Abstract
Introduction:
Leptospirosis is a neglected zoonotic disease caused by a bacteria of the genus Leptospira. In Africa it is frequently mistaken for frequently occurring conditions such as malaria. The aim of this study was to identify rodent species involved in the transmission of the disease, the prevalence of pathogenic Leptospira spp. in selected rodent species and risk factors for human leptospirosis.
Material and Methods:
We conducted a descriptive and exploratory epidemiological and molecular study in Mozambique Island city in 2015. Six neighborhoods, comprising 30 households each were randomly selected. People from the selected 180 households were interviewed regarding their awareness of the disease, the presence of rodents in their houses, chemicals used to eliminate them, sewage disposal, water supply system, and other key issues related to the disease. In each neighborhood we trapped 10 rodents for morphometric study to identify their species and for molecular isolation of Leptospira DNA. We extracted kidneys from 57/60 of rodents trapped, and performed nested polymerase chain reaction targeting rrs 16S ribosomal DNA and lipL32 genes for identification of Leptospira genus and pathogenic Leptospira spp. respectively.
Results:
Of the 180 participants 92 (51%) reported having heard of leptospirosis; 107 (59%) have had the disease; 151 (83%) reported the existence of rats in their house; 100 (56%) had latrines; 118 (66%) used chemicals to kill the rats; 102 (57%) used well water and 114 (63%) used trash containers. The most prevalent rodent species captured was Rattus norvegicus 36/60 (60%), followed by Rattus rattus 19/60 (31.67%) and Mus musculus 3/60 (5%). rrs 16S ribosomal DNA was identified in 20/57 (35.%) rodents. Out these two were positive for lipL32 gene, giving an overall pathogenic Leptospira infection of 3.5% (2/57). The rodent species identified as carriers of pathogenic Leptospira were Rattus norvegicus (1) and R. rattus (1).
Conclusion:
This is the first study in Mozambique to identify the presence of pathogenic species of Leptospira using molecular tools. Leptospirosis risk factors in Mozambique Island city are rodent’s infestation, limited disease awareness, lack of access to clean water, insufficient resources for waste collection, greater clustering of households, poor sanitation environment and degradation of living conditions. Pathogenic Leptospira spp. are present in the area studied and at least two species of rodents, the R. rattus and R. norvegi-cus are potentially involved in the transmission of the causal agents of the disease.
Keywords: Leptospirosis, lipL32, rrs16S, Nested-PCR, Rodents Species, Mozambique
Introduction
Leptospirosis, a disease caused by spirochetes of the genus Leptospira has been described as the most geographically spread and under reported neglected bacterial zoonotic cause of morbidity and mortality disease in the world [1,2]. Pathogenic leptospires persistently colonize the kidneys of asymptomatic reservoir animals that then shed bacteria in urine [1–5]. Rodents are considered to be the natural reservoir and the primary intermediate host responsible for disease transmission to humans in urban environments [6,7]. Pathogenic Leptospira spp. are spread in the environment through rodent urine and can be transmitted to humans through direct or indirect contact with infected urine, food or contaminated water [8,9]. The emergence of leptospirosis is often associated with the growth of informal urban settlements, poor environmental sanitation, climate change [8–12], and both recreational and occupational activities [5]. Human infection can be subclinical or symptomatic and can be associated with a range of clinical manifestations. Many of the non-specific clinical manifestations including fever, headache and myalgia can lead to a misdiagnosis of malaria and other common febrile conditions [4,13].
Worldwide there are more than 250 pathogenic Leptospira serovars classified into 25 serogroups based on their serological phenotype [4,14,15]. Recent species determination by molecular methods identified 22 genomic species of Leptospira with 13 pathogenic Leptospira spp. Among these L. interrogans, L. borgpetersenii, L. santarosai, L. noguchii, L. weilli, L. kirschneri and L. alexanderi, and others are considered to be agents of human and animal disease [13]. Both serological and DNA-based classification systems are currently used for clinical diagnosis and for understanding the pathogenesis and epidemiology of the disease [15].
