Abstract
Introduction:
Pancreatic ductal adenocarcinoma (PDAC) is a disease of near uniform lethality. Invasive tissue biopsies of primary or metastatic lesions remain the gold standard for diagnosis, but repeated sampling is infeasible. Non-invasive liquid biopsies, which better represent the heterogeneity of PDAC, offer new opportunities for early diagnosis during surveillance in high-risk cohorts, and for the longitudinal analysis of tumor evolution and progression in patients on therapy. In particular, liquid biopsies can capture tumor-associated components, such as circulating tumor DNA (ctDNA), extracellular vesicles (EVs), and circulating tumor cells (CTCs), each of which provides genomic and molecular information about the underlying PDAC that can potentially inform clinical decisions.
Areas covered:
Here, we reviewed current knowledge and recent technological advances regarding liquid biopsy in PDAC and mention the pitfalls and benefits in each methodology. We also discuss clinical correlative studies for diagnosis and prognosis in PDAC.
Expert opinion:
Despite the technical or biological challenges mentioned in this review, in pancreatic cancer where tissue samples are limited and repeated tissue biopsies are mostly invasive and infeasible, liquid biopsies opened a new window for tumor diagnosis, molecular stratification, and treatment monitoring. While none of the isolation and analysis methods have gained widespread clinical acceptance, it is imperative that clinical-grade applications of liquid biopsies factor in the advantages and limitations of each assay platform for isolation and analysis of tumor associated components into consideration.
Keywords: Cell-free DNA, circulating tumor cells, circulating tumor clusters, extracellular vesicles, exosomes, liquid biopsies, pancreatic cancer, pancreatic ductal adenocarcinoma
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is currently the third leading cause of cancer-related death in the United States, and is predicted to become the second leading cause within the next decade [1]. PDAC patients have low 5-year survival rates (9% in the United States[2]), which is due to several factors. First, the disease has non-specific symptoms and lacks sensitive or specific biomarkers for early diagnosis. Second, there are no therapies targeting the most prevalent mutations in the disease, such as KRAS, CDKN2A, TP53, and SMAD4 mutations. Finally, although patients with metastatic disease have a couple of different first line therapeutic options, including FOLFIRINOX (leucovorin calcium, fluorouracil, irinotecan hydrochloride, and oxaliplatin), and gemcitabine plus nab-paclitaxel, this cancer is typically prone to intrinsic and acquired chemoresistance [3,4].
Thus, earlier diagnosis of pancreatic cancer can lead to improved patient outcomes. However, currently used circulating biomarkers, such as CA19–9, lack sufficient sensitivity and specificity for diagnostic purposes. Additional biomarkers, including carcinoembryonic antigen (CEA) and CA125 have been investigated as complementary diagnostic strategies to CA19–9 with varying degrees of success [5]. Non-invasive screening methods such as contrast-enhanced computed tomography (CT), magnetic resonance imaging (MRI), and endoscopic ultrasonography (EUS) can detect cancerous lesions and cystic lesions such as intraductal papillary mucinous neoplasias but cannot as readily detect preneoplastic lesions such as pancreatic intraepithelial neoplasms [6]. Owing to the high sensitivity of positron emission tomography/CT and the high specificity of EUS, the combined use of these modalities has shown promising results [6]. However, the general consensus is that these methods are insufficient for the early detection of pancreatic cancer.
The diagnosis and perfunctory molecular analysis of pancreatic cancer may involve EUS-guided fine-needle aspiration or core needle biopsy. Yet due to the retroperitoneal location of the pancreas (Figure 1A) and their invasiveness and expense, repeat biopsies for longitudinal analysis are generally avoided [4,7]. Therefore, using tissue biopsies to longitudinally monitor pancreatic cancer is infeasible, whereas liquid biopsies provide a new opportunity for molecular profiling of the genetic landscapes of pancreatic cancer throughout disease progression.
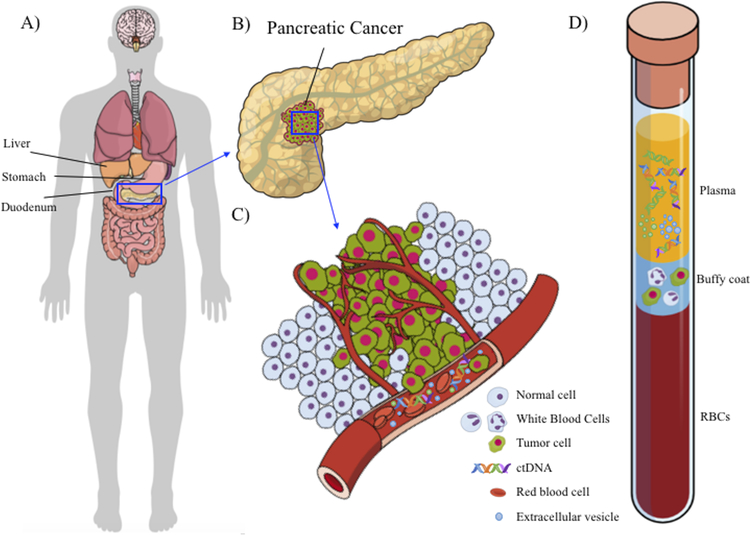
Figure 1. Pancreatic cancer and liquid biopsy (blood).
A) A schematic of the human body and abdominal cavity that shows pancreas is hidden in the abdominal cavity behind stomach, under the liver. B) A schematic of a clump of pancreatic cancer in the pancreas. This is a representative schematic, and the clump could be in any location in the pancreas. C) A schematic of the dissemination of pancreatic cancer components (ctDNA, exosomes, and CTCs) into the blood stream. D) A schematic of a blood tube with three main compartments—plasma, buffy coat, and red blood cells—each containing pancreatic cancer components.
In this article, we review current knowledge and recent technological advances regarding liquid biopsies for pancreatic cancer patients. We focus on blood-derived products, including cell-free DNA (cfDNA), extracellular vesicles (EVs; particularly exosomes), and circulating tumor cells (CTCs). The publications cited here were found using Google Scholar and Pubmed, and most references of the primary and recent studies over last 15 years (2004–2019) unless there were prominent works from a time before this period. Key words used were ctDNA, cfDNA, extracellular vesicles, exosomes, circulating tumor cells, liquid biopsy, pancreatic cancer and microfluidics.
