Abstract
Cancer metastasis is a physical process in which tumor cells break away from the primary tumor, enter, and then exit the blood or lymph vessels, and establish secondary tumors in distant organs. Current clinical studies report a higher risk of cancer metastasis for diabetics than non-diabetics. However, due to complex overlapping risk factors between diabetes and cancer, the mechanism underlying this correlation is largely unknown. Elevated lifetime blood sugar levels in diabetics are known to increase glycation of collagen, causing stiffening of the ECM and connective tissue. In this study, we explored the roles of glycation of 3D collagen matrices in tumor cell invasion and migration. Using time-lapse images, we quantitatively compared the motility behavior of malignant breast tumor cells (MDA-MB-231) and co-culture spheroids (1:1 ratio of MDA-MB-231 cells with normal epithelial MCF-10A cells) embedded in glycated and non-glycated collagen matrices of various concentrations. Experimental results demonstrated that glycation increased tumor invasion within collagen matrices. More specifically, the average speed of MDA-MB-231 cells was higher in glycated collagen gels than in non-glycated collagen gels for all three gel concentrations tested. Cell spreading characterized by its diffusion coefficient or the effective spheroid radii at various time points was significantly greater in glycated collagen than in non-glycated collagen at a concentration of 3.5 mg/mL. This enhancement was moderate and less evident at lower collagen concentrations of 1.0 and 2.0 mg/mL. These results suggest a possible biomechanical link that relates to the high blood sugar level in diabetic patients and the cancer metastatic outcome.
INTRODUCTION
Non-enzymatic glycation is a physiological process that occurs with aging or in the tissue of diabetic patients due to the high level of blood glucose [1–5]. Although glycation is associated with cancer [6–9], the roles of non-enzymatic glycation in tumor invasion are largely unknown. With the recent advancement of biomaterial technology, the mechanical properties of collagen can now be controlled precisely by sugar glycation processes in vitro [10–14]. Type I collagen is the major structural component of the ECM in connective tissues [15]. The basic building blocks of Type I collagen are collagen fibrils with diameters ranging from 25 to 400 nm, which are formed by the cross-linking of tropocollagen molecules. Under physiological conditions, the fibrils form bundles called fibers, and these fibers self-organize into a fiber network [16, 17], which determines the main tissue mechanical properties. The collagen fiber network has been found to stiffen through non-enzymatic glycation of collagen, in which reducing sugars post-translationally cross-link tropocollagen molecules [12, 13, 18]. Work from the Bonassar group and others showed that ribose pre-glycated Type I collagen in a solution formed collagen fibers with larger diameter and pore size upon polymerization when compared to the non-glycated collagen gels with the same collagen concentration [4, 19–21]. Ribose-glycated gels were stiffer and could reach up to a 10-fold increase in bulk modulus when compared with their non-glycated counterparts [22].
In parallel with the advancements made in the field of bio-materials, we now also know that the mechanical properties of extracellular matrices surrounding tumor cells critically regulate tumor cell invasion [20, 23, 24]. It is becoming increasingly apparent that the microenvironment surrounding the tumor cells can play as important a role as the genetic makeup of the tumor cells [25–27]. Tumor cells are architecturally supported by and interact reciprocally with the 3D collagen fiber network. To successfully metastasize, cancer cells must invade through interstitial spaces to gain access to the vasculature or distant organs [28, 29]. Tumor cells migrating through the interstitial space can adhere onto collagen fibers and pull forward (protease-independent mesenchymal motility), squeeze through the pore space of the collagen fiber network (protease-independent amoeboid motility), or enzymatically digest a micro-tunnel through the ECM using Matrix Metalloproteinases (MMPs) (pro-tease-dependent motility) [29–32]. It has been shown that tumor cells interact with ECM reciprocally; for example, they stiffen the collagen fiber network by exerting a force on it, and, in return, the stiffened collagen promotes tumor cell migration and force generation [20, 33]. Although the field of quantitative measurements of force generation in various types of ECMs is still evolving [34–37], it is known that collagen fiber pore size and fiber diameter play important roles in cell force generation and migration [20, 38, 39]. For collagen matrices of a specific concentration, larger fiber diameter and pore size have been found to promote cell force generation and migration [23].
Given the importance of matrix properties in tumor cell invasion and the potential for glycation to alter matrix properties, we have explored the roles of glycation in tumor cell invasion using a ribose-glycated Type I collagen as a model system. In particular, we have analyzed the invasion characteristics of breast tumor cells (MDA-MB-231), either as single cells or spheroids embedded in glycated or non-glycated collagen matrices. Experimental results demonstrated that glycation promoted tumor invasion.
MATERIALS AND METHODS
3D cell culture preparation
Cells
Triple-negative breast tumor cells (MDA-MB-231), provided by the Cornell Center of Microenvironment and Metastasis, were cultured in high glucose Dulbecco’s Modified Eagle Medium (DMEM) (Cat. #: 11965092, Gibco, Life Technologies Corporation, Grand Island, NY) with 10% fetal bovine serum (Cat. #: S11150, Atlanta Biologicals Lawrenceville, GA), 100 units/mL penicillin, 100 μg/mL streptomycin (Cat. #: 15140122, Gibco). Non-tumorigenic breast epithelial cells (cell line MCF-10A), provided by the Cornell Center of Microenvironment and Metastasis, were cultured in DMEM supplemented with nutrient mixture F-12 (DMEM/F-12, Cat. #: 11320033, Gibco) that contains 5% horse serum (Cat. #: S12150, Gibco), 5% EGF (Cat #: PHG0311, Gibco), 0.5 mg/mL hydrocortisone (Cat. #: H0888–1G, Sigma-Aldrich), 100 ng/mL cholera toxin (Cat #: C-8052, Sigma), 10 μg/mL insulin (Cat. #: I-1882, Sigma), 100 units/mL penicillin, and 100 μg/mL streptomycin (Cat. #: 15140122, Introvigen). All cells were cultured in T75 flasks (Cat. #: 10062–860, Corning, Lowell, MA, USA), which were placed in a 5% carbon dioxide, 37°C, and 100% humidity incubator. Cells were passaged every 3–4 days and harvested for experiments when the cell culture reached 70–90% confluency. MDA-MB-231 cells with 20 or fewer passages and MCF-10A cells with 10 or fewer passages were used for the experiments. In the spheroid experiments, MDA-MB-231 cells expressing EGFP and MCF-10A cells expressing dTomato variants, which were kind gifts from Dr Joseph Aslan at the Oregon Health & Science University, were used.