The immunodiagnostic methods for Leptospira spp. are based on the demonstration of serum antibodies or antigens by ELISA or by the microscopic agglutination test (MAT) still considered the gold standard test. These have limited sensitivity and specificity as they can cross react within different serovars [13,16,17]. Molecular diagnosis using different genes and specific primers is becoming more popular and useful in acute phase of the disease. Genes targeted by conventional PCR include (rrs, rrl, flab, gyrB, ompLl, lig, lipL32, lipL21, lipL41, and secY), but only a few (including rRNA and lipL32) have been validated and subJected to clinical evaluation. The 16S rRNA gene is genus-specific for Leptospira while the lipL32 gene encoding the major outer membrane lipoprotein of Leptospira and considered to be the marker of Leptospira spp. pathogenicity [15]. Molecular diagnosis is much more rapid than culture and is a strong indicator of active infection [18].
Meta-analyses indicate that leptospirosis affects 2.3% to 19.8% of patients that seek hospital care for febrile illness on the African continent [11,13,19,20]. In Tanzania, Sri Lanka and Egypt, studies in febrile patients indicate a seroprevalence of 8.8%, 15.5% and 16% of Leptospira spp. respectively [13].
In Mozambique, the impact of leptospirosis in humans is unknown due to the lack of clinical, epidemiological and molecular studies, in both humans and rodents. In two cross sectional serological surveys antibodies to Leptospira spp. were found in 1.3% and 10% of febrile patients [15,21]. In one of these studies, it was also found that most of patients with a presumptive infection lived in a rural setting 32 (84.2%). A significantly higher frequency of contact with rodents was found in patients with confirmed leptospiral infections (5/5, 100%) as compared to those with presumptive infections (15/38, 39.5%) [15].
The presence of Leptospira spp. in R. rattus was demonstrated in rat kidneys on two islands in the Mozambique Channel. One positive specimen was found on each island (2/52) indicating a previously unknown presence of Leptospira spirochetes in these islands [22]. In this manuscript, we describe results of the study that aimed to identify the rodent species that may act as carriers of pathogenic Leptospira species and to identify risk factors for transmission of these spirochetes microorganism. We also wished to determine whether rats are the primary reservoirs of pathogenic Leptospira spp. in Mozambique Island city.
Material and Methods
Study area and population
This exploratory epidemiological and molecular study was undertaken between the 5th and 25th of May, 2015 in the Mozambique Island city (15° 02 ‘S, 40° 44’ E). The island has a surface area of445 Km2 and is located 5 km from the mainland coast of Nampula Province, in the Northeast of Mozambique (See figure 1).
Figure 1:
Map of Mozambique including Mozambique Island city.
The study was approved by the scientific board of the Lúrio University Faculty of Health Sciences. Mozambique Island city has 65,712 inhabitants of which 31,473 are male and 34,239 are female). These inhabitants live in 15,299 households divided into 8 neighborhoods [23]. We randomly selected six neighborhoods including the Esteu, Marangonha, Macaribe, Litine, Museu and Unidade neighborhoods. Within each neighborhood, we interviewed 30 household residents totaling 180 people. Selected participants were informed about the study and methods to be used in order to seek their consent. After consent was obtained we administered a questionnaire to the participants. The questionnaire included questions related to the existence of rats in the surroundings and awareness of the diseases caused by them, the use of chemicals to eliminate them, the water supply and sewage disposal systems and other demographic and sociological questions. In addition we directly observed the sanitary and environmental conditions in order to access leptospirosis risk factors.
Rodent trapping and identification of the species
In each selected neighborhood, we trapped 10 rodents using traditional traps. These traps were placed in the evening and collected in the morning of the next day. Rodents were transported to Lúrio University where the kidneys were collected. Additionally, the morphometric characteristics of the rodents were assessed and recorded for further identification of the species as described elsewhere [24,25]. Briefly morphological identification of the rodents was accomplished by measuring the length and the weight of the rat, and correlating these data with a bibliographic review of manuals containing information for appropriate identification of the species [24,25].
