Abstract
Drosophila oogenesis is a powerful model for the study of numerous questions in cell and developmental biology. In addition to its longstanding value as a genetically tractable model of organogenesis, recently it has emerged as an excellent system in which to combine genetics and live imaging. Rapidly improving ex vivo culture conditions, new fluorescent biosensors and photo-manipulation tools, and advances in microscopy have allowed direct observation in real time of processes such as stem cell self-renewal, collective cell migration, and polarized mRNA and protein transport. In addition, entirely new phenomena have been discovered, including revolution of the follicle within the basement membrane and oscillating assembly and disassembly of myosin on a polarized actin network, both of which contribute to elongating this tissue. This review focuses on recent advances in live-cell imaging techniques and the biological insights gleaned from live imaging of egg chamber development.
Introduction
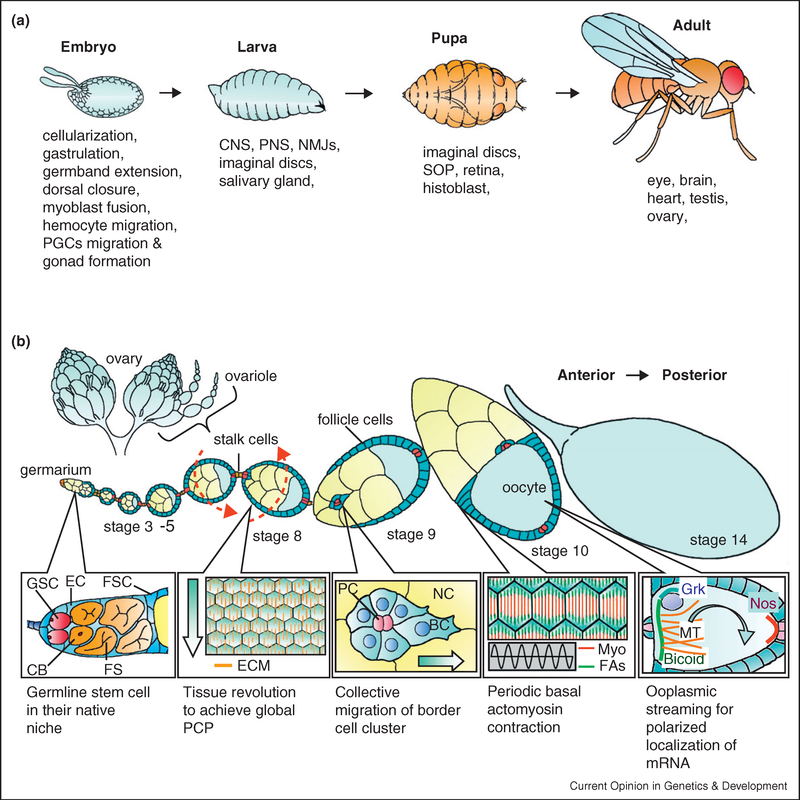
The development of organs and organisms is a dynamic process, a complete understanding of which requires studying living tissue with the highest possible spatial and temporal resolution. The combination of improved culture systems, light-sensitive proteins, and imaging techniques has revolutionized developmental studies over the past decade. Analysis of mutant phenotypes need no longer be limited to end-point evaluation of developmental failure; now investigators can observe how the end result comes about, by monitoring the dynamic behavior of cells and molecules. The ever-expanding arsenal of genetically encoded biosensors and caged proteins further provides opportunities to both monitor and manipulate biological processes in real time. Besides being a renowned genetic model for development and disease, Drosophila melanogaster is becoming more and more amenable to live imaging, as culture conditions are defined that support ex vivo development of larval and adult tissues, most notably the ovary. A few examples of developmental processes studied by live-cell imaging are shown in Figure 1 [1–8]. Here we will review the novel subcellular, cellular, and multicellular dynamics that have been discovered by live imaging studies of egg chamber development in the Drosophila ovary.
Figure 1.
(a) Major stages of the Drosophila life cycle with the tissues and developmental events studied by live-cell imaging listed below. (b) Anatomy of fruit fly ovary and expanded view of egg chambers in a single ovariole. Germline stem cell self-renewal, follicle rotation, border cell migration, periodic actomyosin contraction, and polarized mRNA localization are further illustrated below. Arrows indicate the direction of movement (PGC: primordial germ cell; CNS: central nervous system; PNS: peripheral nervous system; NMJs: neuromuscular junctions; SOP: sensory organ precursor; GSC: germline stem cell; CB: cystoblast; EC: escort cell; FSC: follicle stem cell; FS: fusome; ECM: extracellular matrix; PC: polar cell; BC: border cell; NC: nurse cell; Myo: myosin; FAs: focal adhesion; MT: microtubule; Nos: nanos; Grk: gurken; and PCP: planar cell polarity).