Spheroids
A specially designed array of microwells was used for making co-culture spheroids that are about 100 μm in diameter and contain equal numbers of MDA-MB-231 and MCF-10A cells [40]. Briefly, we first generated a pattern of a 36 × 36 microwell array on a thin layer of agarose gel using soft lithography; each micro-well is cylindrical in shape with a diameter of 200 μm and a depth of 250 μm. The agarose gel surface provides low adhesion to the cells. Each microwell array was then placed in one of the wells of a 12-well plate (Cat. #: 07-200-82, Corning). For cell seeding in each well, we prepare one million MDA-MB-231 cells and one million MCF-10A cells suspended in 2.5 mL of cell media (1:1 ratio mixture of cell media for MDA-MB-231 and MCF-10 cells). The 12-well plate was then kept on a rocker in an incubator (Forma, Thermo Scientific, Asheville, NC, USA) at 37°C, 5% carbon dioxide and 100% humidity for 3 days before harvesting. The average diameter of the spheroids was about 100 μm when collected. More details of the spheroid making process can be found in Ref. [41].
3D cell culture
Type I collagen extracted from rat tail tendon (Cat. #: 354249, Corning) was suspended in 0.1% acetic acid to 5 mg/mL and was stored at 4°C before the experiments. For non-glycated gels, volumes of 40, 80, and 140 μL collagen stock (5 mg/mL) were titrated with 0.88, 1.76, and 3.08 μL 1 N NaOH, respectively, and 20 μL 10× M199 (Cat. #: M0650–100ML, Sigma) was added to approximately yield a final pH of 7.4. Then, the cell culture mixture was added to reach a final volume of 200 μL and a final collagen concentration of 1.0, 2.0, and 3.5 mg/mL. For glycated gel, the collagen stock (5 mg/mL) was pre-glycated by mixing the stock collagen with 250 mM ribose in 4°C for 5 days before being diluted and mixed with the cells for the experiments. The same protocol was followed for the spheroid experiments. For single-cell experiments, a cell concentration of 2 million/mL was used for all experiments. For spheroid experiments, a concentration of about 1600 spheroids/mL was used.
Device preparation
Device setup
A 1× 7 × 8 cm polycarbonate plate with a 3 cm-diameter hole at the center was tightly sealed with a 48 × 60 mm No. 1 cover glass (Cat. #: 3334 VWR) at the bottom using a high-vacuum grease (Cat. #: 1597418, Corning). The cover slide had a thickness of about 150 μm to facilitate high-quality microscopic imaging. Six through holes with a diameter of 4 mm were created using a biopsy punch (Cat. #: 21909–140, Miltex Inc. York, PA) on a PDMS sheet (1.6 × 1.1 cm) with a thickness of 450 μm. Two identical PDMS sheets were bonded to the cover glass side by side (see Fig. 1A) using an oxygen plasma oven (Harrick Plasma Cleaner PDC-001, Harrick Plasma, Ithaca, NY). The assembled device was then sterilized for 15 min in an autoclave before use.
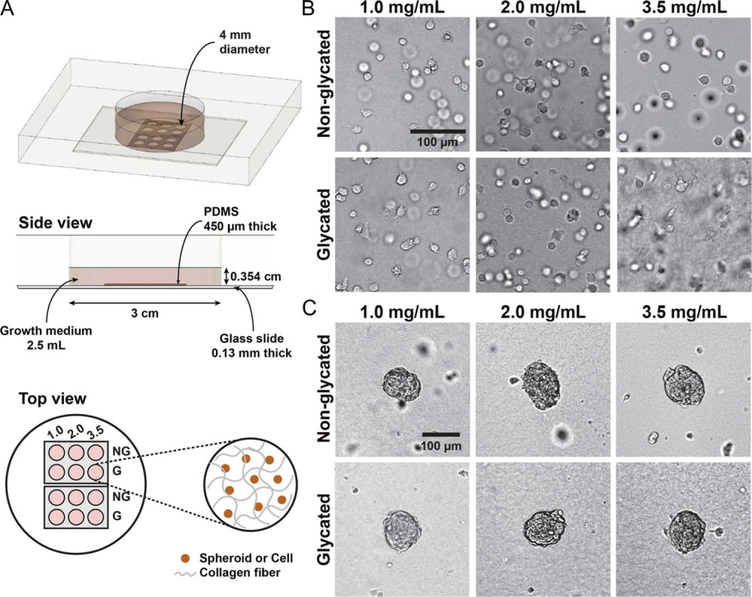
Figure 1.