2. Liquid Biopsies
The liquid biopsy concept is based on the idea that tumors release their components, including cfDNA, EVs (such as exosomes), and CTCs, into bodily fluids such as blood, urine, saliva, and cerebrospinal fluid [8] (Figure 1B–D). Blood biopsies in particular are minimally invasive and can be used for early diagnosis, disease stratification, and longitudinal monitoring of therapeutic response in pancreatic cancer patients. Unlike tissue biopsies, which capture the genetic makeup of only a single piece of tumor, liquid biopsies capture material released from all tumor sources, including metastatic sites, allowing for a better representation of tumor heterogeneity [9].
2.1. cfDNA and Circulating Tumor DNA.
2.1.1. Background knowledge
All cells, including cancer cells, have been reported to secrete DNA into the blood, either actively through the release of DNA fragments [10] or passively through necrosis or apoptosis [11,12]. The relative abundance of tumor-derived cfDNA, also known as circulating tumor DNA (ctDNA), can range from below 0.01% to as high as 93% [13], a wide range quantity that is directly correlated with tumor burden [14,15]. cfDNA can be isolated from either blood plasma or serum; however, the former has been proven to be a more reliable source of nucleic acids, as serum-derived cfDNA can be artifactually secreted from platelets during clotting [8,16]. Nevertheless, the detection of ctDNA in patient serum and plasma after therapy or surgery has been correlated with survival outcomes and early detection in many cancers, including pancreatic cancer [17] [18,19].
2.1.2. Technologies for detection
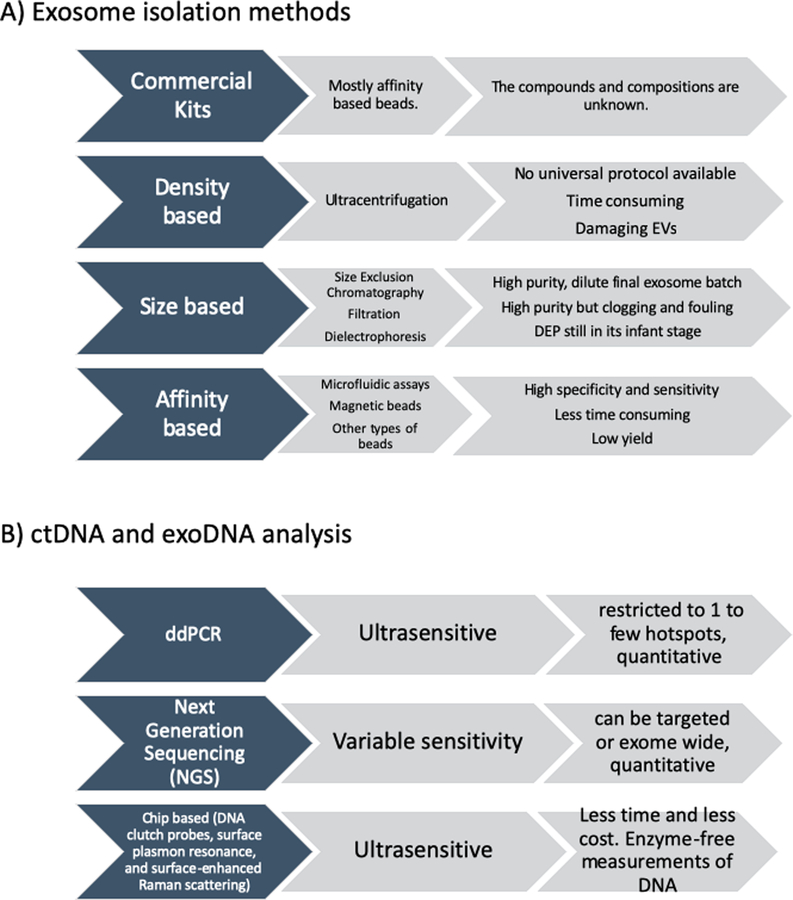
The common and traditional methods for quantification of cfDNA are spectrophotometry [17,20,21], colorimetric DNA quantification, and polymerase chain reaction (PCR)[10]. New advances in sequencing technologies, especially ultrasensitive detection methodologies such as droplet digital PCR (ddPCR) and Next Generation Sequencing (NGS) (Figure 2B), have raised the sensitivity and specificity of ctDNA measurements to levels much higher than those previously achievable [22]. Recently, researchers have developed chip-based technologies, such as DNA clutch probes, surface plasmon resonance, and surface-enhanced Raman scattering, which probe cfDNA in an enzyme-free ultra-sensitive fashion in less time and at lower cost. Nevertheless, isolating and analyzing cfDNA remains challenging owing to its fragmented nature, relatively lower abundance (especially in early or post-treatment cancers), and the vagaries of processing and storage [10,23].
Figure 2. Methods for isolation and analysis of exosomes and DNA in blood.
A) exosome isolation methods and their pros and cons. B) ctDNA and exoDNA analysis methods and their features.
2.1.3. Correlative studies
The study of cfDNA in pancreatic cancer started in the 1980s, when Shapiro et al. [24] first discovered a high level of cfDNA (>100 ng/ml) in 90% of pancreatic cancer patients without testing for ctDNA. Sorenson et al.[17] later performed PCR to detect KRAS mutations in the cfDNA of plasma from 3 pancreatic ductal adenocarcinoma (PDAC) patients. More recently, several groups have attempted to validate the clinical utility of mutation detection in the cfDNA of PDAC patients. For instance, Kinugasa et al. [21], using ddPCR, observed KRAS mutations in 62.5% of serum samples from 75 pancreatic cancer patients in all stages of disease and found that these mutations were correlated with worse overall survival (OS). Sausen et al. [25] applied ddPCR and identified KRAS mutations in the plasma ctDNA of 22 of 51 (43%) patients with early-stage pancreatic cancer. They also found that detectable postoperative ctDNA could predict recurrence an average of 6 months earlier than CT could. Using next-generation sequencing and ddPCR, Peitrasz et al. [26] found that detectable ctDNA was correlated with shorter disease-free survival in patients who had undergone surgery for resectable disease and with shorter OS in patients with advanced PDAC (Table 1).
Table 1.