The generation and development of egg chambers provides a good model for the study of a great spectrum of biological processes required for organogenesis in general, including self-renewal of adult stem cells, cell differentiation, pattern formation, axis specification, cell shape change and migration, tissue elongation, cytoskeleton dynamics, RNA biogenesis, transport, localization and function, and even tumorigenesis. Mosaic analysis and RNAi-mediated gene knockdown are highly effective in this tissue that, unlike the embryo, lacks significant maternally provided RNA or protein. The ovary is also readily permeable to drugs and even molecules as large as antibodies, which can diffuse between cell-cell junctions even in living organs. Direct injection of material into the germline cytosol before live imaging has also been successful. Therefore, ovarian development is not only genetically tractable, but also accessible to various treatments that are more commonly used in cell culture. Yet by maintaining the tissue intact, processes that depend upon ensembles of cells and interactions of multiple cell types, which are impossible to study in simple cell culture, can be observed and manipulated.
A brief introduction to the anatomy of the Drosophila ovary
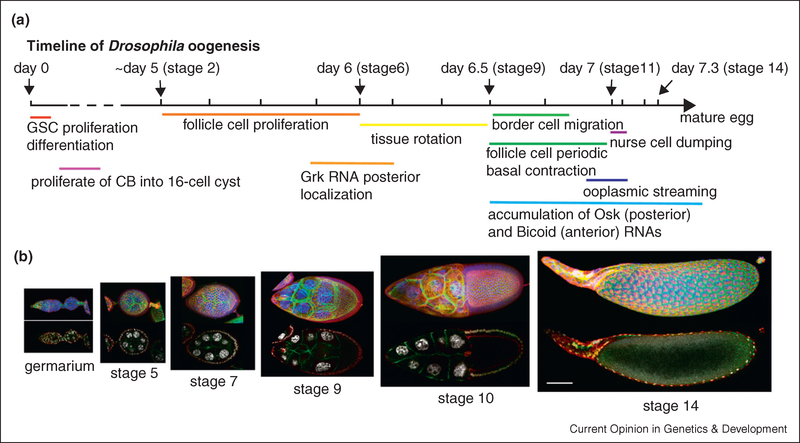
Female flies possess a pair of ovaries, each of which is composed of roughly 15 ovarioles. Each ovariole contains a linear sequence of egg chambers of increasing developmental stages. Germline and somatic stem cells reside near the tip of the ovariole in a region called the germarium. Progeny of the germline and somatic stem cells assemble into egg chambers, which then bud off from the germarium and are linked to adjacent chambers by stalk cells, like beads on a string. Each egg chamber produces a single egg and is composed of 16 germline cells (15 nurse cells and one oocyte), surrounded by a monolayer of roughly 600 epithelial follicle cells. The follicle cells serve several important functions including patterning the oocyte, synthesizing and transporting yolk polypeptides to the oocyte, and secreting the protective layers of the egg shell [9]. The nurse cells produce and transport cytoplasm into the oocyte, which is mostly transcriptionally quiescent. Drosophila ovarian development has been recently reviewed by Horne-Badovinac and Bilder [10], and Bastock and St Johnston [11]. An illustrated developmental timeline of Drosophila oogenesis is shown in Figure 2.
Figure 2.
(a) Timeline of Drosophila oogenesis with major developmental events labeled below. The beginning of each developmental stage is indicated by a mark on the line. The interval between stages was drawn in proportion to estimated development time. (b) Micrographs of major developmental stages of Drosophila oogenesis. Top panels show three-dimensional projections of z-stacks of confocal images with nuclei labeled in blue, E-cadherin labeled in green, and myosin labeled in red. Bottom panels show single-plane confocal images through the middle of the tissue with nuclei labeled in white, E-cadherin in green, and myosin in red. Scale bar is 50 μm.
Ex vivo culture of fly ovaries for live-cell imaging
Ex vivo culture and observation of egg chambers at stage 10B and later has been possible since the founding work of Petri et al. [12], and Gutzeit and Koppa [13•]. It was also known that egg chambers could develop normally following removal of the muscular sheath that normally encases each ovariole, followed by injection into a host female [13•,14]. However long-term exvivo culture of earlier stage egg chambers took another 16 years to achieve [15••]. To approximate normal development, it is crucial to maintain the tissue under physiological conditions, which requires precise control of environmental temperature, nutrients, oxygen, pH, hormones, and growth factors. Techniques to isolate egg chambers for live imaging have been described in detail in several articles [15••,16–18,19••]. The key breakthrough was the discovery that pH of at least 6.9 and insulin supplementation are crucial for egg chamber growth. With these modifications and addition of fetal calf serum, stages 6–9 egg chambers can be cultured in Schneider’s or Grace’s medium for up to 6 h of continuous observation of tissue growth and cell movements [15••]. More recently, proliferation of germline stem cells and production of cysts in the germarium have also been observed in living cultures for up to 14 h [20••]. While tremendous progress has been made, further improvements are still needed since the egg chambers cultured under current best conditions cannot as yet progress from stage 9 to stage 10. To achieve full development of egg chambers ex vivo may require specific combinations or pulses of juvenile hormone, 20-hydroxyecdysone, insulin or perhaps unknown factors.