Detection of pathogenic Leptospira spp.
We collected kidneys of 57 rodents under sterile conditions. These were preserved at −20°C until being sent to the Parasitology Laboratory at the Faculty of Medicine, Eduardo Mondlane University, in Maputo where they were kept at −80°C until processed.
The kidneys from the rodents were assessed by nested-PCR, targeting the DNA encoding the rrs 16S rDNA gene and the lipL32 lipoprotein gene as previously described [26,27].
Genomic DNA extraction
DNA extraction from kidney tissue was performed using the commercial Puregene DNA Tissue kit (Qiagen®, Ilden, Germany), according to the manufacturer’s instructions. Proteinase K (Qiagen®, Ilden, Germany) and Glycogen were used to achieve higher yields of extracted DNA. All procedures were undertaken in a laminar flow chamber to avoid contaminations.
Nested PCR reaction for molecular identification
We used primers manufactured at Integrated DNA Technologies (IDT, Skokie, IL) with the following sequences.
For the amplification of rrs (16S) (289 bp) we used the primers (A - 5’ - GGCGGCGCGTCTTAAACATG - 3’ (forward); B - 5’ - TTCCCCCCATTGAGCAAGATT - 3’ (reverse); C - 5’ - TGCAAGTCAAGCGGAGTAGC - 3’ (forward nested); D - 5’ - TTCTTAACTGCTGCCTCCCG - 3’ (reverse nested) as previously described [26,27]
For amplification of lipL32 (183 bp) we used the following primers (A - 5’ - CGCTTGTGGTGCTTTCGGTGGT - 3’ (forward); B - 5’ - CTCACCGATTTCGCCTGGG - 3’ (reverse); C - 5’ - TTCTGAGCGAGGACACAATCCC - 3’ (forward nested); D - 5’ - CTCCCATTTCAGCGATTACGG - 3’ (reverse nested) as previously described [26,27]. The lyophilized primers were diluted with 1x TE solution pH8 (IDT) to a concentration of 100 μM and kept at −20°C. Further dilutions were performed using PCR grade water (IDT) to a concentration of 20 μM for further use in the PCR reaction according manufacturer’s instructions (MyTaq Mix Bioline London, United Kingdom).
The PCR occurred in a 2x MyTaq Mix (Bioline® London, United Kingdom). The primers (20 μM) for rrs (16S) (289 bp) were added to the mix at a concentration of 0.39 μM each and the extracted DNA corresponding to 9.8% of the final volume. Additionally, the PCR grade water (IDT, Skokie, IL) was used to achieve the final reaction volume. The nested PCR reaction was performed under the same conditions using 4.9% of the volume from the primary reaction product. As a positive control we used DNA of Leptospira interrogans extracted from pathogenic strains kindly provided by one of the Portuguese Reference Leptospirosis laboratories, at the Instituto de Higiene e Medicina Tropical- Universidade Nova de Lisboa (IHMT-UNL). As a negative control, we used PCR grade water. The reaction occurred in a thermocycler (GeneAmp® PCR System 9700, Applied BioSystems - California, USA) and the cycles and temperatures were the following: Initial Denaturation - 3 minutes at 94°C; 30 cycles - Denaturation - 1 minute at 94°C; Annealing - 90 seconds at 55°C; Elongation - 1 minute at 72°C; Final Elongation - 10 minute at 72°C [27]. For the lipL32 amplification we used the same procedures except for the number of cycles was raised to 40.
Detection of PCR products
DNA products (amplicons) were analyzed by electrophoresis in agarose gel (2%) stained with Ethidium Bromide Bio-Rad Laboratories® - California -USA (10 mg/ml) to a final concentration of 0,5 μg/mL and visualized under ultra violet trans illumination [28].