Studies on detection of cancer associated components in blood of pancreatic cancer patients and related.
| Reference | Type of patients | Isolated component | Tests used | Rate of detection (of total patients) | Type of study |
|---|---|---|---|---|---|
| Sorenson et al.[17] | PDAC | cfDNA | PCR | N/A | KRAS mutation detection |
| Kinugasa et al[21]. | All stages | cfDNA | ddPCR | 62.5% had KRAS mutation | KRAS mutation detection |
| Sausen et al.[25] | Early stage | cfDNA | ddPCR | 43% had KRAS mutation | KRAS mutation detection |
| Tjensvoll et al.[27] | Advanced PDAC | ctDNA | PCR | 71% had KRAS mutation in treatment naïve | Longitudinal analysis |
| Cohen et al.[28] | Resectable PDAC |
ctDNA | PCR based Safe-Sequencing System | KRAS mutation detection and biomarkers (including CA19–9, CEA, HGF, OPN) | |
| Lee et al.[29] | Early stage pancreatic cancer patients | ctDNA | PCR based Safe-Sequencing System | 90.5% (22/37 pre And 13/35 post operation) had KRAS mutation | Pre- and post- operation |
| Allenson et al.[54] | Localized locally advanced metastatic PDAC | ctDNA exosomes | ddPCR | 46%, 31%, and 58%, respectively had KRAS mutation in ctDNA (KRAS) 67%, 80%, and 85%, respectively, had KRAS mutation in exoDNA |
Reliability of ctDNA and exoDNA |
| Yang et al.[55] | PDAC Chronic pancreatitis Intraductal papillary mucinous neoplasia |
exosomes | ddPCR | 40% had KRAS mutation 55% (5/9) 14% (1/7) |
KRAS and TP53 detection |
| Castillo et al.[46] | PDAC | exosomes | ddPCR and Liquid chromatography−mass spectrometry | 44% using UC (KRAS) 73 % using pulldown methods (KRAS) |
KRAS detection and Exosome surface proteins |
| Bernard et al.[56] | Localized and metastatic pancreatic cancer | ctDNA exosomes | ddPCR, Next Generation Sequencing | Refer to paper | Prognostic and predictive utility of mutant nucleic acids in patients under therapy, KRAS detection |
| Allard et al.[81] | Pancreatic cancer | CTCs | CellSearch | 37.5% (6/16) had CTCs | CTC detection based on antibodies |
| Nagrath et al.[72] | Pancreatic cancer | CTCs | CTC-Chip | 100 % had CTCs | CTC detection based on antibodies |
| Khoja et al.[84] | PDAC | CTCs | ISET CellSearch |
93% had CTCs 40% had CTCs |
CTC detection based on size |
| Effenberger et al.[86] | PDAC | CTCs | magnetic-activated cell sorting enrichment technology | 33% (23/69) had CTC | CTC detection based on antibodies |
| Kulemann et al.[88] | Localized and advanced PDAC | CTCs | ScreenCell filtration and ddPCR | 72% (44/58) had CTCs 42% (11/26) had KRAS mutation |
CTC detection based on size and genomic analysis of CTCs |
In regard to tumor monitoring, Tjensvoll et al. [27] performed a longitudinal analysis (median, 3.7 months; range, 0.6–12.9 months) of plasma ctDNA in 14 patients with advanced PDAC. In treatment-naïve patients, detectable ctDNA was associated with worse progression-free survival and OS. In addition, for 3 patients, ctDNA levels were closely correlated with therapy response as indicated by CA19–9 levels and CT findings. In a recent study, Cohen et al. [28] combined KRAS mutation detection in ctDNA and a library of cancer-associated protein biomarkers (including CA19–9, CEA, HGF, OPN) in assessing 221 patients with resectable PDAC and 182 healthy donors. Using the “Safe-Sequencing System”, an assay that incorporates molecular barcodes, the researchers could detect tumor-derived material with a sensitivity of 64.0% and specificity of 99.5% in patients with early-stage disease. Moreover, Lee et al.[29] in the same group used PCR based Safe Sequencing System assays to detect KRAS mutations from 42 patients using matched plasma samples prior and after operations from pancreatic cancer patients (in early stages). They found KRAS mutation in 38 patients (90.5%), with detected KRAS (i.e. ctDNA) in 23/37 patients’ prior operation and 13/35 after operation. The detection of ctDNA prior to operation was correlated with worse median recurrence free survival (RFS), 10.3 months versus not reached and after operation was 5.4 months versus 17.1 months. These data suggest ctDNA is a promising prognostic biomarker for early diagnosis of pancreatic cancer (Table 1).
2.2. Extracellular Vesicles and Exosomes
2.2.1. Background knowledge
Extracellular Vesicles (EVs) are another blood component that has proven to be biologically and clinically relevant. EVs include exosomes (30–150 nm; 1.10–1.11 g/ml), which have endosomal origins, and microvesicles (typically 50–500 nm but as large as 1.3 μm; 1.18–1.19 g/ml), which are shed from plasma membranes [30,31]. Exosomes are generated by the inward budding of multivesicular endosomes and the formation of intraluminal vesicles that are released after multivesicular endosomes attach to the plasma membrane, whereas microvesicles are shed through the direct outward budding of the plasma membrane [32,33]. EVs contain a milieu of cargo material that may include proteins, lipids, and nucleic acids (i.e., DNA and RNA components such as microRNA [miRNA], messenger RNA, and long non-coding RNA). EVs are released by all cell types and may serve as mechanisms of waste removal, cell-cell communication, and even the establishment of a pre-metastatic niche in cancer [32,34]. They are heterogeneous, differing in size, composition, morphology, and biogenesis, and generally are named based on these factors.
2.2.2. Technologies for isolation
In general, methods for isolating exosomes fall into 3 categories: density-based separation (ultracentrifugation or centrifugation using density solutions), size-based separation (including chromatography, dielectrophoresis, and filtration), and affinity-based separation (using antibody-coated magnetic beads or microfluidics). Although ultracentrifugation (Figure 2A), which is typically performed at 100,000× g, is time consuming and has a high initial infrastructure expense, it has relatively low long-term costs. Also, protocols for using ultracentrifugation to isolate exosomes vary widely; in tracking 818 experiments, EV-TRACK (a consortium for EV-transparent reporting and centralizing knowledge) found 218 unique combinations of centrifugation steps and pelleting times [35]. The high speeds used in ultracentrifugation may damage exosomes, resulting in the release of contaminating proteins. This release may be resolved by isolating exosomes with a sucrose gradient, although this technique can yield high-density lipoproteins and exosomes of similar densities [36].
Size-based methods of EV separation include size exclusion chromatography (SEC) (Figure 2D), which is achieved with tools such as qEV isolation columns (Izon science), which can fractionate exosomes [33,36]. In addition, SEC can separate EVs from contaminating extracellular proteins, resulting in a more purified fraction [37]; however, since the resulting eluate is diluted, SEC must be followed by an ultracentrifugation step to concentrate the EVs. Another size-based method involves using membranes with nanometer pore sizes to filter EVs from larger particles [38–40]. However, clogging can be an issue, and the membranes may have to be changed several times. Davies et al. [41] developed a microfluidic device equipped with integrated membranes; in that device, electrophoretic forces instead of pressure are applied as the driving force. EV separation can also be achieved by dielectrophoresis, a physical property dependent on membrane rigidity and exosome size. One recently introduced microfluidic device, an alternating current electrokinetic (ACE) microarray device, uses alternating current to generate dielectrophoresis on a microarray, enabling the separation of exosomes from small volumes (30–50 μL) of plasma[42].
