Abstract
In the era of “big data”, we are gaining rich person-specific information about neuroanatomy, neural function, and cognitive functions. However, the optimal ways to create precise approaches to optimize individual function in healthy and disease are unclear. Multimodal analysis and modeling approaches that combine anatomical networks, functional signals, and cognitive neuroscience in single subjects can guide neuromodulation. Our progress could be improved by progressing from statistical fits to mechanistic models. Using transcranial magnetic stimulation as an example, we discuss how integrating methods with a focus on mechanisms could improve our predictions TMS effects within individuals, refine our models of health and disease, and improve our treatments.
1. Introduction
Brain stimulation is being applied at nearly every scale of neural organization to optimize cognition and treat brain dysfunction. Analyses of human brain networks (the connectome, Sporns et al., 2005) provide us with increasingly sophisticated means to describe anatomical and functional human brain network organization. However, it is unclear how to optimally use these data to guide translational approaches. Like many domains of inquiry, brain stimulation studies often rely on cross-sectional analyses at the group level to make inferences at the individual level. However, these types of analyses could be fundamentally flawed in principle (Molenaar, 2004), and the discrepancy between group and individual estimates can be demonstrated using various measures in psychophysical (Fisher et al., 2018) and neural data (Seghier and Price, 2018). Studying processes within individuals over time is justified to understand the nature of neurocognitive functions and how they respond to interventions (Molenaar, 2004; Poldrack et al., 2015). As the drive to achieve “precision medicine” approaches to brain disorders continues (Insel and Cuthbert, 2015), it is crucial to consider strategies to address major challenges in brain stimulation.
While we have long influenced the brain with interventions such as education (Ngandu et al., 2015) and pharmaceuticals (Franke et al., 2014), invasive and noninvasive brain stimulation have been rising in recent decades (Snowball et al., 2013). In clinical neuromodulation, we often aim to study and reduce functional (i.e., cognitive or behavioral) problems within individuals. However, the effects of stimulation are often modest at the group level (Aleman et al., 2007; Berlim et al., 2014; Burt et al., 2002; Chou et al., 2015; Schutter, 2009; Slotema et al., 2010), and variable at the individual level (López-Alonso et al., 2014). Thus, it is important to consider how to use data to enhance treatment at the level of each individual.
We offer a theoretical perspective with the intent to promote personalized neuromodulation with transcranial magnetic stimulation (TMS) from basic to translational contexts. We describe possibilities for use-inspired research that are still in developmental stages. To personalize neuromodulation, we will require a progression from “deep phenotyping” Gordon et al. (2017a), Poldrack et al. (2015) – acquiring many samples and many variables within subjects over time to understand mechanisms – to a reduced set of procedures that are clinically viable. Thus, the basis for this review is to identify an open set of challenges that, if addressed, could identify strategies that develop larger, less variable, and more specific effects in individuals. Identifying data that describe specific mechanisms of target processes within individuals could lead to better “precision” approaches to TMS. After candidate personalizing strategies are found, gold-standard clinical trials should compare the personalized interventions to existing best practices. If personalized approaches are superior, they can be further refined for real-world clinical use by reducing unnecessary expenditures and procedures.
While we focus on TMS as a special case for clarity and brevity, many of the concepts described here can be used widely when considering how to use multimodal data analysis to promote real-world positive outcomes. The spatiotemporal precision and noninvasiveness of TMS carries advantages for some of our suggestions, but it is possible to consider integrating data in the ways described here with various stimulation and recording techniques. The key notion is that there is a natural relationship between seeking to influence brain processes at the individual level and validating mechanistic models that make predictions within single subjects. This idea could apply to TMS as well as any brain stimulation method.
2. Multimodal neuroimaging & neuromodulation
When the goal is to integrate data sources to predict TMS effects, it is appropriate to use neural data to guide brain stimulation. Researchers can use neuroimaging to measure gross brain anatomy (Durston et al., 2001), anatomical white matter networks (Basser and Jones, 2002), metabolism (Bailey et al., 2005), hemodynamic activity (Smith, 2004) and electrophysiology (Pfurtscheller and Da Silva, 1999). In conjunction with these techniques, TMS can be used to induce instantaneous post-synaptic potentials (Bonato et al., 2006) and long term depression or long-term potentiation-like inhibition or excitation at a resolution of 1 cm 2 at the level of the cortex (Valero-Cabré et al., 2017; Wang et al., 1996)1.
It is challenging to understand how locally induced TMS effects influence the brain and behavior. An emerging focus in cognitive neuroscience is how to integrate various data sources to understand and predict cognitive function (Petersen and Sporns, 2015; Medaglia et al., 2015). In Network neuroscience (Bassett and Sporns, 2017), multimodal imaging data can be represented as networks or graphs that typically comprise nodes that represent locations (e.g., brain regions) and edges that represents connections between nodes. At the smallest scale, the nodes of a brain network might represent individual neurons, and the edges in the brain network represent synaptic connections between neurons. At the larger scales relevant to personalized neuromodulation, nodes are often brain regions defined by either anatomy or function, and the edges are either white matter connections between regions measured with diffusion tractography or functional interactions measured with fMRI. Numerous approaches to representing neuroimaging data in graphs have emerged2. For our purposes, network neuroscience forms one important foundation with which to examine complex neural systems. In the context of predicting TMS effects, the challenge is to find the “correct” data and models that optimize predictions.
Thus, a major challenge to personalizing neuromodulation involves how to use various data sources to inform decisions about TMS targeting and parameter selection for a given person (López-Alonso et al., 2014). The difficulties in doing so can be seen as accounting for two important sources of inter-individual variability. First, individual brains vary substantially in cytoarchitectonic and macrostructural anatomy, as well as in functional organization with respect to structural anatomy (Amunts et al., 1999; Mazziotta et al., 2001; Desikan et al., 2006; Langs et al., 2016; Tong et al., 2017). In many cases, it is insufficient only to target anatomically homotopic regions in each brain; instead, it should be to identify and target functionally homologous regions (Sack et al., 2009). Second, because neuromodulation aims to modify cognitive or affective processes and behavior that are dysfunctional in psychiatric or neurologic disorders, it is insufficient to know only the functional topographic organization of an individual’s brain, but we must also know the neurocognitive mapping between systems physiology, mental processes, and behavior.
Obtaining a neurocognitive mapping (i.e., how cognitive architectures (Langley et al., 2009) are implemented in the brain) can be seen as the principle goal of cognitive neuroscience (Gazzaniga, 2004; Petersen and Sporns, 2015), but it has remained elusive over decades of research. To personalize neuromodulation, we must consider that the mapping between brain and cognition can also vary between subjects (Gratton et al., 2018), and that only some of the variability in anatomy and physiology will be important for explaining differences in neurocognitive maps (Stephan et al., 2017). These issues are emerging as an important theme (Drysdale et al., 2017; Dubois and Adolphs, 2016; Seghier and Price, 2018), and will form one basis for our approach to personalized treatments. Integrating these concerns into our discussion, we describe the effort to personalize neuromodulation by progressing from statistical model fitting to mechanistic models.
