusters ranging in size from 2 to 52 individuals (Fig. 2B).
Figure 2.
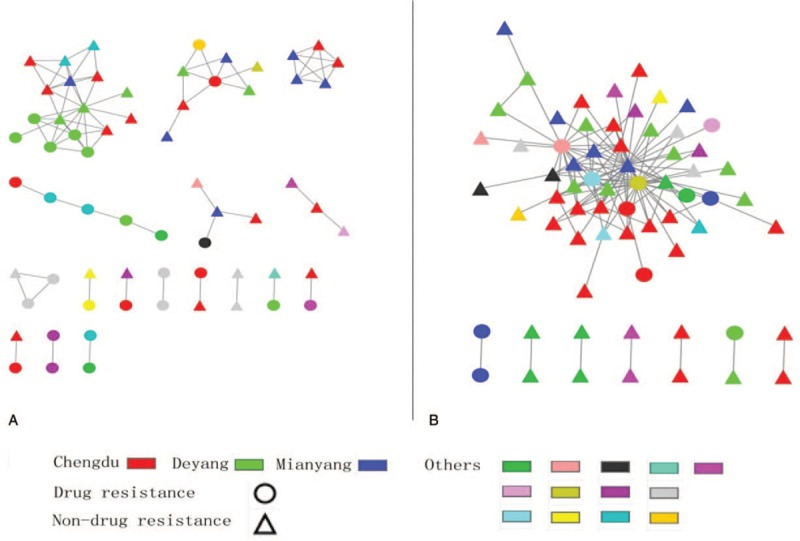
The transmission networks of CRF01_AE and CRF07_BC sequences. (A) A total of 64 CRF01_AE sequences were used for genetic transmission network analysis, and the biggest cluster had 16 sequences. (B) A total of 66 CRF07_BC were used for genetic transmission network analysis, and the biggest cluster had 52 sequences.
We did not find a significant association between genetic linkages in terms of location, age, occupation, marital status, education level, viral load and CD4+. However, individuals who showed drug resistance to ART were less likely to fall into clusters than those who did not (P < .05) (Table 4).
Table 4.
Distributions of the demographic information in MSM within the HIV-1 genetic transmission network.
Among clusters for CRF01_AE pol sequences, 20 MSM (31.25%) were found to have only 1 linked individual, and 44 (68.75%) had >2 linked individuals. Five MSM with drug resistant HIV-1 were in 1 cluster.
Among clusters for CRF07_BC pol sequences, 14 MSM (21.21%) were observed to have only 1 potential linked partner, and 52 (78.79%) had >2 potential linked partners.
We also determined the robustness of genetic linkage by the exclusion of sites associated with drug resistance that may be a confounding factor due to convergent evolution for mutations conferring drug resistance. The agreement rates of partner pairs with or without the sites associated with drug resistance for CRF01_AE and CRF07_BC pol sequences were 94.75% and 93.94%, respectively. If we excluded 35 codons in CRF01_AE pol sequences associated with drug resistance, an additional pair of individuals were linked, while 4 individuals in 2 clusters >2 linked individuals were unlinked (agreement rate: 94.75%). For CRF07_BC pol sequences, one pair of individuals were linked, while 3 individuals in the biggest cluster were unlinked (agreement rate: 93.94%).
4. Discussion
Sichuan province has the highest prevalence of HIV/AIDS in China, and MSM homosexual transmission in this province has gradually become the fastest growing transmission in the recent 10 years.[2] ART has resulted in marked improvements in morbidity and mortality from HIV-1 infection. Analyses of the prevalence of HIV-1 resistant variants and linking-associated factors for genetic transmission networks in MSM with virologic failure in ART would provide great implications for the successful management and treatment options of ART.
In our study, we found that the prevalence of drug resistance was 50.68% in MSM with virological failure in ART. CRF52_01BC and CRF55_01B strains were newly isolated among MSM, which suggested that HIV-1 genetic diversity among MSM had increased in this area. The diversity of HIV subtype was also found in other places of China.[26,27] Moreover, we found that the CRF01_AE and CRF07_BC were the 2 overarching circulating strains in MSM in Sichuan province. The CRF_01AE was transmitted through heterosexual activities in Southeast Asia and Chinese southeast coastal provinces,[28] and was identified among Chinese HIV-infected MSM in 2005. CRF_07BC was firstly detected from IDUs in Guangxi province in 1997, and was mainly circulated among injection drug users in Northwest and Southwest China.[29] CRF01_AE and CRF07_BC strains were also found to be the main circulating strains among MSM in several cities of China,[30–32] which were similar to our results.