Live imaging of germarium development
Imaging the dynamics of stem cells within their native niches has the potential to reveal information inaccessible in fixed samples. Short-term imaging (~30 min) of the Drosophila germarium during stem cell division was used to study asymmetric distribution of Wicked, a component of the U3 snoRNP complex that is important for maintenance of stem cell fate [21]. Long-term imaging (~14 h) of living germaria has recently been achieved by Morris and Spradling [20••]. In their study, full cycles of division and differentiation of germline stem cells were recorded. Interestingly, these live imaging studies revealed that the escort cells, which were thought possibly to migrate along with the germline stem cell daughters and be replenished by escort stem cells, in fact remain largely stationary and are mitotically quiescent. This finding emphasizes the necessity of observing developmental events as they actually occur and the hazards of inferring dynamic behavior from fixed samples.
Another live imaging study in the germarium focused on the fusome, a complex structure of endomembranes, membrane-associated cytoskeleton, and microtubules, which ramifies into the interconnected cells of each germline cyst [22]. For a long time, it was unclear whether the fusome lumen was shared between all the cells of the cyst. Snapp et al. found that photobleaching one portion of the fusome present in one cyst cell caused a rapid depletion of fluorescence in the whole structure, suggesting that the fusome endomembranes are part of a single continuous endoplasmic reticulum (ER) [23•].
Live imaging of egg chamber rotation
What controls the overall shape of an organ? In principle, it could be achieved simply by the sum of the shapes of its component cells. Alternatively, the overall shape of an organ might emerge dynamically from mechanical interactions between different cells and extracellular components. The latter turns out to be the case in Drosophila ovary. Early stage egg chambers are spherical but they gradually elongate as they grow, ultimately producing eggs that are 2.5-fold longer than they are wide. This change of tissue shape coincides with development of dramatically polarized arrays of basal F-actin bundles oriented perpendicular to the long axis of the egg chamber [24]. At the same time, the ECM proteins within the basement membrane surrounding the egg chamber become aligned in the same direction [25]. The polarized basal F-actin and ECM fibers have been proposed to function as a ‘molecular corset’ to constrain radial growth of egg chamber. However, images of fixed tissue did not reveal how the F-actin and ECM polarization was achieved. Using live imaging techniques, Haigo and Bilder made the astonishing observation that follicles rotate within the basement membrane and that this rotation provides a novel mechanism to achieve global alignment and orientation of the F-actin and basement membrane fibers required for elongation of the tissue [26••]. During stages 5–8, egg chambers rotate clockwise or counterclockwise around their long axis at a speed of ~0.5 μm/min and thereby produce circumferentially polarized tracks of collagen IV. This directional rotation depends on integrin-mediated interactions of follicle cells with the ECM, and disruption of collagen IV or integrin expression prevents follicle rotation and results in round eggs. Interestingly, this rotation is similar to follicle rotation found in gall midges by Went in 1977, suggesting that it may be a general phenomenon [27].
Many fascinating questions arise from the observation of egg chamber rotation including for example how this directional movement is achieved. One suggestion is that the follicle cells crawl upon the ECM in a coordinated manner. Since the direction of rotation is random from one follicle to the next, it is probably selected through a stochastic mechanism. The nature of the mechanism is unclear, as is the mechanism by which all the cells within a single egg chamber choose the same direction. What initiates the movement is another mystery. How the correct rotation axis is selected is also an open question. There are multiple possible explanations for how directional rotation might lead to elongation of the tissue. One model is that alignment of the ECM and actin fibers creates a corset that constrains increases in egg chamber volume toward the poles. Another possibility, which is not mutually exclusive, is that the rotation itself creates an anisotropic force. Finally, the polarized actin cytoskeleton and integrin-mediated adhesion to the ECM that occurs as a consequence of the follicle rotation serve as the substrate for periodic myosin contractions, which also contribute to egg chamber elongation, as described in the next section.