At the foundation of the perspective that follows is the fact that aggregated data tend to poorly represent individual psychological (Fisher et al., 2018) and neural processes (Seghier and Price, 2018). Pertinent to precision neuromodulation, intra-subject variables such as TMS thresholds might be relatively consistent over days (Sommer et al., 2002), but neural responses to specific TMS sequences are not (Dyke et al., 2018; Schilberg et al., 2017) . Cognitive networks measured with fMRI vary over days (Gordon et al., 2017a) due to influences such as food intake, caffeine, gene expression, and metabolism (Poldrack et al., 2015). As we treat individuals as the basic unit of measurement, our models will be pressed to accommodate meaningful within-subject variation that predicts responses to TMS.
2.1. Prediction using statistical fits versus mechanistic models
Association does not imply causation. The same is true of covariance-based multivariate techniques. Many contributions from multivariate analysis applied to neuroimaging data, including machine learning, produce strong associations with measurable behavior (Månsson et al., 2015; Nouretdinov et al., 2011; Yarkoni and Westfall, 2017; Varoquaux and Thirion, 2014; Hoeft et al., 2011; Hoexter et al., 2013). However, they do not necessarily clarify the internal mechanisms of the system, which are necessary to make causal claims (Illari and Williamson, 2012; Medaglia et al., 2015; Mill et al., 2017). In this sense, we can understand the problem for personalizing neuromodulation in terms of a distinction between statistical fits and mechanistic models in data analysis.
2.1.1. Statistical model fitting
A “fit” refers to the use of a statistical model to describe the relationship between one or more input and output variables. Breiman described two cultures in statistical model fitting (Breiman et al., 2001). One culture assumes that the data are generated by a given stochastic data model (e.g., regression and its many variants). The other culture uses algorithmic modeling, which developed outside statistics. This latter culture uses techniques such as decision trees, random forests, neural networks, and support vector machines to identify weighted combinations of variables that yield high predictive accuracy, and is most frequently used in the modern field of machine learning. On Breiman’s account, the first culture yields models that are interpretable, but generally less accurate and risk being hammers in search of nails (Adalı et al., 2018). The second culture yields less interpretable models, which may prevent practitioners from endorsing the methods. A key challenge for statistical models is that there often exist potentially many functions that could similarly predict input-output relationships among observed and unobserved independent and dependent variables. This fact results in what is known as the “model selection” problem (Nasrabadi, 2007), and is one primary limitation to fit-based techniques.
2.1.2. Mechanistic models
In contrast, a mechanistic model is one which encodes the internal system components and processes that cause a phenomenon (Illari and Williamson, 2012). Systems engineering is based on the notion that we can find dynamic models that describe the relationships among input variables, the internal states of a system, and the system’s outputs. A dynamic model represents a testable theory for how the system functions in the context of its environment (the input variables). Knowing how a system works provides information from which we can design (Chen, 1998), control (Kalman, 1959; Schiff, 2012), and repair (Schmidt and Leach, 2003) a system’s function.
2.1.3. Moving from statistical fits to mechanistic models
In science at large, the word mechanism can be fraught (Illari and Williamson, 2012). In neuroscience, mechanisms can be invoked to explain brain-brain, brain-cognition, and brain-cognition-input phenomena. However, the exact description of mechanisms varies wildly in content and scope. Some mechanisms in theoretical neuroscience are quite successful at describing information in neural systems and how it is stored and transmitted (Sompolinsky, 2014). However, robust models for cognitive systems, much less cognitive-behavioral responses to stimulation are in relatively short supply. In this sense, the effort to personalize neuromodulation is a distinct arm on the way to validating mechanisms that are generalizable. In the discussion that follows, it is best to think of a framework that is “mechanism-inspired.” That is, it aims to integrate data in a way that is compatible with many views on mechanisms, but with relatively sparse details about each. Instead, we focus on the links between mechanisms across spatiotemporal scales that could be most relevant to personalizing neuromodulation.
Ideally, the distinction between statistically fitted models and mechanistic models should decrease as each type of model becomes more successful at predicting a system’s outputs. In both approaches, prediction accuracy for new (out of sample) data is an important criterion to optimize. In statistical model fitting, the assumption is sometimes made that higher predictive accuracy is associated with more reliable information about the mechanisms for the data (Breiman et al., 2001). However, this is not necessarily the case. For instance, in human neuroimaging, even very basic measures of network organization offer high-accuracy prediction value when applied to single modality or multimodal data (Finn et al., 2015). However, if asked to create a person named Tina from a statistical model that predicted her identity based on a connectomic “fingerprint ” (Finn et al., 2015), it would not be clear how to create Tina herself from the model. That is, the model is useful to predict a label that refers to Tina, but it doesn’t tell us much about how the system (Tina) works, nor what we could do next for Tina should she become a candidate for a brain stimulation intervention.
3. Multimodal informatics for neuromodulation
Nevertheless, we can develop personalized neuromodulation with an equal emphasis on predictive accuracy and interpretive value. We suggest that statistical fits are valuable when the appropriate predictions are defined. Mechanistic model-based forecasts should be a long-run priority to accurately define systems (i.e., the dynamic relationships within the nervous system that describe connectomes and their input-output characteristics), which can help to develop novel hypotheses and potential approaches for treatments.
3.1. The data at hand
We can non-invasively measure an individual’s connectome to estimate local gray matter and white matter volume and integrity (T1-weighted imaging), white matter anatomical connections between regions (diffusion-weighted imaging and tractography), local and inter-regional hemodynamic activity (functional magnetic resonance imaging and functional near-infrared spectroscopy), and millisecond timescale electrophysiological activity (electroencephalography, EEG; and magnetoencephalography, MEG). All of these data can be used to algorithmically fit models that predict the influence of TMS on behaviors of interest They can also be integrated to refine and test dynamic models of the relationships among variables.
The following sections will offer a perspective to integrate and enrich numerous attempts to use complex brain network data to guide TMS. Numerous recent instances have been reported including precision stimulation of individual-specific fMRI connectome hubs (Lynch et al., 2018), white matter tract based targeting (Nummenmaa et al., 2014), resting-state network based prediction using the Human Connectome Project data (Opitz et al., 2016), resting state network connectivity in subjects who have received TMS (Fox et al., 2014, 2013), network-guided targeting of the cerebellum (Esterman et al., 2017), and network and model-guided targeting for depression (Luber et al., 2017). In the perspective that follows, we seek to extend increasing interest in network-guided targeting to the more general challenges in personalizing neuromodulation.
3.2. Fitting models to predict TMS effects
Any statistical model that can predict responses to TMS treatments with high accuracy would be valuable. We can use many variables to predict TMS effects (see Fig. 1). Raw genetic (Kleim et al., 2006; Malaguti et al., 2011), demographic, psychophysiological, and neural data can be used as features when fitting and training models. Using multimodal systems models, we can further quantify the organization of the components of genetic, social, and the neural data sources before, during, and after brain stimulation as independent variables. Expressing relationships among system components in graph theoretic representations allow us to examine the predictive value of associations among features (the network edges) as well as the predictive value of the network’s topology — the specific configuration of edges among nodes. All of these mathematical representations can be used in weighted combinations to predict TMS effects immediately after stimulation (e.g., either at each trial or between administrations of tasks in a cognitive neuroscience experiment) or at longer time scales such as those examined in clinical neuromodulation.