The incidence of drug resistance to NNRTIs (48.64%) was higher than that of NRTIs (36.51%) in patients with ART, which indicated that NRTI would have better therapeutic efficacy in ART when compared with NNRTI. It is reported that drug resistance has been more often occurred in NNRTIs, followed by NRTIs and PIs among HIV-1 infected patient.[33,34] Incomplete suppression of HIV may contribute to drug resistance, and mutations in HIV reverse transcriptase and protease would be the molecular basis of drug resistance.[35] Therefore, the main genetic mutations associated with resistance were identified in NRTIs and NNRTIs, but mutations were less observed in PIs. A similar pattern was also found in other places in China and low- and middle-income countries.[33–36]
M184V/I was the most common NRTI-related mutation, which might be due to the frequent use of Lamivudine (3TC) in ART, and 3TC resistance mutation M184V/I was highly prevalent in HIV-1 infected patients.[37,38] M184V/I may alter the optimal binding of NRTIs and decrease 3TC removal and enzymatic processivity, and results in drug resistance.[39] We observed that K103N/KN, V106M/MA/MV, G190A/AG/S, and Y188L/C are the major NNRTI-related mutations in China and other countries,[40,41] which may be the result of the wide-spread use of NNRTIs. A study in Henan province showed a similar mutation pattern related to NNRTIs with our study.[33] NNRTIs associated drug resistant mutations could greatly impact viral replicative fitness, and therefore cause persistent of virus in the absence of antiretroviral drugs and promote infection to a new host.[42]
With the development of the economy of China, cities have attracted an increasing number of migrant people, and people usually flow between different cities. The transmission networks of HIV-1 among MSM were mainly from different cities, such as the biggest cluster with 52 sequences of CRF07_BC from 16 cities. The main reason might be the wide application of Gay mobile apps, which allows convenient seeking of sexual partners.[43] HIV-1 infected patients in the transmission networks from different cities also reflect the frequent mobility and multiple sexual partners of MSM.
Our studies showed that most (57.81% for CRF01_AE and 62.12% for CRF07_BC) of the MSM in networks were concentrated in Chengdu (the capital city of Sichuan province) and its surrounding cities. MSM is easily recognized by acquaintances and reluctant to stay in their hometowns due to stigma and social discrimination in China.[8,9] Most of the MSM concentrated in urban cities, where they are more inclusive of sexual orientation and more easily constructed for close social and sexual networks.[44] Moreover, we found about 70% MSM had ≥2 potential transmission partners for both CRF01_AE and CRF07_BC sequences, which means individuals with multiple links could potentially have higher transmission risk. Therefore, more attention should be paid to the earlier diagnosed HIV-infected subjects with multiple links, who may be still active in transmission networks and be the key intervention targets. Besides, analyses of genetic transmission networks could investigate the relationships between infected MSM, and understand how HIV is spread within MSM and further deliver interventions for providing more testing services and in time treatment after diagnosis to this population.
The sequences of drug resistance to ART in the genetic transmission networks were less than those without drug resistance. Reconstructed resistant viruses had lower replicative capacity than that of pre-therapy counterparts, and thus the drug-resistant mutations may impair the activity of protease and replicative capacity.[45] The previous study indicated that reconstructed resistant viruses had a lower replicative capacity when compared with wild type strain without ART. However, the genetic mutation of wild type strain would be replaced by drug-resistant mutations without the condition of ART.[46] Therefore, effective and in-time treatment for HIV-1 infected patients could curb the spread of the virus.
There are 4 limitations worth mentioning. First, this study was cross-sectional, so time-based sequence and cause-effect relationships among these variables cannot be established. Second, only several demographic characteristics were collected in our study. There are many other factors associated with drug resistance and transmission networks, such as drug complications, number of sexual partners and unprotected sexual behaviors, and these factors should be taken into consideration in future studies. Third, we did not exclude the sites associated with drug resistance that may be a possible confounding factor due to convergent evolution for mutations conferring drug resistance. That may overestimate the linkage network, but we found the genetic linkage network was robust to the inclusion or exclusion sites associated with drug resistance. Fourth, we did not perform the sequencing analysis among MSM who were not on ART or with a successful ART, which may not catch most transmission network, future studies on HIV-1 sequences in all MSM could better explain the transmission networks.