An array microwell device for studies of 3D tumor cell invasion. (A) Experimental setup. A Plexiglas plate with a cylindrical hole of 3 cm in diameter and 1 cm in depth is sealed at the bottom with a No. 1 glass slide using high vacuum grease. Two PDMS array devices are plasma-bonded to the surface of the glass slide. Each array device contains a 450 μm thick PDMS membrane with 6 wells that are 4 mm in diameter. Cell-embedded collagen is placed in each of the wells. Typically, the collagen thickness is about 400 μm and the medium above the collagen is about 3.14 mm in height. (B) Bright-field images of MDA-MB-231 cells embedded in glycated and non-glycated collagen gels. (C) Bright-field images of MDA-MB-231:MCF-10A (1:1 ratio) spheroids embedded in glycated and non-glycated collagen gels. In each micro-well array device, six different collagen types are used: glycated and non-glycated collagen at concentrations of 1.0, 2.0, and 3.5 mg/mL. These images are taken at t = 0, which is approximately 2 h after the cells or spheroids have been introduced into the collagen.
Surface activation
To make the surface hydrophilic, the PDMS wells were first treated with oxygen plasma (Harrick Plasma Cleaner PDC-001, Harrick Plasma, Ithaca, NY) for 1 min in the high setting. Each well was then treated with 5 μL of 1% polyethyleneimine (PEI) for 10 min and then with 5 μL of 0.1% glutaraldehyde for 30 min. The wells were left in a biohood overnight in sterile distilled water. The device was washed with sterile distilled water and aspirated before the use in the experiment. Surface activation is a crucial step because it prevents the collagen matrices from detaching due to the forces that the cells exert.
Cell seeding, collagen polymerization
For each concentration of collagen, 5.5 μL of the prepared cell/spheroid-embedded collagen was pipetted into each of the 4 mm-diameter microwells with the device placed on ice, which makes an approximate gel thickness of 438 μm. The device was then incubated at 37°C and 5% CO2 for 45 min for collagen polymerization. To prevent the cells and spheroids from settling down at the bottom during the collagen polymerization process, the device was first placed upside-down, where the glass slide was on top. Then, the device was flipped a total of three times at time points 5, 15, and 30 min. Using this protocol, most of the cells or spheroids were located in the bulk of the collagen gel as seen in Movie S3. We note that image of a single cell above, at or below the imaging focal plane forms a bright, gray or dark dot [see Movie S3 and also Ref. [42]]. We use this image characteristics to make sure that all the images were taken in the mid 200 μm section of the sample. Detailed optical explanation for the 3D imaging using an inverted optical microscope can be found in reference [42]. During the entire collagen polymerization process, the glass slide was always in direct contact with a metal block preheated to 37°C for fast and consistent heat transfer. For the single-cell experiments, 2.5 mL of the MDA-MB-231 cell media was added to the device after the collagen polymerization, at which point the cells were ready for imaging. For the spheroid experiments, 2.5 mL of a mixture (1:1 ratio) of MDA-MB-231 and MCF-10A cell media was used.
Imaging and data analysis
All images were taken with a 10× magnification objective lens (NA = 0.25, Olympus America, Center Valley, PA, USA) installed on an epi-fluorescent microscope (IX 81, Olympus America, Center Valley, PA, USA) and a CCD camera (ORCA-R2, Hamamatsu Photonics, Bridgewater, NJ, USA). The light source for fluorescence imaging was provided by the X-Cite series 120PC unit (Excelitas Technologies, Waltham, MA, USA). The scope was surrounded by a stage incubator (Precision Plastics Inc., Beltsville, MD, USA) that maintained a temperature of 37°C, humidity of ~50%, and CO2 level of 5%. On an automated x–y microscope stage (MS-2000, Applied Scientific Instrumentation, Eugene, OR), images were taken every 8 min for 20 h for single-cell experiments and every 8 min for 36 h for spheroid experiments using Metamorph (Molecular Devices, San Jose, CA, USA) or CellSens software (Olympus America, Center Valley, PA, USA). To characterize cell motility, cell positions were manually tracked at each timepoint using the Manual Tracking plugin from ImageJ (National Institute of Health, Bethesda, MD). For the single-cell experiments, 100 MDA-MB-231 cells were tracked for each collagen condition. For the spheroid experiment, all MDA-MB-231 cells that invaded out of the spheroid were tracked, typically about 60–100 cells under each condition. The average speed and the mean squared displacement were computed using the tracked cell positions with an in-house MATLAB program [43]. We note that although single cells were embedded in the bulk of the collagen matrices, cells were tracked in one image plane. The calculated speed is the actual speed in 3D projected onto a 2D plane.
For the spheroid experiment, we used the following protocol to characterize the size of the spheroids. First, the fluorescent images of MDA-MB-231 cells were utilized to generate an azimuthally averaged intensity profile from the center of the spheroid using an in-house MATLAB program. The intensity profile was then fitted to a Gaussian distribution to find the variance, or the sigma value, which represents the distance from the center of the spheroid at which 68.2% of the intensity is included. This sigma value was used as the size (or radius) of the spheroid for tumor spheroid invasion analysis.
RESULTS AND DISCUSSION
Experimental setup
The goal of this study is to investigate how ribose glycation impacts breast tumor cell invasion. For this purpose, we prepared six collagen matrices with different stiffness and pore size using a ribose glycation method previously developed in the Bonassar lab [11, 44] and also by varying the collagen concentration. A detailed preparation method and the mechanical properties of these collagen gels have been described in Ref. [20]. Briefly, we created a total of six collagen types, with three collagen concentrations (1.0 mg/mL, 2.0 mg/mL, and 3.5 mg/mL) either glycated with 250 mM ribose (G) or non-glycated (NG). The differential shear modulus of these three collagen matrices ranges from ~5 to 900 Pas covering that of normal and malignant breast tissues. The architecture of the collagen matrices is shown in Fig. S1. We note that the binding of the ribose to collagen was previously verified using a Fourier Transform Infrared (FTIR) method and detailed in Ref. [11].