Affinity-based approaches for exosome isolation may involve microfluidic devices and/or magnetic or aldehyde/sulfate latex beads that can be coated with antibodies targeting proteins of interest. Antibodies against tetraspanins (e.g., CD9, CD63, CD81) have been used for general exosome isolation [43–45]. However, more specific antibodies believed to have the ability to isolate cancer-specific exosomes have been used for early detection and tumor monitoring in patients with low disease volumes [46,47]. Compared with the exosome isolation approaches described above, affinity-based methods typically result in high-purity samples but a lower total sample yield. For instance, Reaátegui et al [47] introduced a herringbone pattern microfluidic device (EVHB-Chip) with special nano-coating prior to antibody coatings (specific to glioblastoma, such as EGFR, EGFRvIII, ephA2, podoplanin, PDGFR, and MCAM) that enables the sensitive isolation of tumor associated EV-RNA within 3 hours (100 EVs/μl). Also, using a temperature-based approach by Gelatin coating, they were able to dissociate the captured EVs for downstream analysis. Other specialized methods include the use of an external field, such as an acoustic wavelength, to separate exosomes [48]. Also, Zhang et al.[49*] established a 3 dimensional nanopattern herringbone chip coated with anti-CD81, CD63 and EpCAM (mostly with anti-CD81 outperforming other antibodies for ovarian cancer in their study) that can capture exosomes with a high sensitivity of 10 exosomes per μl.
2.2.3. Correlative studies
Pancreatic cancer-associated exosomes have been used as biomarkers for early disease detection and tumor monitoring. For instance, Melo et al. [50], using mass spectrometry to assess cancer cell-derived exosomes and flow cytometry to assess serum from 251 patients, identified a surface proteoglycan, glypican-1, whose specific expression on cancer-associated exosomes had 100% specificity and sensitivity in distinguishing early- and late-stage pancreatic cancer from healthy tissue and benign pancreatic disease. Although this observation has been confirmed by other studies [51,52], Lai et al. [53], in a cohort of 29 patients with pancreatic cancer and 11 patients with chronic pancreatitis, found that glypican-1 lacked reliable diagnostic capability. This observation was further supported by Castillo et al. [46], who found that glypican-1 was expressed on both normal tissues and pancreatic cancer cell lines and thus not diagnostically valuable. However, these discrepancies might be due to differences in the detection assay, patient cohorts, and/or the available antibodies targeting the proteins of interest.
Exosomes also provide a substrate for the molecular profiling of circulating nucleic acids such as exosomal DNA (exoDNA) and exosomal RNA [30]. For instance, hypothesizing that exoDNA can be used as a biomarker for early PDAC detection, Allenson et al. [54] found that ultrasensitive droplet digital PCR (ddPCR) of exoDNA had higher rates of KRAS mutation detection (67%, 80%, and 85%) than did ddPCR of paired ctDNA (46%, 31%, and 58%) in patients with localized, locally advanced, and metastatic PDAC, respectively. This high rate of exosomal KRAS was correlated with lower disease-free survival in these patients. In another study, Yang et al. [55] used ddPCR to detect KRAS and TP53 mutations in patients with PDAC (40% and 4%, respectively), chronic pancreatitis (5/9 KRAS and no TP53), and intraductal papillary mucinous neoplasias (2/7 KRAS and 1/7 both). Both of these studies found circulating mutant molecules in the apparently healthy population, which suggests that there is a “baseline” rate of circulating mutant molecules that is present in the aging population, either from clonal hematopoiesis or possibly, an occult neoplasm (Table 1).
Recently, Castillo et al. [46] performed proteomic and genomic profiling of pancreatic cancer-derived exosomes. Liquid chromatography-mass spectrometry of exosomes derived from pancreatic cancer cell lines and control lines, including normal ductal and fibroblast cells, revealed that PDAC-derived exosomes specifically express 6 surface proteins: CLDN4, EpCAM, CD151, LGALS3BP, HIST2H2BF, and HIST2H2BE. Subjecting 173 samples from 103 PDAC patients to ddPCR, the authors detected KRAS mutations in the exosomes of 44% of patients using ultracentrifugation (UC) versus in 73% of cases using a pull-down method with antibodies against these surface proteins as “bait” for exosome separation. Thus, the exosome pull-down method can be used to enhance the sensitivity of mutation detection, especially in low volume setting, such as detecting minimal residual disease.
Bernard et al.[56] recently reported on interrogating either exoDNA or ctDNA, in a large series of 425 plasma samples from 194 patients with localized or metastatic pancreatic cancer. The authors demonstrated both the prognostic and predictive utility of measuring mutant nucleic acid in pancreatic cancer liquid biopsies. Specifically, they found that in treatment-naïve patients, the detection of ctDNA or an exoDNA KRAS mutation allele frequency (MAF) >5% was significantly correlated with progression-free survival and overall survival. In a cohort of 34 patients with metastatic pancreatic cancer that underwent longitudinal disease monitoring with liquid biopsies, an exoDNA KRAS MAF peak of 1% predicted disease progression with a median of 50 days earlier than radiological progression-based CT scan (P=0.0003). Further, in a separate cohort of 34 patients undergoing neoadjuvant therapy, exoDNA KRAS MAF kinetics before and after the completion of therapy was predictive of progressive disease and thus eligibility for surgical resection (P=0.003) (table 1).
2.3. Circulating tumor cells (CTCs)
2.3.1. Background knowledge
First reported in 1869 [57], CTCs are another cancer related blood component whose biopsy is clinically useful. Thought to be critical in the metastasis cascade [58], CTCs are rare, accounting for approximately 1 of every 1 × 108 blood cells; thus, their detection and isolation remains a challenge [59]. Many groups have tried to isolate and study the role of CTCs in prediction of cancer progression and metastasis, monitoring effectiveness of drug treatment, and evaluation as a surrogate biomarker in clinical trials [60]. CTCs can be 2–4 times the size of capillary pores and can be found as single cells or clusters [61]. Larger CTCs and clusters[62,63] may become trapped in capillaries, whereas smaller ones likely remain in peripheral blood circulation. Thus, the number of CTCs may vary temporally and spatially, and whether isolated CTCs are the true source of systemic metastasis remains unclear [60]. This issue is exacerbated in pancreatic cancer, as blood enters the liver through the portal vein immediately after exiting the pancreas, large CTCs and clusters could become trapped. Thus, peripheral blood CTCs might not be the optimum choice for the diagnosis and prognosis of pancreatic cancers, whereas portal vein samples [64,65] might provide a better representation of the CTC population.