Live imaging of follicle cell basal contractions
Immediately after egg chambers stop rotating, they start to grow dramatically and continue to elongate. Within 10–12 h, the egg chamber increases its volume by 10-fold and elongates 1.6-fold in the absence of cell division. Achieving this elongation in the presence of such dramatic tissue expansion requires anisotropic mechanical forces to constrain the radial volume increase. One of the major generators of forces in tissues is actomyosin contractility. Using time-lapse imaging with fluorescently tagged myosin, we found that contractile myosin began to accumulate at the basal surfaces of a subset of follicle cells beginning in early stage 9. We also observed that the basal surfaces of follicle cells actively contract and relax, shrinking specifically along the short axis. The contractions were strikingly cyclical with an average period of 6.5 min [28••]. These myosin-mediated basal contractions require the Rho-ROCK pathway, cytosolic calcium, integrin-mediated cell-ECM interactions, and E-cadherin-mediated cell-cell adhesion. Both pharmacological and genetic approaches show that interfering with the contractions results in rounder eggs whereas enhancing the contractions leads to longer eggs. Since these contractions do not occur in all follicle cells but rather are mostly confined to follicle cells near the center of the egg chamber, we postulate that the effect of the contractions is to constrain the tremendous increase in tissue volume to the two ends. We envision that this is somewhat similar to squeezing a ball of dough to make it longer, although obviously the elastic properties of cells differ from those of dough. Nevertheless, exertion of an anisotropic force near the middle of the tissue as it expands in volume over the course of 10 h does affect its shape, although it is not clear precisely which cellular and extracellular elements respond to the force and create the lasting change in shape. Live imaging reveals that elongation of this tissue involves much more dynamic molecular and cellular behaviors compared to the static view of a corset that was developed from analysis of fixed tissue. It will be important in the future to decipher the mechanisms that initiate, sustain, and pattern the oscillations. Oscillations in myosin accumulation and cellular contractility have been observed in other epithelia undergoing morphogenetic changes [29,30]. Interestingly the oscillation periods and subcellular locations of myosin accumulation differ in the different tissues, suggesting that the oscillation mechanism can be regulated tissue-specifically to achieve diverse morphogenetic outcomes.
Live imaging of collective border cell migration
At late stage 8 a small group of anterior follicle cells adjacent to the polar cells, referred to as border cells, round up in response to the cytokine Unpaired (Upd), which the polar cells secrete. Upd activates the JAK/STAT pathway in the border cells, causing them to extend protrusions, delaminate from the epithelium and migrate in between the nurse cells. These 4–7 cells surround and carry the two non-motile polar cells from the anterior tip of the egg chamber, reaching the oocyte by stage 10 [31–33]. Genetic screens and analysis of border cell migration in fixed tissue revealed multiple signaling pathways that control distinct features of the movement. Whereas JAK/STAT signaling determines which of the follicle cells acquire the ability to move, receptor tyrosine kinases, PVR and EGFR, determine the direction of movement in response to ligands secreted by the oocyte [34–36]. The steroid hormone ecdysone by contrast controls the timing of border cell migration [32].
Analysis of border cell migration using live imaging has begun to reveal the dynamic features of their movement. For example, it was surprising to find that inhibition of both EGFR and PVR function in border cells did not suppress cell protrusion in the forward direction. On the contrary, multiple cells extend long and random protrusions in all directions, suggesting that the guidance signals may not only promote cell protrusion at the front but also inhibit protrusions in the wrong directions [15••].
A major molecular driver of protrusion in cells is the small GTPase Rac and Rac has long been known to be required for border cell migration [35,37,38]. However, both dominant-negative (DN) and constitutively active (CA) forms of Rac cause strong migration defects, indicating that Rac activity must be spatially and/or temporally regulated. Recently it has become possible to control Rac activity in vivo using genetically encoded and caged forms of Rac, created by Wu et al. [39]. These caged Rac proteins provide a novel approach to activate or inhibit Rac activity with high spatial and temporal resolution in response to flashes of blue laser light. Using this tool, we found that activating or inhibiting Rac activity in one migrating border cell causes dramatic responses of the other cells in the cluster [40••]. Activating Rac in any cell, caused the cluster to migrate in that direction. Inhibition of Rac in the leading cell caused all cells to lose their sense of direction and thus to protrude outward in all directions. In this study, the endogenous pattern of Rac activity was also monitored using a Rac FRET biosensor that was originally generated in Matsuda’s lab and modified by Kardash and co-workers [41,42]. Rac activity is normally higher at the front and lower at the back of border cell clusters, and is higher in the front portion of the leading cell than in the back of the leading cell. Polarity of Rac activity is lost when guidance receptor activity is inhibited, although some uniform Rac activity persists. These findings suggest that a low level of uniform Rac activity promotes protrusion in all directions in the absence of guidance receptor activity, and that in response to asymmetric guidance receptor activation, Rac activity increases at the front, enhancing forward directed protrusion. In addition, via an unknown mechanism enhanced Rac activity at the front inhibits protrusion in other directions of all the cells in the cluster. A key open question is by what mechanism the cells sense relative levels of Rac activity in adjacent cells.