A microwell array device was designed (see Fig. 1A) to allow real-time imaging of tumor cell/spheroid invasion into the collagen matrices with six different collagen architectures (stiffness and pore size) but identical chemical and nutrient conditions. The device contains six micro-wells, which are seeded with cell-embedded collagen matrices with concentrations of 1.0, 2.0, and 3.5 mg/mL that are either glycated (G) or non-glycated (NG). A second identical device is used to increase the throughput of each experiment (see the top view of Fig. 1A). Bright-field images of single cells or spheroids embedded in collagen are shown in Fig. 1B and C, respectively. The imaging is started approximately 2 h after the cells/spheroids are introduced into the collagen, which is defined as t = 0 for the imaging sequences.
Glycated collagen promoted single tumor cell invasion
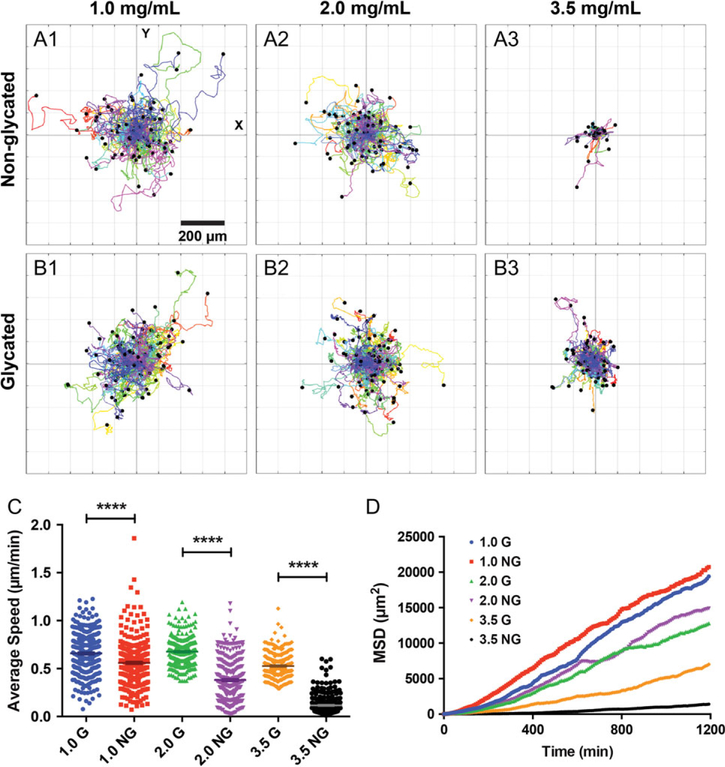
By quantifying tumor cell movement using time series imaging, we found that single MDA-MB-231 cells moved faster and spread further in glycated collagen than in non-glycated collagen (see Fig. 2 and Movie S1). Fig. 2A (non-glycated) and B (glycated) are 20 h-long polar plots of cell trajectories under six different collagen conditions. Visually, one can see that cells move further away from the starting point within 20 h in glycated collagen gels than in non-glycated collagen gels at a collagen concentration of 3.5 mg/mL. At a lower concentration, this change is less evident. Using the cell trajectories, we computed cell speed as shown in Fig. 2C. The results in Fig. 2C showed that the average cell speed was higher in the glycated collagen matrices than in the non-glycated collagen matrices at all three collagen concentrations. The speed increase was the most significant in the 3.5 mg/mL collagen gel, from 0.1 (NG) to 0.58 (G) μm/min. We further computed the mean squared displacement of cells using the cell trajectories under six different collagen concentrations as shown in Fig. 2D. The results showed that glycation promoted cell spreading significantly in the case of 3.5 mg/mL. Because the cells were tracked in 2D, we used a first-order approximate equation for mean squared displacement (MSD),MSD = 4Dt, to compute the diffusion coefficient of cell movement, where MSD is the average of the displacement squared of the cells at various time points, and D is the diffusion coefficient of the cells. We found D = (4.62, 4.46), (3.48, 2.95), and (0.30, 1.44) μm2/min for (NG, G) gels at the three different collagen concentrations of 1.0, 2.0, and 3.5 mg/mL, respectively (Fig. 2D). These diffusion coefficients showed that glycation mildly affected the cell spreading in the low collagen concentration cases and enhanced cell invasion significantly within collagen at a concentration of 3.5 mg/mL. We note that the results presented in Fig. 2C and D were computed from a total of 300 cell trajectories obtained from three repeating experiments. Fig. S2 (see online supplementary material for a color version of this figure) shows cell trajectories, average speed, and MSD from one set of experiment.
Figure 2.
Collagen glycation modulates single tumor cell invasion. (A–B) Polar plots of cell trajectories in non-glycated (A1–3) and glycated (B1–3) collagen at a concentration of 1.0, 2.0, and 3.5 mg/mL. Each colored line represents one cell track, and each track is 20 h long and taken at 8 min time interval. The first data point of each track is centered at (0,0) for comparison purposes. There are 100 cell tracks in each plot. (C) The average speed of MDA-MB-231 cells in glycated and non-glycated collagen. **** represents p-value < 0.0001. The average speed of the MDA-MB-231 cells in non-glycated 1.0, 2.0, and 3.5 mg/mL collagen are 0.56 ± 0.01, 0.38 ± 0.01, and 0.66 ± 0.01, 0.68 ± 0.01, and 0.12 ± 0.01 μm/min, respectively. The average speed of the MDA-MB-231 cells in glycated 1.0, 2.0, and 3.5 mg/mL collagen are 0.53 ± 0.01 μm/min, respectively. A total of 300 cell tracks from three repetitive experiments were used to compute the average speed. D. Mean square displacement of MDA-MB-231 cells in 1.0, 2.0, and 3.5 mg/mL glycated (G) and non-glycated (NG) gels. The MSD was computed using 300 cell tracks from three repetitive experiments.