Fig. 1.
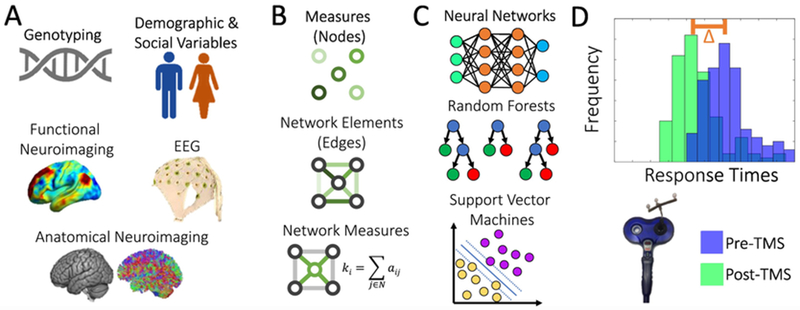
Fitting models to predict TMS effects using complementary methods in connectomics. (A) Data sources used for model fitting can include subject-level information such as genotype, demographics, social status, neuroimaging and neurophysiology. (B) The data can be represented as individual measures (network nodes), pairwise elements (network edges), or more topologically complex measures of organization (network measures). (C) With no prior assumptions about the relationships between the input features and TMS responses, machine learning tools can learn the optimal associations between the data and (D) individual response to TMS.
We can determine the value of a fitted model when we use it to predict new data (i.e., not those used to identify the model). Concerns about what is signal and what is noise are central to evaluating model fits. Here, we mean noise in the signal processing sense, which are the unwanted and typically unknown changes to a signal due to signal recording, storage, transmission, processing, or conversion. The presence of noise means that the results obtained from one data sample might not be duplicated across repeated samples. Noise is fundamentally distinguished from variation resulting from the process of interest. In the case of personalizing TMS, we should ideally be concerned with modeling inter-subject and intra-subject variation above and beyond noise. One essential notion behind within-subjects measurements and deep phenotyping is to distinguish sources of meaningful variation across subjects in the group from unwanted noise sources.
Often, cross-validation approaches train the model on data from part of the sample (e.g., split-half, leave-one-out) and predict the rest of the sample (Arlot et al., 2010). In these cases, the structure of the noise profile of the data often increases the prediction value of the model within the sample. However, if the model were applied to a new sample of data, the performance will tend to decrease (Whelan and Garavan, 2014). This is because the noise profile of the new sample and the variables of interest will differ from the original sample. This limitation to model fitting is highly consequential if we want to personalize neuromodulation.
In contrast, if we want to predict trial-wise or session-wise responses to TMS, we could take two approaches. First, we could fit a model on data from a large sample of individuals to predict whether a new subject would respond to TMS (a between-subjects model predicting within-subject effects). This approach typically requires a sample with more subjects than the number of features used to fit the model. Second, we could fit a model that can determine whether a person will respond to a TMS intervention within the subject (a within-subject model predicting within-subject effects). This approach typically requires a sample with more within-subject responses to TMS (either many trial-level responses or session-level responses) than features.
The first (between-subjects) model’s prediction will be limited by (1) the predictive power of the features in isolation or combination, (2) the relationship of the noise profile of the new subject’s data relative to the training sample’s data and (3) the relationship between the cross-sectional data and the true within-subject responses to TMS. Regarding this latter point, the cross-sectional (between subjects) data model could reveal a different relationship between TMS and the fitted features than the within-subjects data dimension. If this is the case, the predictive power of the model will be limited for a given subject because it does not capture the true statistical relationship between the variables within a given subject (Seghier and Price, 2018). In some cases, the relationship between between-subjects information and within-subjects dynamics is quite poor (Fisher et al., 2018).
The second (within-subjects) model’s prediction will be limited by (1) the predictive power of the features in isolation or combination, (2) the relationship of the noise profile of the subject’s data relative to the training sample’s data and (3) the stationarity of the within-subject process (i.e., how time-varying processes are within the subject). Obtaining a within-subjects training prediction implies that we should stimulate a subject at the trial or session level iteratively to find optimal stimulation parameters given the target cognitive, brain, or behavioral response. To minimize the time burdens to optimize to the subject, we can fit adaptive algorithms from machine learning to find input-output relationships between stimulation parameters and behavior.
Practically speaking, there is a trade-off between inter-subject and intra-subject model-fitting. The former strategy may better capitalize on stable, trait-like features that vary slowly in time (over days, weeks, and years). These can include demographics, social networks, genetics, anatomical neuroimaging, and demonstrably stable functional networks (Gratton et al., 2018). We can often acquire these measures in one or two time-intensive sessions plus additional time for data processing. The latter strategy may be more sensitive to dynamic, state-like features that vary more quickly in time (from milliseconds to hours). These can include recent social interactions, epigenetics (Rando and Verstrepen, 2007), and functional neuroimaging and neurophysiological measures. We can often acquire these measures synchronously or quasi-synchronously with the stimulation (Dowdle et al., 2018; Sack et al., 2007; Thut and Miniussi, 2009; Rogasch and Fitzgerald, 2013) and tools are available to remove TMS artifacts (Morbidi et al., 2007; Sweeney et al., 2012; Rogasch et al., 2014a).
Thus, a prudent strategy to personalize neuromodulation could involve a combination of between-subjects analysis to prospect subjects at the group level, followed by more intensive within-subjects system identification to identify subject-level effects. This can allow us to identify “hybrid” statistically fitted models that include both inter- and intra-subject information to predict individual responses to TMS.
3.3. Budding mechanistic models to predict TMS effects
While statistical fitting can be powerful, it can occur without revealing anything about the internal workings of a system (Bassett et al., 2018). Knowing how a system works and how it can fail can help create better interventions. Fortunately, models are emerging independently across various social (Scott, 2017), genetic (Parikshak et al., 2015), cognitive (Petersen and Sporns, 2015), neurophysiological (Fröhlich, 2015; Breakspear, 2017), and biomechanical (Huang et al., 2017) lines of inquiry. The challenge is how to link the relevant models to predict TMS. Obtaining models that allow us to predict and understand TMS responses implies that we first learn something about how the features of the system operate over time to produce cognition and behavior. Then, we need models that tell us how complex neurocognitive systems respond to electromagnetic perturbation. Here, we offer a schematic overview for how these models could begin to link together to forecast TMS responses to personalized neuromodulation.