5. Conclusions
To our knowledge, we were the first to investigate the genetic diversity of HIV-1, drug resistance and transmission networks among MSM with virologic failure in ART in Sichuan Province. We found that the CRF01-AE and CRF07-BC were the main strains in MSM with virologic failure in ART, and the drug resistance-associated mutations were more frequently occurred in NNRTIs, followed by NRTIs and PIs. Genetic transmission network analyses revealed a complexity of transmission pattern, which suggested early-diagnosis and in-time intervention among MSM.
Acknowledgments
We thank the Center for AIDS/STD Control and Prevention, Sichuan Center for Disease Control and Prevention for supporting data. We also thank Shifan Yang, Bing Yu and Yaru Li from West China School of Public Health, Sichuan University for cleaning the data for analysis.
We thank Dr. Shawna Williams to help polish the English of this manuscript.
Author contributions
Data curation: Dan Yuan, Wenwen Zhai, Shujuan Yang.
Formal analysis: Zonglun Du.
Investigation: Li Ye, Ling Su, Hong Yang, Fengshun Yuan, Yiping Li, Honglu Liu, Shu Liang.
Methodology: Dan Yuan.
Project administration: Shujuan Yang.
Software: Dan Yuan, Zonglun Du.
Writing – original draft: Shujuan Yang.
Writing – review & editing: Zonglun Du, Junmin Zhou, Shujuan Yang.
Footnotes
Abbreviations: 3TC = Lamivudine, ART = virologic failure in antiretroviral therapy, CDC = Center for Disease Control and Prevention, HIV/AIDS = human immunodeficiency virus infection/acquired immune deficiency syndrome, HIV-1 = human immunodeficiency virus type 1, HSS = HIV sentinel surveillance, MSM = men who have sex with men, NGOs = non-government organizations, NNRTIs = non-nucleoside reverse transcriptase inhibitors, NRTIs = nucleoside reverse transcriptase inhibitors, PIs = protease inhibitors, PITC = Provider-Initiated HIV Testing & Counseling, RT-PCR = Reverse Transcription-Polymerase Chain Reaction, VCT = voluntary counseling and testing.
How to cite this article: Yuan D, Du Z, Zhou J, Ye L, Su L, Yang H, Yuan F, Li Y, Liu H, Zhai W, Liang S, Yang S. HIV-1 subtype diversity, drug resistance, and genetic transmission networks in men who have sex with men with virologic failure in antiretroviral therapy in Sichuan, China, 2011 to 2017. Medicine. 2019;98:43(e17585).
DY and ZD contributed equally to this work.
This research was funded by the National Natural Science Foundation of China (81703279) and the National Mega Projects of Science and Technology in the 13th Five-Year Plan of China: Technical Platform for Communicable Disease Surveillance Project (No. 2017ZX10103010-002).
The authors have no conflicts of interests to disclose.
References
- [1].Pang W, Shang P, Li Q, et al. Prevalence of opportunistic infections and causes of death among hospitalized HIV-infected patients in Sichuan, China. Tohoku J Exp Med 2018;244:231–42.. [DOI] [PubMed] [Google Scholar]
- [2].Zeng P, Liu Y, He M, et al. HIV-1 genotypic diversity and prevalence of drug resistance among treatment naïve HIV-infected individuals in Chengdu of China. Virus Genes 2013;47:408–13.. [DOI] [PubMed] [Google Scholar]
- [3].Hu Y, Liu L, Liu YJ, et al. Epidemic characteristics of HIV/AIDS among men who have sex with men from 2011 to 2015 in Sichuan Province. J Prev Med Inf 2017;33:642–7.. [Google Scholar]
- [4].Hei FX, Wang L, Qin QQ, et al. Epidemic characteristics of HIV/AIDS among men who have sex with men from 2006 to 2010 in China. Zhonghua Liu Xing Bing Xue Za Zhi 2012;33:67–70.. [PubMed] [Google Scholar]
- [5].Günthard HF, Aberg JA, Eron JJ, et al. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA 2014;312:410–25.. [DOI] [PubMed] [Google Scholar]
- [6].ingankar NK, Thorat SR, Deshpande A, et al. Initial virologic response and HIV drug resistance among HIV-infected individuals initiating first-line antiretroviral therapy at 2 clinics in Chennai and Mumbai, India. Clin Infect Dis 2012;54Suppl 4:S348–54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Guo H, Hu H, Zhou Y, et al. A Novel HIV-1 CRF01_AE/B recombinant among men who have sex with men in Jiangsu Province, China. AIDS Res Hum Retroviruses 2014;30:706–10.. [DOI] [PubMed] [Google Scholar]