CTCs are not a monolithic entity, and are remarkably heterogeneous. For example, these cells can undergo epithelial-mesenchymal transition (leading to loss of surface epithelial markers), and a subset demonstrate immunophenotypic features consistent with cancer stem cells [60], all of which make it challenging to isolate CTCs with optimal specificity and sensitivity. Moreover, the mere detection of CTCs does not always imply the presence of an invasive cancer. For instance, they have been found in mouse models of pancreatic cancer before the emergence of invasive primary tumors [66] and in 7 of 21 patients with pancreatic cystic lesions without evidence of malignant disease [67]. However, CTCs still offer promising clinical outcomes in early detection, survival prediction, and treatment monitoring for many cancers [68]. Their potential clinical relevance revolves in their ability to be profiled through in-depth genomic and molecular analyses of DNA, RNA, and proteins using emerging technologies [69–71]; and the culture of CTCs in mice or ex vivo for developing “living” preclinical models [70].
2.3.2. Technologies for isolation
Technologies for isolating CTCs vary in concept and outcome. The only technology approved by the U.S. Food and Drug Administration is CellSearch® (Menarini Silicon Biosystems Inc, Huntington Valley, PA), an affinity-based technology (works based on antibody coatings) that uses magnetic beads covered with anti-EpCAM, anti-cytokeratin (for epithelial phenotypes of CTCs), and anti-CD45 (for eliminating contaminating leukocytes). Other affinity-based technologies, such as the so-called CTC-chip[72] and herringbone chip[73] typically use microfluidics-based principles for isolation. The microfluidics coating of the herringbone chip has been expanded beyond epithelial phenotypes for CTC capture, and now includes mesenchymal, stem cell, or other tumor-specific markers[73].
Size-based separation is another type of CTC isolation. For instance, Isolation by size of tumor cells (ISET®) (Rarecells DIAGNOSTICS, Paris, France) and ScreenCell® (Paris, France) use filtration to directly isolate large CTCs. A series of microfluidic chips use inertial focusing and vortex, which is also size- and deformability-dependent, to isolate CTCs [74]. Technologies such as ApoStream® (Precision for Medicine, Texas, USA)[75] and DEPArray™ (Castel Maggiore, Italy) use dielectrophoresis to separate CTCs. Some microfluidic chips such as CTC-iChip[76] and NanoVelcro[77] take advantage of both size and affinity to separate CTCs with higher purity and specificity. One advantage that size-based technologies have over affinity-based technologies is the capability to separate all phenotypes, including epithelial and mesenchymal phenotypes, without discrimination [78]. However, all these technologies must be followed with immunohistochemistry, immunocytochemistry, or immunofluorescence labeling to distinguish CTCs from leukocytes. Genomic analysis with RT-PCR also has been used to reliably stratify CTCs in the captured populations [79]. Very recently, Kim et al[80**]. established an intravascular aphaeretic CTC isolation system with a microfluidic herringbone graphene oxide chip (HBGO) coated with anti-EpCAM that can be indwelled intravascularly on a patient for several hours and detect CTCs from peripheral blood in real time.
2.3.3. Correlative studies
Using CellSearch, Allard et al. [81] were the first to attempt to capture CTCs in pancreatic cancer patients. The authors tried to capture CTCs in 964 patients with all types of solid carcinomas; however, CTCs were detectable in only 6 of 16 (37.5%) pancreatic cancer patients, underscoring the challenges of using CTCs in this tumor type. Other groups that used CellSearch to detect pancreatic cancer CTCs also reported low detection rates (10–50%) [65,82–85]. However, using affinity-based technology, Nagrath et al. [72] built the CTC-Chip, which detected cytokeratin+/CD45- CTCs in 15 out of 15 PDAC patients with a median detection rate of approximately 120 cells/ml (range, 9–832 cells/ml). Also, Khoja et al. [84] reported that ISET detected CTCs in 93% of PDAC patients (n=27) whereas CellSearch detected CTCs in only 40% of patients (n=53) (Table 1).
Other groups that used CellSearch were able to discern direct clinical outcomes from CTC detection and quantification, albeit in relatively modestly sized single institution studies. For instance, Kurihara et al. [82] found that CTC positivity (CTC>1) conveyed worse OS in a cohort of 26 PDAC patients. Also, Bidard et al. [83], in a study of 79 PDAC patients, observed poor tumor differentiation and worse OS among patients with CTCs. Okubo et al. [85], in a recent study of 65 patients with advanced pancreatic cancer, found a correlation between CTC positivity and worse OS among 56 patients with unresectable disease and a higher rate of CTC positivity among patients with liver metastasis. Other studies have shown that low numbers of detected epithelial CTCs in patients are also clinically relevant. Effenberger et al. [86] used magnetic-activated cell sorting enrichment technology (anti-cytokeratin/anti-EpCAM that only captures epithelial phenotypes) followed by immunocytochemical staining with anti-cytokeratin/anti-CD45 and DAPI. They found that 23 of 69 pancreatic cancer patients had detectable CTCs per sample and that CTC positivity was associated with worse progression-free survival and OS (Table 1).
The analysis of isolated pancreatic cancer CTCs is not limited to using immunohistochemistry, immunocytochemistry, and/or immunofluorescence to assess phenotypic markers. Some studies have found that genomic alterations in pancreatic cancer CTCs have clinical relevance. For instance, Kulemann et al. [87] used ScreenCell filtration to isolate CTCs from 58 PDAC patients and used ddPCR to detect KRAS mutations in CTCs of 42/58 patients. For comparison, they also collected hematological cells, original tumor biopsy specimens, and healthy pancreatic tissue as a control. They reported that 42% (11 of 26 patients) of the solid tissues and matched CTCs had a discordant KRAS mutation status. The genomic study of CTCs is still in its infancy, and much more investigation with multiple assays and larger cohorts is needed to elucidate the genomic landscape of pancreatic cancer CTCs (Table 1).
3. Conclusion
Liquid biopsy is a powerful tool for the non-invasive, early diagnosis of many solid cancers. Here, we reviewed the currently available and emerging technologies for isolating tumor-associated circulating components such as ctDNA, EVs, exosomes, and CTCs from blood, focusing on methods that have demonstrated high sensitivity and specificity. We discussed published data that used these technologies in the context of pancreatic cancer, with specific relevance of the findings in the context of diagnostic, prognostic, and predictive utility of liquid biopsies. The current technologies are often time consuming, labor intensive, and potentially costly. The hope in the field is to make liquid biopsy a standard of care in the early diagnosis of pancreatic cancer, as well as for longitudinally monitoring the impact of therapy in established disease.