Each persons’ brain develops from a genetic template interacting with the environment. At the moment of brain stimulation, the TMS is usually administered to a relatively focal location in the brain. At and near this location, field modeling provides estimate for how the electrical influences from TMS are distributed across local brain tissue (Thielscher et al., 2015). These estimates can be used as information about how intensively neuromodulation has interacted with specific parts of the brain at the region and network levels. The TMS site of stimulation has local network cytoarchitectonic characteristics and is positioned in a much larger-scale white matter network. Conceptually, we can envision white matter pathways as the “highways” that mediate communication between regions. The function of the larger-scale network (e.g., that commonly measured with fMRI) is mediated through anatomical white matter inter-regional connectivity. White matter pathways shape some but not all of the observable functional network (Hermundstad et al., 2013; Hermundstad et al., 2014; Becker et al., 2018; Liang and Wang, 2017; Medaglia et al., 2018b; Griffa et al., 2017), simulated functional network responses to stimulation (Muldoon et al., 2016; Gollo et al., 2017), simulated dynamic-related cognition (Medaglia et al., 2018a; Bansal et al., 2018) and the evolution of seizures recorded through direct electrocorticography of the cortex (Stiso et al., 2018; Khambhati et al., 2018).
The functional processes within the brain’s anatomical networks support dynamic cognitive processes. Accordingly, our approach to TMS should include information about how cognition operates within the network. Frequently, investigators aim to use knowledge about the location of cognitive processes and cognitive networks to guide targeting (Opitz et al., 2016). A broader challenge for neuromodulation research is how to guide stimulation in a way that synthesizes network anatomy, neural function, and cognitive function to guide the desired responses over time. Cognitive systems can be represented with cognitive architectures (Langley et al., 2009), which are mechanistic models for how cognition works without necessarily describing its organization in the brain. Identifying the neurocognitive mapping of cognitive architectures to the brain is one foundation and frontier of cognitive neuroscience (Gazzaniga, 2004; Petersen and Sporns, 2015). In neuromodulation, the effort to personalize TMS can at once benefit from using validated neurocognitive mappings to guide stimulation and understand the local effects in the context of the larger cognitive system. In turn, modeling the effects of TMS on neurophysiology and behavior can help us discern between competing theoretical neurocognitive models (Pascual-Leone et al., 2000).
3.3.1. Individual variability at the forefront of personalized neuromodulation
Whereas it is possible to obtain statistical fits that do not clarify mechanisms, it is also possible to define mechanistic models that do not predict individual responses. This phenomenon is most commonly observed when a model derived from aggregated (or “group”, “between subjects”, “cross-sectional”) data is used to describe person-level neural and psychological processes (Molenaar, 2004; Medaglia et al., 2011; Fisher et al., 2018; Seghier and Price, 2018). In general, it is important that the configuration of neuroanatomy, function, and the representation of cognitive processes vary across people. Individualized network analyses can identify how network anatomy and function are related to variation in cognitive function (Medaglia et al., 2018b) and responses to TMS (Medaglia et al., 2018a).
The case of personalized TMS provides the ability to at once leverage and discover the bases of cognitive mechanisms. Focusing on processes within subjects over time reveals a very different set of conclusions than aggregating psychological (Fisher et al., 2018) and neural (Seghier and Price, 2018) data. The distinction between group and individual data bears consequences for how we understand mechanisms. A salient classical example is the debate about the Piagetian stage theory of psychological development. When data are aggregated across individuals, it appears that psychological development occurs as a smooth curve. However, examining each individual at a time reveals a stage process that is distinctly Piagetian (Van der Maas and Molenaar, 1992). Because the onsets of the stages vary across individuals, aggregating the data results in a nonrepresentative smooth curve and precludes analysis about within-subject processes. In other words, if we assumed the aggregate was correct, then we would have made the wrong scientific conclusion about mechanisms. In TMS, a concrete challenge of all experimental and research designs is how to target the correct processes within subjects. Investigators use various functional neuroanatomical conventions to make decisions. However, network organization at a high spatial scale varies across individuals. The relevance for TMS’s role in exploiting and testing mechanisms is clear. If a TMS sequence targeted at a personalized network site linked to a cognitive process outperforms that selected by an aggregate approach, the study will have at once identified the possible location of a personalized mechanistic system and possible basis for clinical use.
To date, methods for mapping individualized functional network organization generally use resting-state and task-based functional connectivity to identify functional anatomy of individuals. These methods can be classified into areal mappers and system mappers. Areal mappers seek to divide the cortex into a set of discrete, contiguous patches (areas) that are divided by abrupt changes in functional connectivity profile (Cohen et al., 2008). These methods typically require large amounts of data (> 25 min) and do not place restrictions on the size or number of the regions which can lead to subject’s having different sets of brain regions, complicating group analysis (Gordon et al., 2017b; Laumann et al., 2015; Xu et al., 2016). Some areal mappers use prior constraints on area size or number to ensure an equal number across subjects (Honnorat et al., 2017; Glasser et al., 2016; Thomas Yeo et al., 2011).
In contrast, system mappers aim to map the organization of large-scale distributed systems over the cortex (Beckmann et al., 2009). The most common system mapping technique is dual-regression independent component analysis (ICA), in which group-ICA is used to find group level set of distributed networks, and then spatial maps of the group-ICA are regressed onto each subject’s data to get subject-specific network time-series. Then, the network time series are regressed onto each subject’s data again to obtain subject-specific spatial maps (Beckmann et al., 2009). Other system mapping techniques have been proposed that use a priori out-of-sample group average templates (Gordon et al., 2017a; Schultz et al., 2014; Wang et al., 2015), or Bayesian methods that model both intra- and inter-subject variability to derive subject-specific functional system maps (Kong et al., 2018).
It is important to consider what will demarcate the most useful approaches for precision neuromodulation. First, it is important that the approaches can prospectively map individual functional anatomy. Both dual regression ICA and the Bayesian method proposed by Kong et al. (2018) first require estimating group level connectivity maps before deriving subject-specific versions. This is not ideal for precision neuromodulation, because we would like to be able to estimate individualized functional anatomy for a given subject without having to acquire a full cohort for comparison. Additionally, if these methods are to be useful for clinical translation, they should be able to estimate individualized functional anatomy with a short amount of data that can reasonably be collected in a single scan session. For this reason, gradient based methods such as those proposed by Laumann et al. (2015) may not be ideal. At present, it seems that system mapping methods that use a priori out-of-sample system templates best meet these criteria. Ultimately, for these methods to become widely adopted, they must prove useful for predicting the response to neuromodulation. These responses could be induced brain responses in regions distal to the site-of-stimulation (Chen et al., 2013) or behavioral effects (Lynch et al., 2018).
Whichever approach is used, variability in neurocognitive mapping should be integrated into the foundation of our strategy rather than examined strictly post hoc. To pursue this goal, we should perform brain mapping at the level of individuals by estimating the configuration of individual neuroanatomical networks, functional network organization and its relationship to underlying neuroanatomy, and how cognitive architectures are organized with respect to the anatomically embedded functional networks within each person. See Fig. 2 for a schematic approach to integrating data for mechanism-based forecasting within individuals.
Fig. 2.