Statistical analysis
GraphPad Prism v7 software was used for the statistical analysis. Analysis of quantitative variables was undertaken using the SPSS program version 21. Proportions were compared using the X2 test or the Fisher’s exact test. A 2-tailed P value of < 0.05 was Judged to be significant.
Multiple comparison were performed using ANOVA (two and one way) with Bonferroni’s correction.
Results and Discussion
To our knowledge, this is the first exploratory study done in Mozambique in rodents using a combination of sociological and molecular methods to evaluate the risk factors for associated with human acquisition of the pathogen as well as the presence of pathogenic Leptospira spp in different species of rodents.
Community awareness and environment risk factors for human leptospirosis
Of the 180 participants, 92 (51%) reported having heard of leptospirosis and 107 (59%) reported that they have had the disease in the past based on a description of the disease provided to them and the association of that disease with rodent exposure. One hundred fifty one (83%) reported the existence of rats in their houses. One hundred (51%) had latrines but 17 of these were of poor quality. One hundred eighteen (66%) reported using chemicals to kill rats in their houses. One hundred two (57%) used wells as their water source and 78 reported using piped water or fountains. Most study participants (63%) deposited trash in dumpsters (See table 1).
Table 1:
Risk factors associated with leptospirosis in Mozambique Island city.
| Esteu n = 30 |
Marangonha n = 30 |
Macaribe n = 30 |
Litine n = 30 |
Museu n = 30 |
Unidade n = 30 |
Total n = 180 |
||
|---|---|---|---|---|---|---|---|---|
| Awareness regarding Leptospirosis | Yes | 14 (47%) | 15 (50%) | 16 (53%) | 18 (60%) | 17 (57%) | 12 (40%) | 92 (51%) |
| No | 16 (53%) | 15 (50%) | 14 (47%) | 12 (40%) | 13 (43%) | 18 (60%) | 88 (49%) | |
| Presence of Leptospirosis | Yes | 21 (70%) | 27 (90%) | 15 (50%) | 9 (30%) | 19 (63%) | 16 (53%) | 107 (59%) |
| No | 9 (30%) | 3 (10%) | 15 (50%) | 21 (70%) | 11 (37%) | 14 (47%) | 73 (41%) | |
| Rodents presence in the house |
Yes | 28 (93%) | 26 (87%) | 27 (90%) | 25 (83%) | 21 (70%) | 24 (80%) | 151 (83%) |
| No | 2 (7%) | 4 (13%) | 3 (10%) | 5 (17%) | 9 (30%) | 6 (20%) | 29 (17%) | |
| Latrine | Yes | 23 (77%) | 11 (37%) | 15 (50%) | 12 (40%) | 30 (100%) | 9 (30%) | 100 (56%) |
| No | 7 (23%) | 19 (63%) | 15 (50%) | 18 (60%) | 0 (0%) | 21 (70%) | 80 (49%) | |
| State of the Latrine n = 100 |
Precarious | 3 (13%) | 4 (36%) | 1 (7%) | 2 (17%) | 5 (17%) | 2 (22%) | 17 (9%) |
| Improved | 7 (30%) | 2 (18%) | 5 (33%) | 3 (25%) | 2 (7%) | 4 (44%) | 23 (13%) | |
| Very Improved | 13 (57%) | 5 (46%) | 9 (60%) | 7 (58%) | 23 (76%) | 3 (34%) | 60 (78%) | |
| Water Source | Well | 17 (57%) | 23 (77%) | 21 (70%) | 25 (83%) | 5 (10%) | 11 (37%) | 102 (57%) |
| Piped | 9 (30%) | 5 (10%) | 7 (23%) | 4 (13%) | 21 (70%) | 14 (47%) | 60 (33%) | |
| Fountain | 4 (13%) | 2 (13%) | 2 (7%) | 1 (4%) | 4 (20%) | 5 (16%) | 18 (10%) | |
| Waste Disposal | Open Dumpster | 13 (43%) | 21 (70%) | 25 (83%) | 24 (80%) | 5 (17%) | 26 (87%) | 114 (63%) |
| Beach | 5 (17%) | 3 (10%) | 2 (7%) | 4 (13%) | 1 (3%) | 3 (10%) | 18 (10%) | |
| Containers | 12 (40%) | 6 (20%) | 3 (10%) | 2 (7%) | 24 (80%) | 1 (3%) | 48 (27%) | |