The differences in the cell trajectory and average speed observed between the glycated and the non-glycated gels were most likely caused by the differences in the pore size and the fiber diameter of the collagen gels. Due to the size and morphology limitations, tumor cell invasion is critically regulated by the architecture of the collagen gel that surrounds them [45]. Friedl et al. proposed that the size of the nucleus limits cell movement within the ECM since the nucleus is the stiffest part of the cell body. During migration, the cell cytoplasm is able to penetrate through pores that are less than a micron in size. However, cells are not able to go through the pores that are significantly smaller than their nucleus size due to the rigid nucleus envelope [46–49]. To explore the interrelation between collagen pore size and cell nucleus size in the context of tumor cell invasion, we compared the pore size of the gels and the MDA-MB-231 nucleus size here. Using the confocal image of the collagen gels (Fig. S1, see online supplementary material for a color version of this figure), we have previously measured the pore diameter of the non-glycated 1.0, 2.0, and 3.5 mg/mL collagen matrices. They were 3.1 μm, 2.1 μm, and 1.8 μm, respectively, and the pore diameter of the glycated 1.0, 2.0, and 3.5 mg/mL collagen matrices were 5.1 μm, 3.0 μm, and 3.0 μm, respectively [20]. Using the confocal images of MDA-MB-231 cells embedded in collagen, we quantified the shapes of MDAMB-231 nuclei (see Fig. S3 and Supplementary materials, see online supplementary material for a color version of this figure for details). The shape of the MDA-MB-231 nucleus was ellipsoid in shape, and the major and the minor axes were about 4.55 ± 0.27 and 2.74 ± 0.17 μm, respectively (see Fig. S3, see online supplementary material for a color version of this figure). Because the pore sizes of both the glycated and the non-glycated 1.0 mg/mL collagen gels were greater than the length of the minor axis of the MDA-MB-231 nucleus, the cells navigated quite freely showing a minor difference in the migration pattern. In the case of 3.5 mg/mL non-glycated gel, the minor axis of the cell nucleus, 2.74 μm, was significantly larger than the pore size of 1.8 μm. Therefore, cell migration is likely decreased due to the restriction of the pore size. This is consistent with the observation that the speed decrease is restored in the glycated 3.5 mg/mL collagen gel where the pore size was about 3.0 μm.
We note that the pore size of the collagen alone cannot explain the speed increase due to collagen glycation and that the collagen fiber diameter is also an important parameter. During migration, cells need to adhere to the collagen fiber and migrate through the interstitial space. The architectural support, or the stiffness of the fiber (which is defined by the fiber diameter), clearly plays an important role [39], which was reflected in our data. Cells had greater average speed in 2.0 mg/mL G gel than in 1.0 mg/mL NG gel. Although the pore sizes of these two matrices were nearly identical (3.0 μm and 3.1 μm, respectively), the 2.0 mg/mL G gel with 75 nm had a larger fiber diameter than the 1.0 mg/mL NG gel with 50 nm [20].
Collagen glycation promoted tumor spheroid invasion
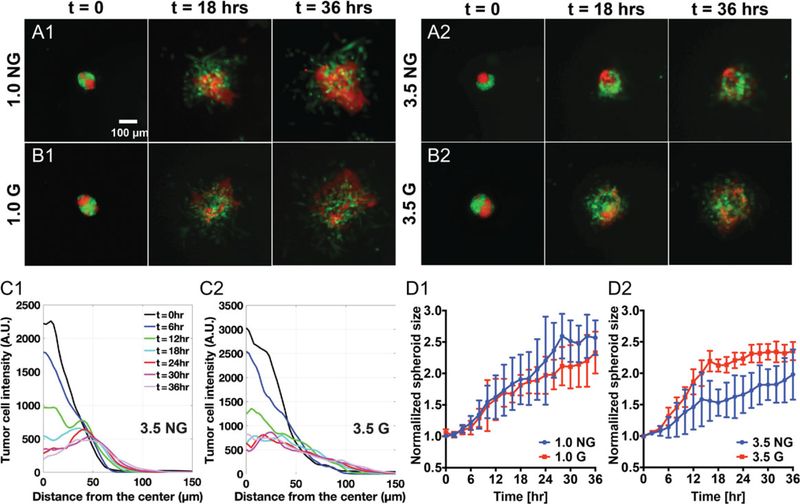
Using the same experimental setup, we carried out the invasion assays of tumor spheroids within glycated and non-glycated collagen matrices. We used a mixture of malignant breast tumor cells (MDA-MB-231) and non-tumorigenic breast epithelial cells (MCF-10A) in 1:1 ratio to make the co-culture spheroids. This is motivated by the complex tumor microenvironment where both malignant and non-malignant cells co-exist. Time sequence images of the co-culture spheroids at t = 0, 18, and 36 h are shown in Movie S2 and Fig. 3A and B. Here, the green cells are the MDA-MB-231 cells constitutively expressing EGFP, and the red cells are the MCF-10A cells constitutively expressing dTomato. The timepoint t = 0 was approximately 2 h after the spheroid had been seeded in the collagen matrices. A visual inspection of the tumor spheroid time evolution in Fig. 3A and B showed that mainly the highly metastatic breast tumor cells (MDA-MB-231) invaded into the collagen gel in all four collagen types. For the 3.5 mg/mL collagen gels, tumor spheroids were clearly more invasive in the glycated collagen (Fig. 3B2) than in the non-glycated collagen (Fig. 3A2). For 1.0 mg/mL collagen gels, the impact of glycation on tumor spheroid invasion was minute. We note that the spheroids invaded more predominantly in the lower collagen concentration (1.0 mg/mL) gels than in the higher collagen concentration (3.5 mg/mL) gels for both the non-glycated (Fig. 3A1 and A2) and the glycated cases (Fig. 3B1 and B2).