- [8].Liu H, Yang H, Li X, et al. Men who have sex with men and human immunodeficiency virus/sexually transmitted disease control in China. Sex Transm Dis 2006;33:68–76.. [DOI] [PubMed] [Google Scholar]
- [9].Koblin BA, Chesney MA, Husnik MJ, et al. High-risk behaviors among men who have sex with men in 6 US cities: baseline data from the EXPLORE study. Am J Public Health 2003;93:926–32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Taniguchi T, Nurutdinova D, Grubb JR, et al. Transmitted drug-resistant HIV type 1 remains prevalent and impacts virologic outcomes despite genotype-guided antiretroviral therapy. AIDS Res Hum Retroviruses 2012;28:259–64.. [DOI] [PubMed] [Google Scholar]
- [11].Castro H, Pillay D, Cane P, et al. Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis 2013;208:1459–63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gupta R, Hill A, Sawyer AW, et al. Emergence of drug resistance in HIV type 1-infected patients after receipt of first-line highly active antiretroviral therapy: a systematic review of clinical trials. Clin Infect Dis 2008;47:712–22.. [DOI] [PubMed] [Google Scholar]
- [13].Wittkop L, Günthard H, de Wolf F, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis 2011;11:363–71.. [DOI] [PubMed] [Google Scholar]
- [14].Machnowska P, Meixenberger K, Schmidt D, et al. Prevalence and persistence of transmitted drug resistance mutations in the German HIV-1 Seroconverter Study Cohort. PLoS One 2019;14:e0209605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Leigh Brown AJ, Lycett SJ, Weinert L, et al. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis 2011;204:1463–9.. doi: 10.1093/infdis/jir550jir550 [pii] [published Online First: 2011/09/17]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lewis F, Hughes GJ, Rambaut A, et al. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med 2008;5:e50.doi: 10.1371/journal.pmed.005005007-PLME-RA-1340 [pii] [published Online First: 2008/03/21]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Burger H, Weiser B, Flaherty K, et al. Evolution of human immunodeficiency virus type 1 nucleotide sequence diversity among close contacts. Proc Natl Acad Sci U S A 1991;88:11236–40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Balfe P, Simmonds P, Ludlam CA, et al. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J Virol 1990;64:6221–33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gibson KM, Steiner MC, Kassaye S, et al. A 28-year history of HIV-1 drug resistance and transmission in Washington, DC. Front Microbiol 2019;10:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wertheim JO, Kosakovsky Pond SL, Forgione LA, et al. Social and genetic networks of HIV-1 transmission in New York city. PLoS Pathog 2017;13:e1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mourad R, Chevennet F, Dunn DT, et al. A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK. AIDS 2015;29:1917–25.. [DOI] [PubMed] [Google Scholar]
- [22].Chen M, Ma Y, Su Y, et al. HIV-1 genetic characteristics and transmitted drug resistance among men who have sex with men in Kunming, China. PLoS One 2014;9:e87033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].WHO. Scaling up Antiretroviral Therapy in Resource-Limited Settings: Treatment Guidelines for a Public Health Approach 2006 Revision. Geneva: WHO; 2006. [Google Scholar]
- [24].Yuan D, Su L, Liu H, et al. Drug-resistance characteristics of CRF01_AE and CRF07_BC subtypes of HIV-1 strains in Sichuan province. Zhonghua Yu Fang Yi Xue Za Zhi 2015;49:901–6.. [PubMed] [Google Scholar]
- [25].Poon AF, Joy JB, Woods CK, et al. The impact of clinical, demographic and risk factors on rates of HIV transmission: a population-based phylogenetic analysis in British Columbia, Canada. J Infect Dis 2015;211:926–35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu Y, Ren X, Yin D, et al. Characterization of a novel HIV-1 unique recombinant form between CRF07_BC and CRF55_01B in men who have sex with men in Guangzhou, China. PLoS One 2017;12:e0175770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang J, Wang XQ, Yin HF, et al. Gene mutations of human immunodeficiency virus drug resistance from men who have sex with men in Shanghai, 2013 Chinese. J Infect Dis 2016;34:23–6.. [Google Scholar]