Live imaging of transport and polarization of RNA and protein in germline cells
Initially discovered in developing oocytes, the polarized localization of mRNAs turns out to be a widely adapted mechanism to establish cell polarity in germ cells, neurons, and cells undergoing asymmetric division [43]. Localization of numerous mRNAs to distinct regions of the fly oocyte is crucial for normal patterning of the embryo following fertilization. One advantage of using egg chambers to study the subcellular localization of molecules is the large size of the germline cells. During development, the oocyte grows from a ~20 μm diameter at stage 3 to more than 100 μm at stage 10. Live imaging of RNA movement in the fly ovary was first achieved by microinjection of fluorescently labeled molecules into late stage egg chambers. An active, long-range, microtubule-dependent cytoplasmic flow termed ooplasmic streaming occurs between late stage 10 and stage 13 [44,45]. Streaming is inhibited earlier in egg chamber development by actin polymerization, kinesin and dynein motor activities and by the two proteins Capucccino (Capu) and Spire [46,47].
Forrest and Gavis pioneered the use of a less invasive labeling technique to track in vivo movement of nanos RNA and found that it translocated along microtubules and then became anchored at the posterior region by an actin-dependent mechanism [48•]. This RNA labeling system takes advantage of the binding between bacteriophage MS2 coat protein (MCP) and a specific RNA sequence that forms a stem-loop structure, and was first used by Bertrand et al. to study mRNA localization in living yeast [49]. When MCP is tagged with fluorescent protein, and co-expressed with an RNA tagged with the MS2-binding sequence, the position of the RNA is revealed by the fluorescent signal from the MCP bound to it [50]. This mRNA labeling method was later used to localize many other important mRNAs in the oocyte including Gurken, Bicoid (bcd), and Oskar [51–53]. A more detailed review of mRNA localization in the Drosophila oocyte can be found in Becalska and Gavis [54].
Localized mRNAs are typically found in ribonucleoprotein (RNP) complexes, so in addition to mRNAs, many mRNA-binding proteins, including Exuperentia (Exu), Staufen (Stau), Vasa, Aubergine and Yps, also exhibit specific localizations within the oocyte. Live imaging has also been used to study the mechanisms responsible for their transport and localization. For example, fluorescently labeled Exu, an RNA binding protein required for proper localization of bcd mRNA, exhibits a dynein-dependent directional movement on polarized microtubules (MTs) and travels through the ring canals that connect the nurse cells to the oocyte [55]. Imaging of Staufen (Stau), a protein associated with RNPs containing oskar mRNA, revealed that oskar mRNA is randomly transported on MTs in all directions with a weak posterior bias [53]. Interestingly, Shimada et al. recently discovered that the mRNA-binding protein Ypsilon Schachtel (Yps) is transported via both MT-dependent and MT-independent mechanisms and this transport is regulated by nutrient availability and insulin signaling, possibly as part of a mechanism to preserve oocytes during periods of nutrient deprivation and allowing for rapid resumption of reproduction when conditions improve [56•].
Live imaging of epithelium morphogenesis during dorsal appendage formation
Complex epithelial movements also occur during late stages of oogenesis (from stage 10B to stage 14). Live culture of late stage egg chambers was first reported by William H. Petri in 1979 [12]. The culture of late stage egg chambers may be less demanding because the egg chamber does not grow much and starts to form a vitelline membrane and thus to separate itself from the environment. The formation of the dorsal respiratory appendages during these stages had been analyzed live by Dorman et al. [57]. Three phases of morphogenesis were revealed, and two cell types that form the roof and floor of the structure exhibit different morphological behaviors.
Future prospects
In the past few years, live imaging of earlier stages of oogenesis has revealed patterning mechanisms that were unimaginable based on analysis of fixed tissue, such as the rotation of follicles within the basement membrane and oscillating myosin contractions. The rapid development of new genetically encoded biosensors, caged proteins and microscopy technology provides unprecedented opportunities to address biological questions using live imaging. Biosensors have been engineered to reveal changes in pH, ion concentrations, protein activities, and even the distribution of mechanical forces [58–60]. The possibilities for manipulating protein activities with high spatial and temporal resolution are also likely to expand tremendously in the near future. Recently many photoactivatable proteins have been engineered using different approaches to cage a broad spectrum of signaling molecules, including a cation channel [61], adenylyl cyclase [62], G protein-coupled receptors [63], transcription factors [64–66], and multiple protein kinases [67]. Combining these tools with innovations in whole organ cultures will allow investigators to both manipulate these pathways and monitor the immediate effects, not only on individual cells or cell types but also on the complex interactions between different cell types and ECM.