Figure 3.
Collagen glycation modulates tumor spheroid invasion. (A and B) Time sequence fluorescence images of co-culture spheroids embedded in non-glycated (A1, A2) and glycated (B1, B2) collagen matrices with concentrations of 1.0 mg/mL and 3.5 mg/mL. Images shown are at time points 0, 18, and 36 h. Here, t = 0 is defined as about 2 h after spheroids were introduced into the collagen matrices. The co-culture spheroid consists of MCF-10A cells (red) and MDA-MB-231 cells (green), with a cell number ratio of 1:1. (C) Time evolution of radial MDA-MB-231 cell density distribution in 3.5 mg/mL non-glycated (C1) and 3.5 mg/mL glycated (C2) collagen matrices. (D) Time evolution of normalized spheroid size for MDA-MB-231 cells in glycated collagen matrices than in non-glycated collagen matrices with collagen concentration of 1.0 mg/mL (D1) and 3.5 mg/mL (D2). Each error bar is computed from N = 2 to 3 spheroids.
Time sequences of the positions of the malignant breast tumor cells (MDA-MB-231) were used to perform a quantitative invasion analysis. Note that the non-tumorigenic epithelial cell line (MCF-10A) invaded into the collagen in a sheet in the 36-h time window. Using the recorded green fluorescence signal from the MDA-MB-231 cells, we calculated azimuthally averaged fluorescence intensity profile (see Fig. 3C1 and 2). This pro-file represents the radial cell density distribution from the center of the spheroid. We then fitted the radial cell density pro-file to a Gaussian function (see Fig. S4, see online supplementary material for a color version of this figure) and used the sigma value as the radius of the spheroid. The normalized spheroid radius (divided by the initial spheroid size) are shown in Fig. 3D1 for the 1.0 mg/mL collagen condition and Fig. 3D2 for the 3.5 mg/mL collagen condition. Fig. 3D2 clearly shows that glycation significantly promoted spheroid invasion in the 3.5 mg/mL collagen, while this enhancement was less evident in the 1.0 mg/mL collagen.
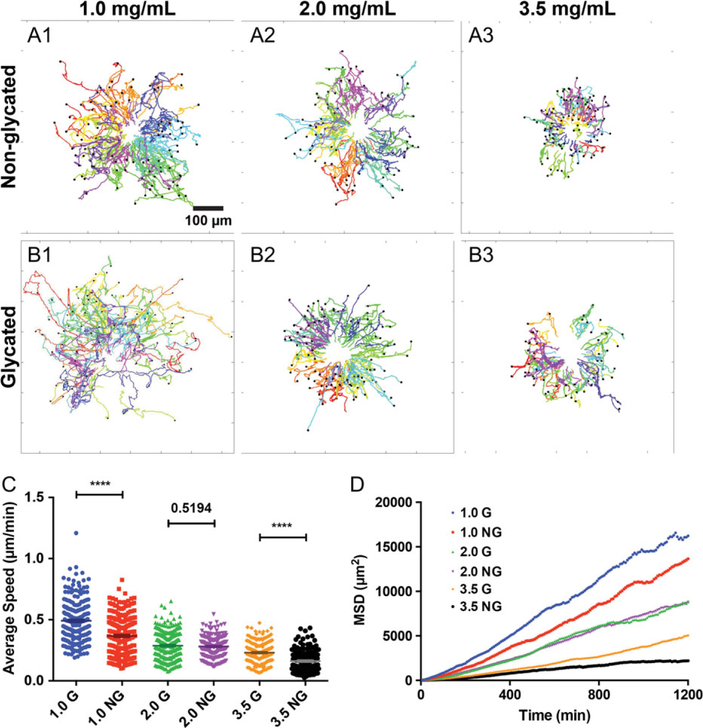
The invasive behavior of the malignant tumor cells was further characterized through the tracking of the positions of the MDA-MB-231 cells invading out of the co-culture spheroids (see Fig. 4). The total duration of the tracks is 36 h and is plotted in Fig. 4A and B for non-glycated and glycated gel, respectively. Note that Fig. 4A and B differs from the polar plots in Fig. 2A and B in that the start of each track is not centered at (0,0). Rather, the tracks are the actual spatial coordinates of the cell positions. Here, we see that the cells were mostly moving away from the center of the spheroids and that the glycation significantly enhanced the MDA-MB-231 tumor cell spreading for the 3.5 mg/mL collagen case. The average speed of the MDA-MB-231 cells (Fig. 4C) in the co-culture spheroids was largely higher in glycated collagen than in non-glycated collagen. The diffusion coefficients, which was calculated from the MSD of MDA-MB-231 cells in Fig. 4D, were (2.97, 3.73), (1.97, 1.95), and (0.52, 1.04) μm2/min for 1.0, 2.0, and 3.5 mg/mL collagen gel (NG, G), respectively. These diffusion coefficients showed that glycation enhanced tumor cell spreading in all three gel concentrations. The average speed and MSD in Fig. 4 were calculated using data from three repeatable experiments. Data from one experiment is shown in Fig. S5, see online supplementary material for a color version of this figure. We note here that the average persistence length of the cells from three repetitive experiments is shown in Figs. S6 and 7, see online supplementary material for a color version of this figure.
Figure 4.