- [28].Hai-Long H, Jian Z, Ping-Ping Y, et al. Genetic characterization of CRF01_AE full-length human immunodeficiency virus type 1 sequences from Fujian, China. AIDS Res Hum Retroviruses 2007;23:569–74.. [DOI] [PubMed] [Google Scholar]
- [29].Xing H, Ruan Y, Li J, et al. HIV drug resistance and its impact on antiretroviral therapy in Chinese HIV-infected patients. PLoS One 2013;8:e54917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang XL, Jia L, Li HP, et al. Transmission cluster and network of HIV-1 CRF01_AE strain in China, 1996-2014. Zhonghua Liu Xing Bing Xue Za Zhi 2019;40:84–8.. [DOI] [PubMed] [Google Scholar]
- [31].Hao M, Wang J, He S, et al. Identification of a Novel HIV-1 second-generation recombinant form (CRF01_AE/07_BC) in men who have sex with men in Beijing, China. AIDS Res Hum Retroviruses 2019;35:500–4.. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- [32].Ma P, Ge Z, Feng Y, et al. Near full-length genome sequence of a novel HIV-1 second-generation recombinant form (CRF01_AE/CRF07_BC) detected among men who have sex with men in Tianjin, China. AIDS Res Hum Retroviruses 2019;35:205–12.. [DOI] [PubMed] [Google Scholar]
- [33].Hou LJ, Wang HW, Duan SP, et al. The prevalence and determinants of drug-resistance-associated mutations in the HIV-1-infected MSM population of Henan Province in China. Arch Virol 2015;160:2051–61.. [DOI] [PubMed] [Google Scholar]
- [34].Iyidogan P, Anderson KS. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses 2014;6:4095–139.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Menéndez-Arias L. Targeting HIV: antiretroviral therapy and development of drug resistance. Trends Pharmacol Sci 2002;23:381–8.. [DOI] [PubMed] [Google Scholar]
- [36].Cunningham E, Chan YT, Aghaizu A, et al. Enhanced surveillance of HIV-1 drug resistance in recently infected MSM in the UK. J Antimicrob Chemother 2017;72:227–34.. [DOI] [PubMed] [Google Scholar]
- [37].Miller V, Larder BA. Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure. Antivir Ther 2001;6Suppl 3:25–44.. [PubMed] [Google Scholar]
- [38].Naeger LK, Margot NA, Miller MD. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184 V and zidovudine-associated mutations: analysis of enzyme processivity, chain-terminator removal and viral replication. Antivir Ther 2001;6:115–26.. [PubMed] [Google Scholar]
- [39].White KL, Margot NA, Wrin T, et al. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184 V and their effects on enzyme function and viral replication capacity. Antimicrob Agents Chemother 2002;46:3437–46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hamers RL, Siwale M, Wallis CL, et al. HIV-1 drug resistance mutations are present in six percent of persons initiating antiretroviral therapy in Lusaka, Zambia. J Acquir Immune Defic Syndr 2010;55:95–101.. [DOI] [PubMed] [Google Scholar]
- [41].Gatanaga H, Ibe S, Matsuda M, et al. Drug-resistant HIV-1 prevalence in patients newly diagnosed with HIV/AIDS in Japan. Antiviral Res 2007;75:75–82.. [DOI] [PubMed] [Google Scholar]
- [42].Barth RE, Wensing AM, Tempelman HA, et al. Rapid accumulation of nonnucleoside reverse transcriptase inhibitor-associated resistance: evidence of transmitted resistance in rural South Africa. AIDS 2008;22:2210–2.. [DOI] [PubMed] [Google Scholar]
- [43].Cao B, Liu C, Stein G, et al. Faster and riskier? Online context of sex seeking among men who have sex with men in China. Sex Transm Dis 2017;44:239–44.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chen M, Ma Y, Chen H, et al. HIV-1 genetic transmission networks among men who have sex with men in Kunming, China. PLoS One 2018;13:e0196548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zennou V, Mammano F, Paulous S, et al. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol 1998;72:3300–6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Martinez-Picado J, Savara AV, Sutton L, et al. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol 1999;73:3744–52.. [DOI] [PMC free article] [PubMed] [Google Scholar]