Although phototoxicity is a factor that limits long-term 3D imaging in many tissues including the ovary, advances in microscopy are likely to continue to improve our ability to see deeper, with higher resolution, and over longer periods of time. For example, light sheet microscopy or selective plane illumination microscopy (SPIM) provides a solution to image large and deep samples with greatly reduced light exposure [68]. For this technique the sample has to be suspended in a tube of transparent gel to allow imaging from multiple directions, which may require special protocol modifications. In addition, the quantity of data collected using SPIM challenges current software and hardware available in most labs. Perfusion systems may enhance survival of tissues that requires constant nutrient supply [69]. In addition, super-resolution techniques such as structured illumination microscopy (SIM), stimulated emission depletion microscopy (STED), 4Pi, and photo-activated localization microscopy/stochastic optical reconstruction microscopy (PALM/STORM) have the potential to reveal protein dynamics at the single molecule level [70,71]. Tradeoffs for the increased resolution offered by these approaches include limitations in imaging speed, depth, and requirements for specific fluorescent probes, such as photoactivatable or photoconvertible fluorescent proteins. The depth limitation can be overcome by combining PALM/STORM with TIRF for some applications, such as imaging the basal myosin oscillations. It is very likely that with the continued improvements in culture conditions, biosensor development, and microscopy techniques, live imaging will greatly advance our understanding of the dynamic molecular, cellular and supracellular mechanisms that control Drosophila oogenesis.
Footnotes
This review comes from a themed issue on Developmental mechanisms, patterning and evolution Edited by Sean Megason, Shankar Srinivas, Mary Dickinson and Anna-Katerina Hadjantonakis
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Mavrakis M, Rikhy R, Lilly M, Lippincott-Schwartz J: Fluorescence imaging techniques for studying Drosophila embryo development. Curr Protoc Cell Biol 2008, Chapter 4:Unit 4.18. [DOI] [PubMed] [Google Scholar]
- 2.Parton RM, Valles AM, Dobbie IM, Davis I: Live cell imaging in Drosophila melanogaster. Cold Spring Harb Protoc 2010, 2010: pdb.top75. [DOI] [PubMed] [Google Scholar]
- 3.Aldaz S, Escudero LM, Freeman M: Live imaging of Drosophila imaginal disc development. Proc Natl Acad Sci USA 2010, 107:14217–14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng J, Hunt AJ: Time-lapse live imaging of stem cells in Drosophila testis. Curr Protoc Stem Cell Biol 2009, Chapter 2:Unit 2E.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin N, Badie N, Yu L, Abraham D, Cheng H, Bursac N, Rockman HA, Wolf MJ: A method to measure myocardial calcium handling in adult Drosophila. Circ Res 2011,108:1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson WR, Hiesinger PR: Preparation of developing and adult Drosophila brains and retinae for live imaging. J Vis Exp (37)2010. doi: 10.3791/1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao J, Zobeck KL, Lis JT, Webb WW: Imaging transcription dynamics at endogenous genes in living Drosophila tissues. Methods 2008, 45:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parton RM, Valles AM, Dobbie IM, Davis I: Drosophila larval fillet preparation and imaging of neurons. Cold Spring Harb Protoc 2010, 2010: pdb.prot5405. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Tanwar PS, Raftery LA: Drosophila follicle cells: morphogenesis in an eggshell. Semin Cell Dev Biol 2008, 19:271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horne-Badovinac S, Bilder D: Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn 2005, 232:559–574. [DOI] [PubMed] [Google Scholar]
- 11.Bastock R, St Johnston D: Drosophila oogenesis. Curr Biol 2008, 18:R1082–R1087. [DOI] [PubMed] [Google Scholar]
- 12.Petri WH, Mindrinos MN, Lombard MF, Margaritis LH: In vitro development of the Drosophila chorion in a chemically defined organ culture medium. Dev Genes Evol 1979, 186:351–362. [DOI] [PubMed] [Google Scholar]
- 13.Gutzeit H, Koppa R: Time-lapse film analysis of cytoplasmic streaming during late oogenesis of Drosophila. J Embryol Exp Morph 1982, 67:101–111.•The first report about microinjecting labeled RNA into late stage egg chambers and discovery of the cytoplasmic streaming that is an important process for polarization of transported RNA.
- 14.Montell DJ, Keshishian H, Spradling AC: Laser ablation studies of the role of the Drosophila oocyte nucleus in pattern formation. Science 1991, 254:290–293. [DOI] [PubMed] [Google Scholar]
- 15.Prasad M, Montell DJ: Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev Cell 2007, 12:997–1005.••This paper is one of the first reports of capturing full length border cell migration by live-cell imaging and discovers the increase of membrane protrusion after inhibiting EGF and PVF signaling in border cells.