Collagen glycation modulates tumor spheroid invasion. (A and B) MDA-MB-231 cell trajectories in non-glycated (A1–3) and glycated (B1–3) collagen at concentrations of 1.0, 2.0, and 3.5 mg/mL, respectively. Each colored line represents the actual spatial coordinates of one cell track. All MDA-MB-231 cells that invaded out of the spheroid are tracked every 8 min for 36 h under each condition. A total of 100, 75, and 100 cells are tracked for 1.0, 2.0, and 3.5 mg/mL non-glycated condition, respectively. A total of 80, 100, and 66 cells are tracked for 1.0, 2.0, and 3.5 mg/mL glycated condition, respectively. (C) Average MDA-MB-231 cell speed in glycated and non-glycated matrices with p-value shown. The average speed of the MDA-MB-231 cells in non-glycated 1.0, 2.0 and 3.5 mg/mL collagen are 0.37 ± 0.01, 0.28 ± 0.01, and 0.16 ± 0.01 μm/min, respectively. The average speed of the MDA-MB-231 cells in glycated 1.0, 2.0, and 3.5 mg/mL collagen are 0.49 ± 0.01, 0.29 ± 0.01, and 0.23 ± 0.01 μm/min, respectively. Results are computed using cell tracks ranging from 143 to 239, which were obtained from the three repetitive experiments. (D) Mean square displacement of MDA-MB-231 cells in 1.0, 2.0, and 3.5 mg/mL glycated and non-glycated gels. Results are computed using cell tracks ranging from 143 to 239, which were obtained from the three repetitive experiments.
While glycation was shown to enhance tumor cell invasion within collagen matrices in both single-cell and spheroid as-says, we do find one notable difference in the results between these two types of assays. We see that tumor cells moved significantly slower in the spheroid assay than in the single-cell assay. This difference can potentially be explained by the collagen compaction around the spheroid caused by cell-generated traction forces. In both the single-cell and the spheroid cases, tumor cells exerted forces onto the collagen matrices and reorganized the fiber network. However, the traction force was stronger in the spheroid case than in the single-cell case. Thus, we anticipate more collagen compaction around spheroids than around individual cells. In vivo, it has been reported that tumors compact collagen matrices in their vicinity and the collagen fibers have been found to align parallel to the tumor boundary [50, 51]. It is thus possible that the collagen compaction in the spheroid assay lowers the tumor cell speed when compared to the single-cell assay.
Future perspective
The collagen glycation assay here presents a useful platform for addressing roles of mechanical properties in tumor cell invasion and also points to a potential mechanical basis for the clinically observed correlation between cancer and diabetes. The next level of inquiry will be to dissect the underlying molecular mechanisms that govern the tumor cell invasion within sugar-glycated collagen gels, particularly the impact of sugar glycation on the mechano-sensing activities ranging from focal adhesion to PI3 Kinase. We note that our approach does not completely separate the roles of biomechanical and biochemical influences of glycation on tumor cell invasion. It has been discussed extensively in the literature that the receptor for advanced glycation end products (RAGE) is an important biomarker for breast tumor cell metastasis [52]. Future work will be needed to elucidate the biochemical impact of glycation in tumor cell invasion.
Supplementary Material
Insight, innovation and integration.
The incidence of cancer metastasis is clinically known to be higher in diabetics than in non-diabetics. However, mechanistic understanding of this correlation is lacking because of many overlapping factors encountered in the clinical setting. Integrating a sugar glycation biomaterial engineering method with a 3D cell and spheroid motility assay, we found that sugar glycation of the 3D collagen matrices stiffened and altered the architecture of the matrices and promoted breast tumor cell invasion. This work points to a possible mechanical link between diabetes and metastatic cancer.
ACKNOWLEDGMENT
The authors wish to thank Asmita Bhatta, Min Seo Kang and Subin Yun for their assistance in tracking the cells.
FUNDING
This work was supported by a grant from the National Cancer Institute (Grant no. R01CA221346), also partial support from the Cornell NanoScale Science & Technology Facility (CNF).
Footnotes
SUPPLEMENTARY DATA
Supplementary data is available at INTBIO online.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1.Andreassen TT, Seyer-Hansen K, Bailey AJ. Thermal stability, mechanical properties and reducible cross-links of rat tail tendon in experimental diabetes. Biochim Biophys Acta 1981;677:313–7. [DOI] [PubMed] [Google Scholar]
- 2.Schnider SL, Kohn RR. Effects of age and diabetes mellitus on the solubility and nonenzymatic glucosylation of human skin collagen. J Clin Invest 1981;67:1630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goh SY, Cooper ME. The role of advanced glycation end products in progression and complications of diabetes. J Clin Endocr Metab 2008;93:1143–52. [DOI] [PubMed] [Google Scholar]
- 4.Gautieri A, Passini FS, Silvan U et al. Advanced glycation end-products: mechanics of aged collagen from molecule to tissue. Matrix Biol 2017;59:95–108. [DOI] [PubMed] [Google Scholar]
- 5.Singh VP, Bali A, Singh N et al. Advanced glycation end products and diabetic complications. Korean J Physiol Pharmacol 2014;18:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015;15:540–55. [DOI] [PubMed] [Google Scholar]
- 7.Kailemia MJ, Park D, Lebrilla CB. Glycans and glycoproteins as specific biomarkers for cancer. Anal Bioanal Chem 2017; 409:395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munkley J, Elliott DJ. Hallmarks of glycosylation in cancer. Oncotarget 2016;7:35478–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannucci E, Harlan DM, Archer MC et al. Diabetes and cancer: a consensus report. CA Cancer J Clin 2010;60:207–21. [DOI] [PubMed] [Google Scholar]
- 10.Roy R, Boskey AL, Bonassar LJ. Non-enzymatic glycation of chondrocyte-seeded collagen gels for cartilage tissue engineering. J Orthop Res 2008;26:1434–9. [DOI] [PubMed] [Google Scholar]
- 11.Roy R, Boskey A, Bonassar LJ. Processing of type I collagen gels using nonenzymatic glycation. J Biomed Mater Res A 2010;93:843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girton TS, Oegema TR, Tranquillo RT. Exploiting glycation to stiffen and strengthen tissue equivalents for tissue engineering. J Biomed Mater Res 1999;46:87–92. [DOI] [PubMed] [Google Scholar]