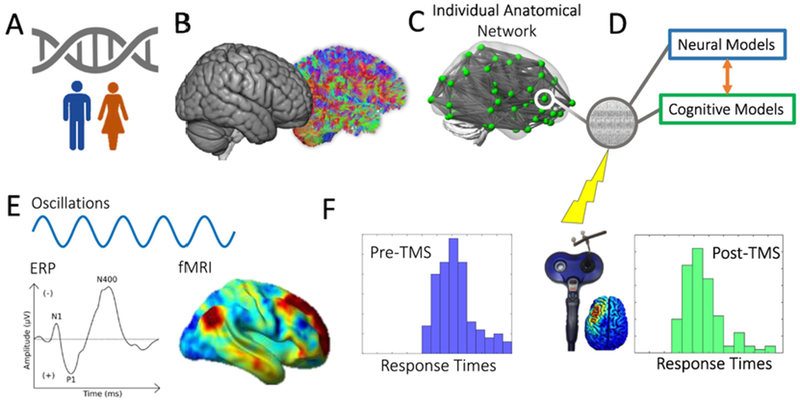
Using data to test mechanisms and forecast TMS effects within individuals. (A) A person’s genotype, demographics, and social context are moderators for TMS effects that may manifest as features at the brain level. (B) Network anatomy forms a person-specific “fingerprint” that we can represent as (C) a graph of the anatomical network that (D) has dynamics embedded within nodes across the network that facilitate local computations and long-distance inter-regional communication. (E) This person-specific network-embedded neurocognitive model is responsible for ERPs, oscillations, and more indirect activity measurable by e.g., fMRI. (F) There is presumably a mapping from a person’s individual dynamics to behavior in the absence of TMS (Left). Once TMS is administered to the brain, its influences are mediated through (Middle) the local field (e.g., those estimated using finite element methods (Opitz et al., 2013)) and modulate neurocognitive processes, resulting in Post-TMS effects at the trial or session level (Right).
In this manner, we can aim to understand TMS responses as the consequences of neuromodulation applied to the individual’s brain. Genetics and environment provide long-timescale variables that may moderate TMS effects. TMS responses may be moderated by the specific anatomical configuration of the stimulated region (e.g., represented by graph theoretic measures) in the context of distributed networks. How the TMS interacts with the individual’s cognitive architecture at and downstream from the site of stimulation determine the temporally proximal effects on the brain and behavior. The exact neurocognitive state of the system during stimulation can moderate the TMS effect (Silvanto et al., 2007; Silvanto and Pascual-Leone, 2008). Behavioral, neuroimaging and neurophysiological data acquired during and after stimulation provide indirect evidence about the effects of TMS for use in adaptive algorithm fitting (i.e., “closed-loop” neuromodulation, Berényi et al., 2012; Kraus et al., 2016). Finally, these data and attempts to build forecasting models could help rule out theories of neurocognitive function when some forecasting models are more successful than others.
3.4. Trial-wise experimentation supports discovery science and closed-loop therapeutics
Efforts to personalize TMS can accelerate by combining traditional neuroscience methods, cognitive theory, and neuroengineering. Broadly, formalisms from control theory can aid these efforts (Kraus et al., 2016; Schiff, 2012). However, the nature of validation experiments will form a gatekeeper to our success. As with cognitive neuroscience, a key challenge in cognitive neuroengineering is to forecast and test phenomenon at the appropriate spatiotemporal scale. Here, we focus on TMS-EEG as a special case where neuroengineering, neuromodulation, and cognitive neuroscience can converge.
TMS-EEG combines modern developments in a hundred year old noninvasive technique with modern, relatively spatiotemporally precise TMS. Recently, new value has been found in EEG data that is sensitive and specific to behavioral performance at the single-trial level (Bayer et al., 2017; Collins and Frank, 2018; Delorme et al., 2015; Mullen et al., 2015; Zrenner et al., 2017). Paired with TMS-shielded EEG equipment and adaptive artifact-reduction algorithms, the notion of trial-level behavioral prediction that informs TMS is an intriguing possibility. Novel EEG noise concerns emerge due to TMS electromagnetic and physical displacement artifacts. However, careful positioning of the EEG leads (Sekiguchi et al., 2011) and artifact cleaning techniques (Rogasch et al., 2014b; Wu et al., 2018) can attenuate these problems. Thus, once investigators have established relevant spatial targets, the methodology is available to probe trial-level research designs to identify neurocognitive mechanisms.
In clinics, cognition is often measured with an assessment that provides a coarse estimate of an individual’s ability against a normative population (Lezak et al., 2004). However, we view the cognitive problem as a measurable consequence of problems with finer scale cognitive processes. If this is the case, we could improve treatment efficacy by addressing the specific mechanism of dysfunction at a finer spatiotemporal scale. Here, we could use cognitive modeling and approaches that combine real-time stimulation and brain monitoring to great effect. For example, executive dysfunction is a transdiagnostic issue that often includes problems in shifting attention3. Shifting attention has reasonably good cognitive models (Posner and Rothbart, 2007), recruits relatively well-known networks (Fan et al., 2005), and can be measured using well-validated tasks at the trial level (Posner, 1980).
Thus, a deficit in shifting attention is a clinical problem with good prerequisites for personalizing neuromodulation. TMS can be used to influence specific regions in relatively focally and with millisecond temporal precision in attention networks estimated at the individual level. The cognitive paradigm provides real-time measurements of performance that result from specific cognitive processes and can be measured concurrently with brain monitoring (e.g., EEG). The pairing of the task with EEG can allow us to indirectly measure brain states immediately prior to a task stimulus and their association with performance on a trial (Makeig et al., 2004; Thut and Miniussi, 2009). Adaptive real-time source localization (Delorme and Makeig, 2004; Hsu et al., 2016) and causal estimates for influences between estimated sources (Delorme et al., 2011; Mullen et al., 2013) could increase the power to identify behaviorally-relevant neural signals and changes due to TMS.
The goal is then to identify an algorithm that adaptively improves performance using TMS. An algorithm can be fit to use information about the pre-stimulus and post-stimulus brain state plus prior trial performance to shape TMS stimulation (i.e., whether to administer TMS, the frequency and amplitude of TMS, the shape of TMS etc.). TMS can be applied in the anticipatory period immediately prior to a trial or concurrent with the trial to try and modify performance. See Fig. 3 for a schematic for refining trial-level control in a TMS-EEG context.
Fig. 3.