- 16.Prasad M, Jang AC, Starz-Gaiano M, Melani M, Montell DJ: A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat Protoc 2007, 2:2467–2473. [DOI] [PubMed] [Google Scholar]
- 17.Parton RM, Valles AM, Dobbie IM, Davis I: Isolation of Drosophila egg chambers for imaging. Cold Spring Harb Protoc 2010, 2010: pdb.prot5402. [DOI] [PubMed] [Google Scholar]
- 18.Tekotte H, Tollervey D, Davis I: Imaging the migrating border cell cluster in living Drosophila egg chambers. Dev Dyn 2007, 236:2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rorth P: Two distinct modes of guidance signalling during collective migration of border cells. Nature 2007, 448:362–365.••This study first documents of the two different migration stages of border cells and relates them to difference in EGF and PVF signal.
- 20.Morris LX, Spradling AC: Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development 2011, 138:2207–2215.••For the first time the germarium is imaged long enough to record the full cycle of stem cell self-renew and differentiation in their native niche.
- 21.Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant JA, Bellaiche Y, Huynh JR: Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol 2009, 11:685–693. [DOI] [PubMed] [Google Scholar]
- 22.de Cuevas M, Lilly MA, Spradling AC: Germline cyst formation in Drosophila. Annu Rev Genet 1997, 31:405–428. [DOI] [PubMed] [Google Scholar]
- 23.Snapp EL, Iida T, Frescas D, Lippincott-Schwartz J, Lilly MA: The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol Biol Cell 2004, 15:4512–4521.• This paper applies fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) to study the connectivity of important subcellular structure fusome.
- 24.Gutzeit HO: Organization and in vitro activity of microfilament bundles associated with the basement membrane of Drosophila follicles. Acta Histochem Suppl 1991, 41:201–210. [PubMed] [Google Scholar]
- 25.Gutzeit HO, Eberhardt W, Gratwohl E: Laminin and basement membrane-associated microfilaments in wild-type and mutant Drosophila ovarian follicles. J Cell Sci 1991, 100(Pt 4):781–788. [DOI] [PubMed] [Google Scholar]
- 26.Haigo SL, Bilder D: Global tissue revolutions in a morphogenetic movement controlling elongation. Science 2011, 331:1071–1074.•• This paper for the first time characterizes global tissue revolution and discovers its function in aligning ECM molecule that eventually leads to tissue elongation.
- 27.Went DF: Pulsating oocytes and rotating follicles in an insect ovary. Dev Biol 1977, 55:392–396. [DOI] [PubMed] [Google Scholar]
- 28.He L, Wang X, Tang HL, Montell DJ: Tissue elongation requires oscillating contractions of a basal actomyosin network. Nat Cell Biol 2010, 12:1133–1142.•• This study first shows the periodic contraction of basal actomyosin in follicle epithelium and discovers the linkage between this contraction and tissue elongation.
- 29.Martin AC, Kaschube M, Wieschaus EF: Pulsed contractions of an actin-myosin network drive apical constriction. Nature 2009, 457:495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauzi M, Lenne PF, Lecuit T: Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature 2010, 468:1110–1114. [DOI] [PubMed] [Google Scholar]
- 31.Montell DJ: Border-cell migration: the race is on. Nat Rev Mol Cell Biol 2003, 4:13–24. [DOI] [PubMed] [Google Scholar]
- 32.Jang AC, Chang YC, Bai J, Montell D: Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat Cell Biol 2009, 11:569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rorth P: Collective guidance of collective cell migration. Trends Cell Biol 2007, 17:575–579. [DOI] [PubMed] [Google Scholar]
- 34.Duchek P, Rorth P: Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science 2001,291:131–133. [DOI] [PubMed] [Google Scholar]
- 35.Duchek P, Somogyi K, Jekely G, Beccari S, Rorth P: Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 2001, 107:17–26. [DOI] [PubMed] [Google Scholar]
- 36.McDonald JA, Pinheiro EM, Montell DJ: PVF1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with Taiman. Development 2003, 130:3469–3478. [DOI] [PubMed] [Google Scholar]
- 37.Murphy AM, Montell DJ: Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J Cell Biol 1996, 133:617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geisbrecht ER, Montell DJ : A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell 2004, 118:111–125. [DOI] [PubMed] [Google Scholar]
- 39.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM: A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 2009, 461:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, He L, Wu YI, Hahn KM, Montell DJ: Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol 2010, 12:591–597.•• This paper is one of the first in vivo applications of photoactivation techniques and reveals novel mechanism of Rac in collective perception of guidance signal.