- 13.Verzijl N, DeGroot J, Ben ZC et al. Crosslinking by advanced glycation end products increases the stiffness of the collagen network in human articular cartilage: a possible mechanism through which age is a risk factor for osteoarthritis. Arthritis Rheum 2002;46:114–23. [DOI] [PubMed] [Google Scholar]
- 14.Mason BN, Starchenko A, Williams RM et al. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater 2013;9:4635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelse K, Pöschl E, Aigner T. Collagens—structure, function, and biosynthesis. Adv Drug Deliv Rev 2003;55:1531–46. [DOI] [PubMed] [Google Scholar]
- 16.Storm C, Pastore JJ, MacKintosh FC et al. Nonlinear elasticity in biological gels. Nature 2005;435:191–4. [DOI] [PubMed] [Google Scholar]
- 17.Muenster S, Jawerth LM, Broedersz C et al. Non-linear mechanical properties of collagen networks during cyclic loading. Biophys J 2010;98:558A–9A. [Google Scholar]
- 18.Depalle B, Qin Z, Shefelbine SJ et al. Influence of cross-link structure, density and mechanical properties in the meso-scale deformation mechanisms of collagen fibrils. J Mech Behav Biomed Mater 2015;52:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross VL, Zheng Y, Choi NW et al. Dense type I collagen matrices that support cellular remodeling and microfabrication for studies of tumor angiogenesis and vasculogenesis in vitro. Biomaterials 2010;31:8596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall MS, Alisafaei F, Ban E et al. Fibrous nonlinear elasticity enables positive mechanical feedback between cells and ECMs. Proc Natl Acad Sci USA 2016;113:14043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason BN, Reinhart-King CA. Controlling the mechanical properties of three-dimensional matrices via non-enzymatic collagen glycation. Organogenesis 2013;9:70–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul RG, Bailey AJ. Glycation of collagen: the basis of its central role in the late complications of ageing and diabetes. Int J Biochem Cell Biol 1996;28:1297–310. [DOI] [PubMed] [Google Scholar]
- 23.Levental KR, Yu H, Kass L et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009;139:891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu HM, Mouw JK, Weaver VM. Forcing form and function: biomechanical regulation of tumor evolution. Trends Cell Biol 2011;21:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 2011;17:320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaman MH, Trapani LM, Siemeski A et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA 2006;103:10889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M, Swartz MA. Modeling Tumor Microenvironments In Vitro. J Biomech Eng 2014;136:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002;2:563–72. [DOI] [PubMed] [Google Scholar]
- 29.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003;3:362–74. [DOI] [PubMed] [Google Scholar]
- 30.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer 2011;11:573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lammermann T, Bader BL, Monkley SJ et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 2008;453:51–5. [DOI] [PubMed] [Google Scholar]
- 32.Wolf K, Mazo I, Leung H et al. Compensation mechanism in tumor cell migration: mesenchymal–amoeboid transition after blocking of pericellular proteolysis. J Cell Biol 2003;160: 267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinwachs J, Metzner C, Skodzek K et al. Three-dimensional force microscopy of cells in biopolymer networks. Nat Methods 2016;13:171–6. [DOI] [PubMed] [Google Scholar]
- 34.Legant WR, Miller JS, Blakely BL et al. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods 2010;7:969–U113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch TM, Muenster S, Bonakdar N et al. 3D traction forces in cancer cell invasion. PLoS One 2012;7:e33476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall MS, Long R, Feng X et al. Toward single cell traction microscopy within 3D collagen matrices. Exp Cell Res 2013; 319:2396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesman A, Notbohm J, Tirrell DA et al. Contractile forces regulate cell division in three-dimensional environments. J Cell Biol 2014;205:155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf K, te Lindert M, Krause M et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol 2013; 201:1069–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pathak A, Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc Natl Acad Sci USA 2012;109:10334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song W, Tung CK, Lu YC et al. Dynamic self-organization of microwell-aggregated cellular mixtures. Soft Matter 2016;12: 5739–46. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y-C, Song W, An D et al. Designing compartmentalized hydrogel microparticles for cell encapsulation and scalable 3D cell culture. J Mater Chem B 2015;3:353–60. [DOI] [PubMed] [Google Scholar]
- 42.Wu MM, Roberts JW, Buckley M. Three-dimensional fluores-cent particle tracking at micron-scale using a single camera. Exp Fluids 2005;38:461–5. [Google Scholar]
- 43.Kim BJ, Hannanta-anan P, Chau M et al. Cooperative roles of SDF-1alpha and EGF gradients on tumor cell migration revealed by a robust 3D microfluidic model. PLoS One 2013;8:e68422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mason BN, Starchenko A, Williams RM et al. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater 2013;9:4635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003;3: 362–74. [DOI] [PubMed] [Google Scholar]
- 46.Wolf K, te Lindert M, Krause M et al. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol 2013; 201:1069–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denais CM, Gilbert RM, Isermann P et al. Nuclear envelope rupture and repair during cancer cell migration. Science 2016; 352:353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davidson PM, Sliz J, Isermann P et al. Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr Biol 2015;7:1534–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isermann P, Lammerding J. Consequences of a tight squeeze: nuclear envelope rupture and repair. Nucleus 2017; 8:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Provenzano PP, Eliceiri KW, Campbell JM et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med 2006;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conklin MW, Eickhoff J, Riching K et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Cancer Res 2013;73:1221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwak T, Drews-Elger K, Ergonul A et al. Targeting of RAGE-ligand signaling impairs breast cancer cell invasion and metastasis. Oncogene 2017;36:1559–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.