4. Expert opinion
In this review, we have discussed the application of liquid biopsies to pancreatic cancer, with a focus on three distinct compartments (ctDNA, exosomes and CTCs), and the prevalent technologies that have been reported in the context of isolating relevant reagents from each of these compartments for assessing the underlying tumor. As the field has evolved from the academic realm to application in patients (through implementation within a so-called “Clinical Laboratory Improvement Amendments” or CLIA environment), we have reached a stage where each technology has to be assessed through the prism of reproducibility, repeatability, feasibility and cost effectiveness. It takes on average ten years for a therapeutic to be approved for an oncology indication, after being tested in hundreds of patients in randomized clinical trials, where significant demonstration of efficacy over current standard has been shown. For liquid biopsy tests, and many laboratory tests in general, similar levels of stringency have simply not been applied even when the tests are offered commercially, or through an academic environment, within a CLIA setting. It is imperative that the oncology community is fully aware of the challenges and limitations inherent to each technology, such that an informed decision can be made about how best to leverage the potential of liquid biopsies for pancreatic cancer.
In the field of liquid biopsy at large, and specifically in the application to pancreatic cancer, there are challenges, both technical and biological. For instance, for the isolation of exosomes, density-based technologies, such as ultracentrifugation, are time-consuming procedures, with a timeframe that varies between 2–20 hours. Further, the high speed of centrifugation might lead to exosome damage and release of contaminating proteins inside the pellet. Also, size-based technologies such as chromatography and filtration provide highly contaminated solutions with low specificity and concentration of exosomes in the eluates, making these technologies insufficient for stand-alone isolation of exosomes, limiting their use mostly as preparatory steps prior to ultracentrifugation. Moreover, both of these technologies lack in the ability to isolate only tumor-associated exosomes, and typically extract both non-tumor and tumor-associated exosomes. The affinity-based technologies have shown some promising results for isolation of exosomes with high specificity and purity, however their reliance on antibodies against exosomal surface proteins and the lack of a universal “bait” protein causes these technologies to remain within the academic purview, and far from clinical applications. Recently, new approaches to use a cocktail of tumor-associated antibodies [46,47] have garnered some enthusiasm for clinical translation. Ultimately, these methods have to yield exosomes that are of sufficient quantity and quality required to perform reliable downstream assessments, such as targeted next generation sequencing (NGS) assays within a CLIA environment, in order to interrogate the molecular landscapes of the tumor from which these exosomes have been released.
Other tumor-associated components from liquid biopsies also suffer from inherent limitations of processing and analyses. For instance, ctDNA can be relatively low abundant in post-surgery samples, and can vary depending on the time of sample retrieval (days or hours before or after surgery) and method of storage (frozen vs. fresh sample). Also, the harsh conditions of sample transportation from clinics to laboratory, their storage or isolation process (the chemical used for cytogenesis) can cause DNA fragmentation/loss and/or new release of DNA from the blood cell residues in the sample, leading to spurious mutation calls. To circumvent these issues, all of these parameters have to be considered before interpretation of the data and extrapolating to parameters such as tumor load or treatment responses. Finally, in the context of CTCs, the number of circulating tumor cells is typically low (1–100 cells/ml blood), an issue that is exacerbated in pancreatic cancer, due to the location of pancreas and drainage of its blood through the portal vein first into the liver and then the lung. This causes the lodging and filtration of some CTCs (usually the larger or more stiff ones), inside the liver and lung and underestimation of the number of CTCs in the peripheral blood. The use of the portal vein samples [64,65] and development of technologies such as the temporary vascular indwelled aphaeretic CTC device developed recently [80] might circumvent this issue and obtain a more realistic number of CTCs in pancreatic cancer for downstream molecular assessments.
In summary, we strongly believe that, despite the aforementioned technical or biological challenges, in a disease like pancreatic cancer where tissue samples are limited and repeated invasive tissue biopsies are expensive or infeasible, liquid biopsies provide an important avenue for tumor diagnosis, molecular stratification, and treatment monitoring. As input requirements for profiling assays diminish, the molecular assessment of liquid biopsy derivatives will become increasingly more sophisticated. While none of the isolation and analysis methods have gained a widespread clinical acceptance, it is imperative that clinical applications of liquid biopsies factor the advantages and limitations of each assay platform into consideration.
Article highlights.
Standard clinical methods for diagnosing pancreatic cancer include assessment of tissue biopsies and aspiration cytology specimens, and non-invasive screening methods (e.g., computed tomography [CT], magnetic resonance imaging, endoscopic ultrasonography [EUS]). However, these are typically used in symptomatic patients and not in the context of early detection.
Liquid biopsies involve isolation of tumor-released components from bodily fluids, including circulating tumor DNA (ctDNA), extracellular vesicles (EVs), and circulating tumor cells (CTCs), which can be beneficial for diagnosis and prognosis of PDAC.
Methods for isolating EVs and CTCs include size-, density-, affinity-, or external field-based separation principles.
Droplet digital polymerase chain reaction (ddPCR) and next generation sequencing (NGS) for DNA using targeted panels are the two most commonly used methods for assessment of the mutational events in liquid biopsies.
Many challenges exist for reliable isolation of tumor associated components such as circulating tumor cells and EVs from blood of pancreatic cancer patients.
Funding
This paper was supported by the Cancer Prevention and Research Institute of Texas research programme (RP140106 and RP170067) and the National Cancer Institute (T32CA217789–03, U54CA096297, U01CA196403, and U01CA200).