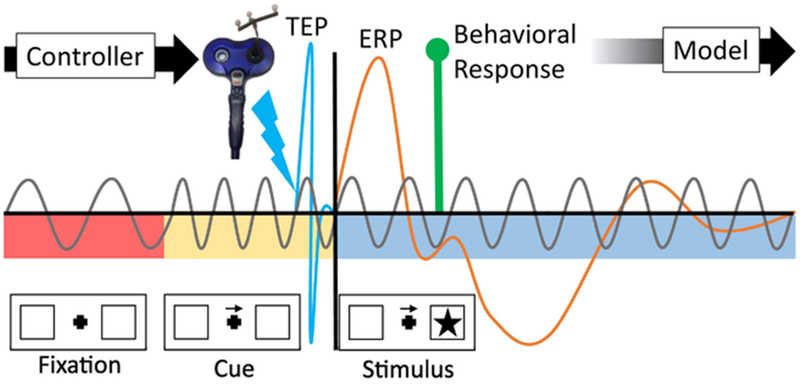
Personalizing TMS at the trial level. Cognitive events can be controlled to identify specific cognitive, behavioral, and neural signatures. In this schematic, a traditional Posner cued attention paradigm (Posner, 1980) proceeds from a fixation (red epoch) to a cue (yellow epoch) to a stimulus (blue epoch). In a TMS-EEG paradigm (for instance, if the TMS is administered to the frontal eye fields (Grosbras and Paus, 2002)), an algorithm could make use of the timing, spectral content, and amplitude of the TMS-evoked potential (TEP), ERP (and its sources []) due to the stimulus, and use data before or during the current stimulus to train a behavior-predictive statistical model. Ideally, these measures will be selected for their role in a mechanistic model, but can also be fitted when a good dynamic model is unavailable. The model can adaptively update over trials and use the controller (e.g., a computer script) to modify the timing, amplitude, frequency, and shape of TMS to improve performance on subsequent trials. If the statistically fitted model relates better to a given “ground truth” model for cued attention, it can help us distinguish among models that link the brain and behavior. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Some concepts from physics and engineering have already revealed principles that can guide our neuromodulation algorithms and forecasting models. For instance, cross-frequency coupling refers to the inter-frequency relationships between phases, phases and amplitudes, and amplitudes, and is thought to coordinate neural dynamics across spatiotemporal scales in the brain (Aru et al., 2015; Belluscio et al., 2012; Canolty and Knight, 2010). Cross-frequency coupling has been demonstrated in the hippocampus (Axmacher et al., 2010) and from scalp EEG during working memory maintenance (Friese et al., 2013), across dorsal and ventral streams during working memory performance (Popov et al., 2018), during reward processing in the nucleus accumbens (Cohen et al., 2009) and thalamo-cortical interactions based on attention demands (Saalmann et al., 2012). Cross-frequency phase-amplitude coupling often involves a high frequency signal (e.g., gamma, or >24 Hz) related to the phase of a much slower signal (e.g., theta, or 4–8 Hz) (Aru et al., 2015). Determining mechanistically significant cross-frequency relationships from spurious correlations is a challenging problem (Aru et al., 2015). However, noninvasive brain stimulation can entrain the phases of natural oscillations to influence behavior (Thut et al., 2011; Ozen et al., 2010; Raco et al., 2016; Spaak et al., 2014; Helfrich et al., 2014), suggesting that there is some mechanistic relevance for cross frequency coupling that could potentially be utilized to influence cognition4. More broadly, the repertoire of neural dynamics estimable at the source level (Makeig and Onton, 2011) may be highly useful in our models, and the simultaneous used of TMS may further support causal inferences for the roles of specific trial-level dynamics.
Linking TMS-induced performance within a task to clinical outcomes is crucial to evaluate the utility of a TMS approach. After a session that personalizes TMS with an experimental session, the individual’s clinical scores can be monitored over a longer timescale for clinically significant change. If a model fitting procedure leads to a successful clinical outcome, we can attempt to develop better forecasting models as an iterative process. As described above, the success of this strategy in a given individual may be moderated by their genotype, demographic status, and anatomical and functional network organization. The intuitions behind trial-wise adaptive TMS-EEG are generalizable to other paired cognitive, neuromodulatory, and brain monitoring approaches.
4. Technical frontiers
Significant scientific progress can occur using current techniques used in the appropriate combination. Using advanced imaging can provide individualized anatomical (Medaglia et al., 2018a; Bansal et al., 2018; Stiso et al., 2018; Khambhati et al., 2018) and functional (Kong et al., 2018; Bijsterbosch et al., 2018; Gordon et al., 2017a; Wang et al., 2015; Xu et al., 2016; Gordon et al., 2017b) network estimates. MEG and EEG both offer noninvasive fast signal recording. TMS coils can be designed to introduce deeper (Zangen et al., 2005) (but see Deng et al., 2014 ; Guadagnin et al., 2016), quieter (Peterchev et al., 2015), and more focal (Huh et al., 2018) stimulation. However, diffusion tractography algorithms produce connectome that lack ground truth and produce many systematic false positive tracts (Maier-Hein et al., 2017) and do not estimate fiber directionality (van den Heuvel et al., 2015). MEG is not cost-effective and can be difficult to configure with synchronous brain stimulation. As with MEG, EEG is susceptible to the source localization problem (Malioutov et al., 2005) and a relative lack of sensitivity to high frequency content, much of which is thought to be related to momentary cognitive performance (Delorme et al., 2015; Herrmann and Demiralp, 2005; Jerbi et al., 2009; Heusser et al., 2016; Schirrmeister et al., 2017; Delorme et al., 2015; Collins and Frank, 2018) and thus crucial to trial-level closed loop control. Thus, the effort to personalize neuromodulation should develop alongside ongoing efforts to improve the precision of neural measurements and stimulation devices.
5. Clinical trials as the gold standard for personalizing neuromodulation
Many of the general approaches to precision neuromodulation described above can be costly, time-intensive, and difficult to implement in clinical contexts. It is doubtless that many potential procedures will suffer limitations due to technology and sample characteristics. In addition, even if some within-subjects predictions and behavioral control are feasible, they might not be clinically useful due either to their expense, complexity, or minimal improvements on measures of patient outcomes. However, it is worth noting that precision approaches using control engineering in invasive contexts to treat Parkinson’s symptoms with deep brain stimulation have become part of routine care (Schiff, 2012). In addition, meta-analyses of randomized controlled trials for TMS in depression report a 13.6% remission rate relative to sham treatments (Berlim et al., 2014). Even if this number were only doubled to 27.2% of patients, it would be a substantial gain with real-world value. The costs of continuing to deliver standard of care at a 13.6% success rate should be weighed against investing in innovative precision procedures. In the end, new procedures should be tested in randomized controlled trials against approved practices to objectively determine the value for patients.
6. Conclusion
To personalize neuromodulation, we will need to confront hard problems using the tools available from cognitive neuroscience, neuropsychology, and systems engineering. Some approaches will only be possible with improvements in brain monitoring and stimulation technology. However, many approaches may be available to us given the right combination of tools and data for precise approaches. As a community, we should proceed with models that both fit and mechanistically forecast TMS responses, and move from cross-sectional predictions to truly personalized TMS procedures.
Acknowledgments
JDM acknowledges support from the Office of the Director at the National Institutes of Health and the National Institute of Mental Health through grant number DP5-OD-021352-01,R01-DC014960-01, and the Perelman School of Medicine.
Footnotes
Readers are referred to Valero-Cabré et al. (2017) for a comprehensive primer on principles of TMS.
Readers are referred to Medaglia et al. (2015), Bassett and Sporns (2017), and Rubinov and Sporns (2010) for primers on network analysis and Bassett et al. (2011), Blondel et al. (2008), Menichetti et al. (2014), Muldoon and Bassett (2016), and De Domenico et al. (2016) for examples of “multiplex” or “multilayer” graphs in human brain networks.