| Usage of chemical agents to eliminate rodents |
Yes | 27 (90%) | 21 (70%) | 17 (57%) | 19 (63%) | 16 (53%) | 18 (60%) | 118 (66%) |
| No | 3 (10%) | 9 (30%) | 13 (43%) | 11 (37%) | 14 (47%) | 12 (40%) | 62 (34%) |
We concluded that several risk factors for leptospirosis are present in Mozambique Island city. These include infestation by rodents, limited disease awareness, lack of access to clean water, insufficient resources for waste collection, high density clustering of households, a poor sanitation environment and degradation of living conditions. There was no statistically significant association between risk factors for leptospirosis and participant reports of previously having had the disease. With the existence of rats and the level of knowledge of the disease in Mozambique Island city, it is important that health authorities take appropriate measures to educate the population about the disease and its risk factors, to improve sanitation, and to improve rodent control. Such measures have reduced other poverty related diseases in other settings [29–32].
Rodents morphometric identification
We captured 60 rodents belonging to 3 different species and distributed as follow: 36 (60%) were Rattus norvegicus, 19 (32%) were R. rattus and 3 (5%) were Mus musculus. We were not able to identify the species of two rodents using the morphometric tools we had at our disposal. The differences amongst proportions of rodents captured in each neighborhood were not statistically significant. The same species of rodents were also captured in a similar studies and settings performed in Angola and we would expect that the profile of the disease in Mozambique will be similar to that of Angola [27]. Future studies of rodent speciation should include molecular approaches since morphological criteria cannot always discriminate among genera [12].
Leptospira spp. molecular detection
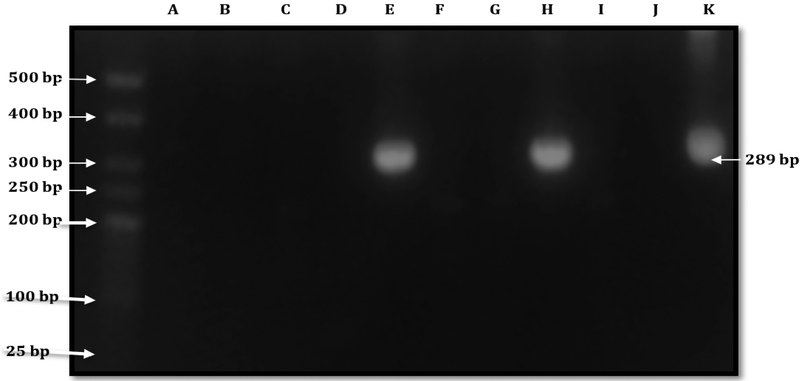
PCR analysis showed that 20/57 (35.08%) of rodent kidney samples were positive for the presence of DNA coding the rrs 16S gene (See figure 2).
Figure 2:
Agarose gel electrophoresis showing the Nested PCR amplification targeting the DNA coding rrs 16S. Bands were observed in two samples (Lanes E and H) and in the positive control (pathogenic Leptospira DNA) in Lane K. PCR grade water (Lane J) served as the negative control, Weight of the bands is around 300 bp (289 bp). A 25 bp ladder was used.
The rrs 16S gene DNA sequence has proven to be specific for the detection and identification of Leptospira genus, although it cannot differentiate pathogenic species [14]. Of these 20 rodents 11 (55%) were of the R. norvegicus species, 6 (30%) the R. rattus species, two (10%) the M. musculus species. One positive DNA reaction occurred in one of the rats whose specie we did not identify. In this study, R. norvegicus was the most common specie infected with Leptospira spp. However, no significant differences were found amongst positive and negative rodents for the species captured. This finding is consistent with several studies indicating rats as the main natural reservoir of Leptospira spp. and strongly associated with human leptospirosis [7,12,33]. In addition, our findings are consistent with a study in Southeast Asia supporting the premise that rodent infestation is driven by habitat, rather than rodent species [14].