Footnotes
Declaration of interest
Anirban Maitra has received royalties from Cosmos Wisdom Biotechnology Company LTD for being a co-inventor on a license related to pancreatic cancer early detection, and this financial relationship is managed by the MD Anderson Conflict of Interest Committee. Editorial assistance was provided by Joseph Munch, Department of Scientific Publications, UT MD Anderson Cancer Center. There are no other conflicts of interests to declare. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Ionescu-Tirgoviste C, Gagniuc PA, Gubceac E, et al. A 3D map of the islet routes throughout the healthy human pancreas. Sci Rep. 2015. Sep 29;5:14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019. January;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 3.Gillen S, Schuster T, Meyer Zum Buschenfelde C, et al. Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010. Apr 20;7(4):e1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet. 2016. July 2;388(10039):73–85. [DOI] [PubMed] [Google Scholar]
- 5.Chan A, Prassas I, Dimitromanolakis A, et al. Validation of biomarkers that complement CA19.9 in detecting early pancreatic cancer. Clin Cancer Res. 2014. Nov 15;20(22):5787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016. April 21;2:16022. [DOI] [PubMed] [Google Scholar]
- 7.Tang S, Huang G, Liu J, et al. Usefulness of 18F-FDG PET, combined FDG-PET/CT and EUS in diagnosing primary pancreatic carcinoma: a meta-analysis. Eur J Radiol. 2011. Apr;78(1):142–50. [DOI] [PubMed] [Google Scholar]
- 8.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013. August;10(8):472–84. [DOI] [PubMed] [Google Scholar]
- 9.Glantz MJ, Cole BF, Glantz LK, et al. Cerebrospinal fluid cytology in patients with cancer: minimizing false-negative results. Cancer. 1998. February 15;82(4):733–9. [DOI] [PubMed] [Google Scholar]
- 10.Gorgannezhad L, Umer M, Islam MN, et al. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018. Apr 17;18(8):1174–1196. [DOI] [PubMed] [Google Scholar]
- 11.Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 1975. September;35(9):2375–82. [PubMed] [Google Scholar]
- 12.Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001. Nov;313(1–2):139–42. [DOI] [PubMed] [Google Scholar]
- 13.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011. June;11(6):426–37. [DOI] [PubMed] [Google Scholar]
- 14.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001. Feb 15;61(4):1659–65. [PubMed] [Google Scholar]
- 15.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005. Nov 8;102(45):16368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]; • [One of the pioneering studies establishing the value of ctDNA detection in cancer patients]
- 16.Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010. Oct;38(18):6159–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorenson GD, Pribish DM, Valone FH, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994. Jan-Feb;3(1):67–71. [PubMed] [Google Scholar]
- 18.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017. Apr 26;545(7655):446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014. May 15;20(10):2643–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008. Sep;14(9):985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinugasa H, Nouso K, Miyahara K, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015. Jul 1;121(13):2271–80. [DOI] [PubMed] [Google Scholar]
- 22.Jiang P, Lo YMD. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet. 2016. Jun;32(6):360–371. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q, Zheng J, Qing Z, et al. Detection of Circulating Tumor DNA in Human Blood via DNA-Mediated Surface-Enhanced Raman Spectroscopy of Single-Walled Carbon Nanotubes. Anal Chem. 2016. May 3;88(9):4759–65. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro B, Chakrabarty M, Cohn EM, et al. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983. Jun 1;51(11):2116–20. [DOI] [PubMed] [Google Scholar]
- 25.Sausen M, Phallen J, Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015. Jul 7;6:7686. [DOI] [PMC free article] [PubMed] [Google Scholar]; • [An early comprehensive study demonstrating the implications of ctDNA monitoring in pancreatic cancer patients]
- 26.Pietrasz D, Pecuchet N, Garlan F, et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin Cancer Res. 2017. Jan 1;23(1):116–123. [DOI] [PubMed] [Google Scholar]
- 27.Tjensvoll K, Lapin M, Buhl T, et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol Oncol. 2016. Apr;10(4):635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen JD, Javed AA, Thoburn C, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci U S A. 2017. Sep 19;114(38):10202–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• [The seminal paper describing the CancerSEEK assay that combines ctDNA with multianalyte proteins]
- 29.Lee B, Lipton LR, Cohen J, et al. Circulating tumor DNA as a prognostic biomarker in early stage pancreatic cancer. Journal of Clinical Oncology. 2018;36(15_suppl):e16206–e16206. [Google Scholar]
- 30.Xu R, Rai A, Chen M, et al. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018. May 23. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Zhang S, Yao J, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015. Nov 5;527(7576):100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018. Apr;19(4):213–228. [DOI] [PubMed] [Google Scholar]
- 33.Witwer KW, Buzas EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017. Mar;27(3):172–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium E-T, Van Deun J, Mestdagh P, et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017. Feb 28;14(3):228–232. [DOI] [PubMed] [Google Scholar]
- 36.Pariset E, Agache V, Millet A. Extracellular Vesicles: Isolation Methods. Advanced Biosystems. 2017;1(5):1700040. [DOI] [PubMed] [Google Scholar]
- 37.Coumans FA, van der Pol E, Boing AN, et al. Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J Extracell Vesicles. 2014;3:25922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo HK, Sunkara V, Park J, et al. Exodisc for Rapid, Size-Selective, and Efficient Isolation and Analysis of Nanoscale Extracellular Vesicles from Biological Samples. ACS Nano. 2017. Feb 28;11(2):1360–1370. [DOI] [PubMed] [Google Scholar]
- 39.Liu F, Vermesh O, Mani V, et al. The Exosome Total Isolation Chip. ACS Nano. 2017. Nov 28;11(11):10712–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant R, Ansa-Addo E, Stratton D, et al. A filtration-based protocol to isolate human plasma membrane-derived vesicles and exosomes from blood plasma. J Immunol Methods. 2011. Aug 31;371(1–2):143–51. [DOI] [PubMed] [Google Scholar]
- 41.Davies RT, Kim J, Jang SC, et al. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. 2012. Dec 21;12(24):5202–10. [DOI] [PubMed] [Google Scholar]
- 42.Ibsen SD, Wright J, Lewis JM, et al. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano. 2017. Jul 25;11(7):6641–6651. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, Skog J, Hsu CH, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010. Feb 21;10(4):505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanwar SS, Dunlay CJ, Simeone DM, et al. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014. Jun 7;14(11):1891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ko J, Bhagwat N, Yee SS, et al. Combining Machine Learning and Nanofluidic Technology To Diagnose Pancreatic Cancer Using Exosomes. ACS Nano. 2017. Nov 28;11(11):11182–11193. [DOI] [PubMed] [Google Scholar]
- 46.Castillo J, Bernard V, San Lucas FA, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol. 2018. Jan 1;29(1):223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]; • [Original study describing the compendium of surface proteins on extracellular vesicles in pancreatic cancer patients that can be used for capture and enrichment of cancer specific exosome for genomic studies]