Consider also psychosocial treatments for phobia, where treatments are efficacious because the momentary stimulus-fear associations are known and can be manipulated for clinical gains.
Other measures of time series dynamics (e.g., multiscale entropy, Costa et al., 2002 ; nonstationarity, Cao and Slobounov, 2011; Kennel, 1997) have been explored in clinical and experimental settings, perhaps offering similar opportunities for model fitting and mechanism discovery.
References
- Adalı T, Trussell HJ, Hansen LK, Calhoun VD, 2018. The dangers of following trends in research: sparsity and other examples of hammers in search of nails. Proc. IEEE 106 (6), 1014–1018. [Google Scholar]
- Aleman A, Sommer IE, Kahn RS, 2007. Efficacy of slow repetitive transcranial magnetic stimulation in the treatment of resistant auditory hallucinations in schizophrenia: a meta-analysis. J. Clin. Psychiatry 68 (3), 416–421. [DOI] [PubMed] [Google Scholar]
- Amunts K, Schleicher A, Bürgel U, Mohlberg H, Uylings HB, Zilles K, 1999. Broca’s region revisited: cytoarchitecture and intersubject variability. J. Comp. Neurol 412 (2), 319–341. [DOI] [PubMed] [Google Scholar]
- Arlot S, Celisse A, et al. , 2010. A survey of cross-validation procedures for model selection. Stat. Surv 4, 40–79. [Google Scholar]
- Aru J, Aru J, Priesemann V, Wibral M, Lana L, Pipa G, Singer W, Vicente R, 2015. Untangling cross-frequency coupling in neuroscience. Curr. Opin. Neurobiol 31, 51–61. [DOI] [PubMed] [Google Scholar]
- Axmacher N, Henseler MM, Jensen O, Weinreich I, Elger CE, Fell J, 2010. Cross-frequency coupling supports multi-item working memory in the human hippocampus. Proc. Natl. Acad. Sci 200911531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DL, Maisey MN, Townsend DW, Valk PE, 2005. Positron Emission Tomography. Springer. [Google Scholar]
- Bansal K, Medaglia JD, Bassett DS, Vettel JM, Muldoon SF, 2018. Data-driven Brain Network Models Predict Individual Variability in Behavior. arXiv preprint arXiv: 1802.08747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Jones DK, 2002. Diffusion-tensor MRI: theory, experimental design and data analysis-a technical review. NMR Biomed. Int. J. Devoted Dev. Appl. Magn. Reson. Vivo 15 (7-8), 456–467. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Sporns O, 2017. Network neuroscience. Nat. Neurosci 20 (3), 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST, 2011. Dynamic reconfiguration of human brain networks during learning. Proc. Natl. Acad. Sci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Zurn P, Gold JI, 2018. On the nature and use of models in network neuroscience. Nat. Rev. Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M, Rubens MT, Johnstone T, 2017. Simultaneous EEG-FMRI reveals attention-dependent. Proc. Natl. Acad. Sci 104 (26), 11073–11078. [Google Scholar]
- Becker CO, Pequito S, Pappas GJ, Miller MB, Grafton ST, Bassett DS, Preciado VM, 2018. Spectral mapping of brain functional connectivity from diffusion imaging. Sci. Rep 8 (1), 1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Mackay CE, Filippini N, Smith SM, 2009. Group comparison of resting-state FMRI data using multi-subject ICA and dual regression. Neuroimage 47 (Suppl 1), S148. [Google Scholar]
- Belluscio MA, Mizuseki K, Schmidt R, Kempter R, Buzsáki G, 2012. Cross-frequency phase-phase coupling between theta and gamma oscillations in the hippocampus. J. Neurosci 32 (2), 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berényi A, Belluscio M, Mao D, Buzsáki G, 2012. Closed-loop control of epilepsy by transcranial electrical stimulation. Science 337 (6095), 735–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlim M, Van den Eynde F, Tovar-Perdomo S, Daskalakis Z, 2014. Response, remission and drop-out rates following high-frequency repetitive transcranial magnetic stimulation (RTMS) for treating major depression: a systematic review and meta-analysis of randomized, double-blind and sham-controlled trials. Psychol. Med 44 (2), 225–239. [DOI] [PubMed] [Google Scholar]
- Bijsterbosch JD, Woolrich MW, Glasser MF, Robinson EC, Beckmann CF, Van Essen DC, Harrison SJ, Smith SM, 2018. The relationship between spatial configuration and functional connectivity of brain regions. Elife 7, e32992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E, 2008. Fast unfolding of communities in large networks. J. Stat. Mech: Theory Exp. 2008 (10), P10008. [Google Scholar]
- Bonato C, Miniussi C, Rossini P, 2006. Transcranial magnetic stimulation and cortical evoked potentials: a TMS/EEG co-registration study. Clin. Neurophysiol 117 (8), 1699–1707. [DOI] [PubMed] [Google Scholar]
- Breakspear M, 2017. Dynamic models of large-scale brain activity. Nat. Neurosci 20 (3), 340. [DOI] [PubMed] [Google Scholar]
- Breiman L, et al. , 2001. Statistical modeling: the two cultures (with comments and a rejoinder by the author). Stat. Sci 16 (3), 199–231. [Google Scholar]
- Burt T, Lisanby SH, Sackeim HA, 2002. Neuropsychiatric applications of transcranial magnetic stimulation: a meta analysis. Int. J. Neuropsychopharmacol 5 (1), 73–103. [DOI] [PubMed] [Google Scholar]
- Canolty RT, Knight RT, 2010. The functional role of cross-frequency coupling. Trends Cogn. Sci. 14 (11), 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Slobounov S, 2011. Application of a novel measure of eeg non-stationarity as ‘Shannon-entropy of the peak frequency shifting’ for detecting residual abnormalities in concussed individuals. Clin. Neurophysiol 122 (7), 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Oathes DJ, Chang C, Bradley T, Zhou Z-W, Williams LM, Glover GH, Deisseroth K, Etkin A, 2013. Causal interactions between fronto-parietal central executive and default-mode networks in humans. Proc. Natl. Acad. Sci 110 (49), 19944–19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-T, 1998. Linear System Theory and Design. Oxford University Press, Inc.. [Google Scholar]
- Chou Y.-h., Hickey PT, Sundman M, Song AW, Chen N.-k., 2015. Effects of repetitive transcranial magnetic stimulation on motor symptoms in Parkinson disease: a systematic review and meta-analysis. JAMA Neurol. 72 (4), 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NU, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE, 2008. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage 41 (1), 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MX, Axmacher N, Lenartz D, Elger CE, Sturm V, Schlaepfer TE, 2009. Good vibrations: cross-frequency coupling in the human nucleus accumbens during reward processing. J. Cogn. Neurosci 21 (5), 875–889. [DOI] [PubMed] [Google Scholar]
- Collins AG, Frank MJ, 2018. Within-and across-trial dynamics of human EEG reveal cooperative interplay between reinforcement learning and working memory. Proc. Natl. Acad. Sci 201720963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Goldberger AL, Peng C-K, 2002. Multiscale entropy analysis of complex physiologic time series. Phys. Rev. Lett 89 (6), 068102. [DOI] [PubMed] [Google Scholar]