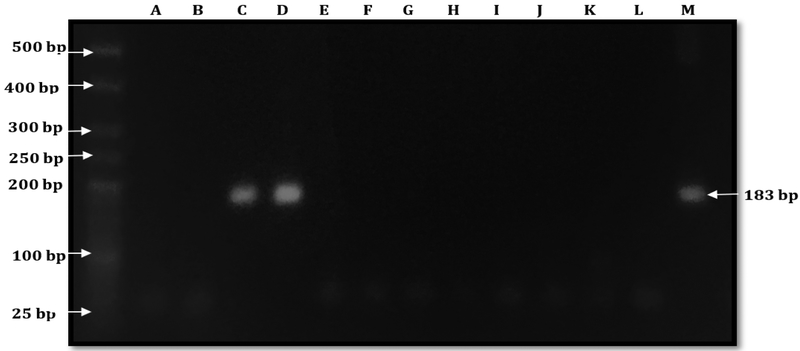
Out of 20 samples positive for DNA coding the rrs 16S gene, two (10%) were positive to LipL32 gene (See figure 3) These were found in one R. norvegicus and R. rattus, respectively.
Figure 3:
Agarose gel electrophoresis showing the Nested PCR amplification targeting the DNA coding the LipL32 lipoprotein. Bands were observed both in the samples (Lanes C and D) and in the positive control (pathogenic Leptospira DNA) in Lane M. PCR Grade Water (Lane L) served as the negative control. Lanes A, B, E-K show results for 9 samples whose Leptospira did not demonstrate the pathogenic strain sequence. The weight of the bands is approximately 200 bp (183 bp). A 25 bp ladder was used.
Thus, the overall prevalence of pathogenic species of Leptospira in kidneys was of 3.5% (2/57). These results must be interpreted with caution since this was an exploratory work done in a small sample aimed to detect Leptospira spp. in rodents and to optimize molecular protocols in our laboratory.
In addition, and based on a literature review, it is unlikely to find non-pathogenic Leptospira spp. in rodent’s kidneys, as pathogenic Leptospira spp. are found in several mammals, while the saprophytic species are found mostly in the environment [7,14,33,34]. Optimization of the protocols for IipL32 detection should be improved in our laboratory, including the evaluation of the amount of the DNA extracted.
This study was conducted during the dry season and it is possible that the frequency of infected rats might be higher if the study had been done during the rainy season when there is more probability for rat infestation, as has been observed in several studies [1,5,15].
Despite the high exposure rate of the population studied to rodents and the awareness of the disease among health professionals, it is expected that some febrile patients are misdiagnosed with malaria, as observed in a number of studies in Mozambique and elsewhere. Further hampering diagnosis are the lack of epidemiological and clinical studies aiming to define the epidemiology and the profile of this zoonotic and neglected disease, the lack of environmental surveillance systems and limited laboratory tools to confirm the diagnosis [2,14,15,21].
Although these results are of exploratory nature, they will help to define future research priorities, which should be targeted to humans, rodents and the environment, and to define educational and promotional strategies directed at behavioral change designed to decrease risk factors for transmission and to improve sanitation.
Conclusion
Pathogenic Leptospira spp. are present in Mozambique Island city and at least two species of rodents, the R. rattus and R. norvegicus are the reservoirs of the bacteria. Leptospirosis should be suspected in cases of febrile disease, to avoid misdiagnosis with malaria, a most common cause of disease in Mozambique
Further studies both on rodents and on humans should be carried out in order to identify the circulating species of Leptospira, so that appropriate prevention, diagnostic and treatment measures can be taken to control the disease and thus contribute to the achievement of millennium development goals.