- 41.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM: Localized Rac activation dynamics visualized in living cells. Science 2000, 290:333–337. [DOI] [PubMed] [Google Scholar]
- 42.Kardash E, Reichman-Fried M, Maitre JL, Boldajipour B, Papusheva E, Messerschmidt EM, Heisenberg CP, Raz E: A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat Cell Biol 2010, 12:47–53 sup pp 41–11. [DOI] [PubMed] [Google Scholar]
- 43.StJohnston D: Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol 2005, 6:363–375. [DOI] [PubMed] [Google Scholar]
- 44.Glotzer JB, Saffrich R, Glotzer M, Ephrussi A: Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr Biol 1997, 7:326–337. [DOI] [PubMed] [Google Scholar]
- 45.Cha BJ, Koppetsch BS, Theurkauf WE: In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell 2001, 106:35–46. [DOI] [PubMed] [Google Scholar]
- 46.Dahlgaard K, Raposo AA, Niccoli T, St Johnston D: Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell 2007, 13:539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theurkauf WE: Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science 1994, 265:2093–2096. [DOI] [PubMed] [Google Scholar]
- 48.Forrest KM, Gavis ER: Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 2003, 13:1159–1168.• This is the first application of MS2-MCP RNA labeling technique in study of mRNA dynamics in oocyte.
- 49.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM: Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998, 2:437–445. [DOI] [PubMed] [Google Scholar]
- 50.Weil TT, Parton RM, Davis I: Making the message clear: visualizing mRNA localization. Trends Cell Biol 2010, 20:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaramillo AM, Weil TT, Goodhouse J, Gavis ER, Schupbach T: The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J Cell Sci 2008, 121:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weil TT, Forrest KM, Gavis ER: Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell 2006, 11:251–262. [DOI] [PubMed] [Google Scholar]
- 53.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D: In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 2008, 134:843853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becalska AN, Gavis ER: Lighting up mRNA localization in Drosophila oogenesis. Development 2009, 136:2493–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mische S, Li M, Serr M, Hays TS: Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol Biol Cell 2007, 18:2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimada Y, Burn KM, Niwa R, Cooley L: Reversible response of protein localization and microtubule organization to nutrient stress during Drosophila early oogenesis. Dev Biol 2011,2:250–262.• For the first time, the transport of RNPs is found to be regulated by ovary nutrient status.
- 57.Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA: bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev Biol 2004, 267:320–341. [DOI] [PubMed] [Google Scholar]
- 58.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T et al. : Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 2010, 466:263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herbst KJ, Ni Q, Zhang J: Dynamic visualization of signal transduction in living cells: from second messengers to kinases. IUBMB Life 2009, 61:902–908. [DOI] [PubMed] [Google Scholar]
- 60.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA: Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev 2009, 90:1103–1163. [DOI] [PubMed] [Google Scholar]
- 61.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K: Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 2005, 8:1263–1268. [DOI] [PubMed] [Google Scholar]
- 62.Schroder-Lang S, Schwarzel M, Seifert R, Strunker T, Kateriya S, Looser J, Watanabe M, Kaupp UB, Hegemann P, Nagel G: Fast manipulation of cellular cAMP level by light in vivo. Nat Methods 2007, 4:39–42. [DOI] [PubMed] [Google Scholar]
- 63.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K: Temporally precise in vivo control of intracellular signalling. Nature 2009, 458:1025–1029. [DOI] [PubMed] [Google Scholar]
- 64.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE: Induction of protein-protein interactions in live cells using light. Nat Biotechnol 2009, 27:941–945. [DOI] [PubMed] [Google Scholar]
- 65.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL : Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 2010, 7:973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drepper T, Krauss U, Meyer zu Berstenhorst S, Pietruszka J, Jaeger KE: Lights on and action! Controlling microbial gene expression by light. Appl Microbiol Biotechnol 2011, 90:23–40. [DOI] [PubMed] [Google Scholar]
- 67.Karginov AV, Zou Y, Shirvanyants D, Kota P, Dokholyan NV, Young DD, Hahn KM, Deiters A: Light regulation of protein dimerization and kinase activity in living cells using photocaged rapamycin and engineered FKBP. J Am Chem Soc 2010, 133:420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santi PA: Light sheet fluorescence microscopy: a review. J Histochem Cytochem 2011, 59:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huisken J, Stainier DY: Selective plane illumination microscopy techniques in developmental biology. Development 2009, 136:1963–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toomre D, Bewersdorf J: A new wave of cellular imaging. Annu Rev Cell Dev Biol 2010, 26:285–314. [DOI] [PubMed] [Google Scholar]
- 71.Schermelleh L, Heintzmann R, Leonhardt H: A guide to super-resolution fluorescence microscopy. J Cell Biol 2010, 190: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]