- 47.Reategui E, van der Vos KE, Lai CP, et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat Commun. 2018. Jan 12;9(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu M, Ouyang Y, Wang Z, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci U S A. 2017. Oct 3;114(40):10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang P, Zhou X, He M, et al. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nature Biomedical Engineering. 2019 2019/02/25. [DOI] [PMC free article] [PubMed] [Google Scholar]; • [This paper introduces an ultrasensitive device for isolation of exosomes from low amount of plasma (~100 μl). This device will be potentially usable for pediatric patients children or for preclinical studies given that the volume of blood draw is limited]
- 50.Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015. Jul 9;523(7559):177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu J, Sheng Y, Kwak KJ, et al. A signal-amplifiable biochip quantifies extracellular vesicle-associated RNAs for early cancer detection. Nat Commun. 2017. Nov 22;8(1):1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis JM, Vyas AD, Qiu Y, et al. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. ACS Nano. 2018. Apr 24;12(4):3311–3320. [DOI] [PubMed] [Google Scholar]
- 53.Lai X, Wang M, McElyea SD, et al. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017. May 1;393:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017. Apr 1;28(4):741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang S, Che SP, Kurywchak P, et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol Ther. 2017. Mar 4;18(3):158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernard V, Kim DU, San Lucas FA, et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology. 2019. Jan;156(1):108–118 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.T. A. A Case of Cancer in Which Cells Similar to Those in the Tumors Were Seen in the Blood after Death. Australasian Medical Journal. 1869;14:146–149. [Google Scholar]
- 58.O’Flaherty JD, Gray S, Richard D, et al. Circulating tumour cells, their role in metastasis and their clinical utility in lung cancer. Lung Cancer. 2012. 4//;76(1):19–25. [DOI] [PubMed] [Google Scholar]
- 59.DiPardo BJ, Winograd P, Court CM, et al. Pancreatic cancer circulating tumor cells: applications for personalized oncology. Expert Rev Mol Diagn. 2018. Sep;18(9):809–820. [DOI] [PubMed] [Google Scholar]
- 60.Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science. 2013. Sep 13;341(6151):1186–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011. Mar 25;331(6024):1559–64. [DOI] [PubMed] [Google Scholar]
- 62.Kamyabi N, Huang J, Lee JJ, et al. A microfluidic device for label-free isolation of tumor cell clusters from unprocessed blood samples. Biomicrofluidics. 2019. Jul;13(4):044111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabisiewicz A, Grzybowska E. CTC clusters in cancer progression and metastasis. Med Oncol. 2017. Jan;34(1):12. [DOI] [PubMed] [Google Scholar]
- 64.Arnoletti JP, Fanaian N, Reza J, et al. Pancreatic and bile duct cancer circulating tumor cells (CTC) form immune-resistant multi-cell type clusters in the portal venous circulation. Cancer Biol Ther. 2018. Aug 1:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gall TM, Jacob J, Frampton AE, et al. Reduced dissemination of circulating tumor cells with no-touch isolation surgical technique in patients with pancreatic cancer. JAMA Surg. 2014. May;149(5):482–5. [DOI] [PubMed] [Google Scholar]
- 66.Rhim AD, Mirek ET, Aiello NM, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012. Jan 20;148(1–2):349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhim AD, Thege FI, Santana SM, et al. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology. 2014. Mar;146(3):647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013. Jan;59(1):110–8. [DOI] [PubMed] [Google Scholar]
- 69.Blassl C, Kuhlmann JD, Webers A, et al. Gene expression profiling of single circulating tumor cells in ovarian cancer - Establishment of a multi-marker gene panel. Mol Oncol. 2016. Aug;10(7):1030–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu M, Bardia A, Aceto N, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014. Jul 11;345(6193):216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu M, Ting DT, Stott SL, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012. Jul 26;487(7408):510–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007. Dec 20;450(7173):1235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stott SL, Hsu CH, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010. Oct 26;107(43):18392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khojah R, Stoutamore R, Di Carlo D. Size-tunable microvortex capture of rare cells. Lab Chip. 2017. Jul 25;17(15):2542–2549. [DOI] [PubMed] [Google Scholar]
- 75.O’Shannessy DJ, Davis DW, Anderes K, et al. Isolation of Circulating Tumor Cells from Multiple Epithelial Cancers with ApoStream((R)) for Detecting (or Monitoring) the Expression of Folate Receptor Alpha. Biomark Insights. 2016;11:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013. Apr 3;5(179):179ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jan YJ, Chen JF, Zhu Y, et al. NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Adv Drug Deliv Rev. 2018. Feb 1;125:78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poruk KE, Valero V 3rd, Saunders T, et al. Circulating Tumor Cell Phenotype Predicts Recurrence and Survival in Pancreatic Adenocarcinoma. Ann Surg. 2016. Dec;264(6):1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Albuquerque A, Kubisch I, Breier G, et al. Multimarker gene analysis of circulating tumor cells in pancreatic cancer patients: a feasibility study. Oncology. 2012;82(1):3–10. [DOI] [PubMed] [Google Scholar]
- 80.Kim TH, Wang Y, Oliver CR, et al. A temporary indwelling intravascular aphaeretic system for in vivo enrichment of circulating tumor cells. Nat Commun. 2019. Apr 1;10(1):1478. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• [First manuscript to introduce a device for real time monitoring of circulating tumor cells by intravenously implanting the device on patients]
- 81.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004. Oct 15;10(20):6897–904. [DOI] [PubMed] [Google Scholar]
- 82.Kurihara T, Itoi T, Sofuni A, et al. Detection of circulating tumor cells in patients with pancreatic cancer: a preliminary result. J Hepatobiliary Pancreat Surg. 2008;15(2):189–95. [DOI] [PubMed] [Google Scholar]
- 83.Bidard FC, Huguet F, Louvet C, et al. Circulating tumor cells in locally advanced pancreatic adenocarcinoma: the ancillary CirCe 07 study to the LAP 07 trial. Ann Oncol. 2013. Aug;24(8):2057–61. [DOI] [PubMed] [Google Scholar]
- 84.Khoja L, Backen A, Sloane R, et al. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. Br J Cancer. 2012. Jan 31;106(3):508–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okubo K, Uenosono Y, Arigami T, et al. Clinical impact of circulating tumor cells and therapy response in pancreatic cancer. Eur J Surg Oncol. 2017. Jun;43(6):1050–1055. [DOI] [PubMed] [Google Scholar]
- 86.Effenberger KE, Schroeder C, Hanssen A, et al. Improved Risk Stratification by Circulating Tumor Cell Counts in Pancreatic Cancer. Clin Cancer Res. 2018. Jun 15;24(12):2844–2850. [DOI] [PubMed] [Google Scholar]
- 87.Kulemann B, Rosch S, Seifert S, et al. Pancreatic cancer: Circulating Tumor Cells and Primary Tumors show Heterogeneous KRAS Mutations. Sci Rep. 2017. Jul 3;7(1):4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kulemann B, Pitman MB, Liss AS, et al. Circulating tumor cells found in patients with localized and advanced pancreatic cancer. Pancreas. 2015. May;44(4):547–50. [DOI] [PubMed] [Google Scholar]