Acknowledgment
The Medical Education Partnership Initiative (MEPI) provided financial resources to create master’s degree programs at Universi-dade Lúrio through Grant Numbers R24TW008908 and R24TW008910 from the Fogarty International Center. The manuscript writing and publication was supported by Fogarty International Center, Office of the Director, Eunice Kennedy Shriver National Institute of Child Health and Human Development and National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Number D43TW010135. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Fogarty International Center or the National Institutes of Health. Furthermore, we thank Professor Virgilio E Do Rosário from IHMT-UNL for his contribution on designing, recruiting and coordinating all IHMT-UNL team involved in the establishment and implementation of the Master program and to the IHMT_UNL for providing the purified Leptospira DNA used as a positive control in the nested PCR reaction.
Abbreviations
- ELISA
Enzyme Linked Immunoassay
- IHTM-UNL
Instituto de Higiene e Medicina Tropical- Universidade Nova de Lisboa
- IDT
Integrated DNA Technologies
- lipL32
Lipoprotein L32 Gene
- MAT
Microscopic Agglutination Test
- PCR
Polymerase Chain Reaction
- rrs 16S
Subunity 16 ribosomal RNA
Footnotes
Conflict of Interest
Authors declare no conflict of interest.
Bibliography
- 1.Levett PN. Leptospirosis”. Clinical Microbiology Reviews 142 (2001): 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. “Report of the Second Meeting of the Leptospirosis Burden”. E.R. Group, Editor. WHO: Geneva: (2011). [Google Scholar]
- 3.Barragan V, et al. “High Leptospira Diversity in Animals and Humans Complicates the Search for Common Reservoirs of Human Disease in Rural Ecuador”. PLOS Neglected Tropical Diseases 109 (2016): e0004990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halliday JE., et al. “Urban leptospirosis in Africa: a cross-sectional survey of Leptospira infection in rodents in the Kibera urban settlement, Nairobi, Kenya”. American Journal of Tropical Medicine and Hygiene 896 (2013): 1095–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau CL., et al. “Climate change, flooding, urbanisation and leptospirosis: fuelling the fire?” Transactions of the Royal Society of Tropical Medicine and Hygiene 10410 (2010): 631–638. [DOI] [PubMed] [Google Scholar]
- 6.Haake DA and Levett PN. “Leptospirosis in humans”. Current Topics in Microbiology and Immunology 387 (2015): 65–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.KroJgaard LH., et al. “High prevalence of Leptospira sp in sewer rats (Rattus norvegicus)”. Epidemiology and Infection 13711 (2009): 1586–1592. [DOI] [PubMed] [Google Scholar]
- 8.Loffler SG., et al. “Genotypes of pathogenic Leptospira spp isolated from rodents in Argentina”. Memórias do Instituto Oswaldo Cruz 1092 (2014): 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bharti ARNJ., et al. “Peru-United States Leptospirosis Consortium. Leptospirosis: a zoonotic disease of global importance”. Lancet Infectious Diseases 312 (2003): 757–771. [DOI] [PubMed] [Google Scholar]
- 10.Fraga TR., et al. “Leptospirosis: aspects of innate immunity, immunopathogenesis and immune evasion from the complement system”. Scandinavian Journal of Immunology 735 (2011): 408–419. [DOI] [PubMed] [Google Scholar]
- 11.Pui CF., et al. “Diversity of Leptospira sp in Rats and Environment from Urban Areas of Sarawak, Malaysia”. Journal of Tropical Medicine (2017): 3760674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortes-Gabriel E, et al. “First Isolates of Leptospira spp., from Rodents Captured in Angola”. American Journal of Tropical Medicine and Hygiene 945 (2016): 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allan KJ., et al. “Epidemiology of Leptospirosis in Africa: A Systematic Review of a Neglected Zoonosis and a Paradigm for ‘One Health’ in Africa”. PLOS Neglected Tropical Diseases 99 (2015): e0003899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosson JF., et al. “Epidemiology of leptospira transmitted by rodents in southeast Asia”. PLOS Neglected Tropical Diseases.86 (2014): e2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribeiro P, et al. “Seroepidemiology of leptospirosis among febrile patients in a rapidly growing suburban slum and a flood-vulnerable rural district in Mozambique, 2012–2014: Implications for the management of fever”. International Journal of Infectious Diseases 64 (2017): 50–57. [DOI] [PubMed] [Google Scholar]
- 16.Brown PD., et al. “Evaluation of the polymerase chain reaction for early diagnosis of leptospirosis”. Journal of Medical Microbiology 432 (1995): 110–114. [DOI] [PubMed] [Google Scholar]
- 17.Levett PN and Branch SL. “Evaluation of two enzyme-linked immunosorbent assay methods for detection of immunoglobulin M antibodies in acute leptospirosis”. American Journal of Tropical Medicine and Hygiene 666 (2002): 745–748. [DOI] [PubMed] [Google Scholar]
- 18.Waggoner JJ and Pinsky BA. “Molecular diagnostics for human leptospirosis”. Current Opinion in Infectious Diseases 295 (2016): 440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerreiro H, et al. “Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans”. Infection and Immunity 698 (2001): 4958–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko AI., et al. “Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen”. Nature Reviews Microbiology 710 (2009): 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collares-Pereira M, et al. “Preliminary survey of Leptospirosis and Lyme disease amongst febrile patients attending community hospital ambulatory care in Maputo, Mozambique”. Central African Journal of Medicine 438 (1997): 234–238. [PubMed] [Google Scholar]
- 22.Freulon M, et al. “[Detection of Leptospira organisms in Rattus rattus of two islands in the Mozambique Channel: Europa and Juan-de-Nova]”. Bulletin de la Société de Pathologie Exotique 1031 (2010): 48–50. [DOI] [PubMed] [Google Scholar]
- 23.Censos Moçambique I. 2007 (2007).
- 24.Carvalho C “Controle de Roedores”. In Manual Prático da Biologia e Controle de Roedores. CIBA-GEIGY (1986): 295. [Google Scholar]
- 25.Quinn R “Comparing rat’s to human’s age: how old is my rat in people years?” Nutrition 216 (2005): 775–777. [DOI] [PubMed] [Google Scholar]
- 26.Merien F, et al. “Polymerase chain reaction for detection of Leptospira sp in clinical samples”. Journal of Clinical Microbiology 309 (1992): 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jouglard SD., et al. “Nested polymerase chain reaction for detection of pathogenic leptospires”. Canadian Journal of Microbiology 528 (2006): 747–752. [DOI] [PubMed] [Google Scholar]
- 28.Lee PY., et al. “Agarose gel electrophoresis for the separation of DNA fragments”. Journal of Visualized Experiments 62 (2012): 3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allan KJ., et al. “Renewing the momentum for leptospirosis research in Africa”. Transactions of the Royal Society of Tropical Medicine and Hygiene 10910 (2015): 605–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabiee MH., et al. “Rodent-borne diseases and their public health importance in Iran”. PLOS Neglected Tropical Diseases 124 (2018): e0006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minter A, et al. “A model for leptospire dynamics and control in the Norway rat (Rattus norvegicus) the reservoir host in urban slum environments”. Epidemics (2018). [DOI] [PubMed] [Google Scholar]
- 32.Carvalho-Pereira T, et al. “The helminth community of a population of Rattus norvegicus from an urban Brazilian slum and the threat of zoonotic diseases”. Parasitology 1456 (2018): 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scialfa E, et al. “Isolation of Leptospira interrogans from suburban rats in Tandil, Buenos Aires, Argentina”. Revista Argentina de Microbiología 422 (2010): 126–128. [DOI] [PubMed] [Google Scholar]
- 34.Costa F, et al. “Global Morbidity and Mortality of Leptospirosis: A Systematic Review”. PLOS Neglected Tropical Diseases 99 (2015): e0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]