- De Domenico M, Sasai S, Arenas A, 2016. Mapping multiplex hubs in human functional brain networks. Front. Neurosci 10, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme A, Makeig S, 2004. Eeglab: an open source toolbox for analysis of single-trial eeg dynamics including independent component analysis. J. Neurosci. Methods 134 (1), 9–21. [DOI] [PubMed] [Google Scholar]
- Delorme A, Miyakoshi M, Jung T-P, Makeig S, 2015. Grand average ERP-image plotting and statistics: a method for comparing variability in event-related single-trial EEG activities across subjects and conditions. J. Neurosci. Methods 250, 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme A, Mullen T, Kothe C, Acar ZA, Bigdely-Shamlo N, Vankov A, Makeig S, 2011. Eeglab, sift, nft, bcilab, and erica: new tools for advanced EEG processing. Comput. Intell. Neurosci 2011, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z-D, Lisanby SH, Peterchev AV, 2014. Coil design considerations for deep transcranial magnetic stimulation. Clin. Neurophysiol 125 (6), 1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. , 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31 (3), 968–980. [DOI] [PubMed] [Google Scholar]
- Dowdle LT, Brown TR, George MS, Hanlon CA, 2018. Single pulse TMS to the dlpfc, compared to a matched sham control, induces a direct, causal increase in caudate, cingulate, and thalamic bold signal. Brain Stimul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Fetcho RN, Zebley B, Oathes DJ, Etkin A, et al. , 2017. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat. Med 23 (1), 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J, Adolphs R, 2016. Building a science of individual differences from FMRI. Trends Cogn. Sci. 20 (6), 425–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durston S, Pol HEH, Casey B, Giedd JN, Buitelaar JK, Van Engeland H, 2001. Anatomical MRI of the developing human brain: what have we learned?. J. Am. Acad. Child Adolesc. Psychiatry 40 (9), 1012–1020. [DOI] [PubMed] [Google Scholar]
- Dyke K, Kim S, Jackson GM, Jackson SR, 2018. Reliability of single and paired pulse transcranial magnetic stimulation parameters across eight testing sessions. Brain Stimul. Basic Transl. Clin. Res. Neuromodulation [DOI] [PubMed] [Google Scholar]
- Esterman M, Thai M, Okabe H, DeGutis J, Saad E, Laganiere SE, Halko MA, 2017. Network-targeted cerebellar transcranial magnetic stimulation improves attentional control. NeuroImage 156, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, McCandliss BD, Fossella J, Flombaum JI, Posner MI, 2005. The activation of attentional networks. Neuroimage 26 (2), 471–479. [DOI] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT, 2015. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci 18 (11), 1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AJ, Medaglia JD, Jeronimus BF, 2018. Lack of group-to-individual generalizability is a threat to human subjects research. Proc. Natl. Acad. Sci 201711978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Buckner RL, Liu H, Chakravarty MM, Lozano AM, Pascual-Leone A, 2014. Resting-state networks link invasive and noninvasive brain stimulation across diverse psychiatric and neurological diseases. Proc. Natl. Acad. Sci 111 (41), E4367–E4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Liu H, Pascual-Leone A, 2013. Identification of reproducible individualized targets for treatment of depression with TMS based on intrinsic connectivity. NeuroImage 66, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke AG, Bagusat C, Rust S, Engel A, Lieb K, 2014. Substances used and prevalence rates of pharmacological cognitive enhancement among healthy subjects. Eur. Arch. Psychiatry Clin. Neurosci. 264 (1), 83–90. [DOI] [PubMed] [Google Scholar]
- Friese U, Köster M, Hassler U, Martens U, Trujillo-Barreto N, Gruber T, 2013. Successful memory encoding is associated with increased cross-frequency coupling between frontal theta and posterior gamma oscillations in human scalp-recorded EEG. NeuroImage 66, 642–647. [DOI] [PubMed] [Google Scholar]
- Fröhlich F, 2015. Experiments and models of cortical oscillations as a target for noninvasive brain stimulation In: Progress in Brain Research. 222, Elsevier, pp. 41–73. [DOI] [PubMed] [Google Scholar]
- Gazzaniga MS, 2004. The Cognitive Neurosciences. MIT Press. [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. , 2016. A multi-modal parcellation of human cerebral cortex. Nature 536 (7615), 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollo LL, Roberts JA, Cocchi L, 2017. Mapping how local perturbations influence systems-level brain dynamics. NeuroImage 160, 97–112. [DOI] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Gilmore AW, Nelson SM, Dosenbach NU, Petersen SE, 2017. Individual-specific features of brain systems identified with resting state functional correlations. NeuroImage 146, 918–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, Ortega M, Hoyt-Drazen C, Gratton C, Sun H, et al. , 2017. Precision functional mapping of individual human brains. Neuron 95 (4), 791–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C, Laumann TO, Nielsen AN, Greene DJ, Gordon EM, Gilmore AW, Nelson SM, Coalson RS, Snyder AZ, Schlaggar BL, et al. , 2018. Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron 98 (2), 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffa A, Ricaud B, Benzi K, Bresson X, Daducci A, Vandergheynst P, Thiran J-P, Hagmann P, 2017. Transient networks of spatio-temporal connectivity map communication pathways in brain functional systems. NeuroImage 155, 490–502. [DOI] [PubMed] [Google Scholar]
- Grosbras M-H, Paus T, 2002. Transcranial magnetic stimulation of the human frontal eye field: effects on visual perception and attention. J. Cogn. Neurosci 14 (7), 1109–1120. [DOI] [PubMed] [Google Scholar]
- Guadagnin V, Parazzini M, Fiocchi S, Liorni I, Ravazzani P, 2016. Deep transcranial magnetic stimulation: modeling of different coil configurations. IEEE Trans. Biomed. Eng. 63 (7), 1543–1550. [DOI] [PubMed] [Google Scholar]
- Helfrich RF, Schneider TR, Rach S, Trautmann-Lengsfeld SA, Engel AK, Herrmann CS, 2014. Entrainment of brain oscillations by transcranial alternating current stimulation. Curr. Biol 24 (3), 333–339. [DOI] [PubMed] [Google Scholar]
- Hermundstad AM, Bassett DS, Brown KS, Aminoff EM, Clewett D, Freeman S, Frithsen A, Johnson A, Tipper CM, Miller MB, et al. , 2013. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc. Natl. Acad. Sci 110 (15), 6169–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermundstad AM, Brown KS, Bassett DS, Aminoff EM, Frithsen A, Johnson A, Tipper CM, Miller MB, Grafton ST, Carlson JM, 2014. Structurally-constrained relationships between cognitive states in the human brain. PLoS Comput. Biol. 10 (5), e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C, Demiralp T, 2005. Human EEG gamma oscillations in neuropsychiatric disorders. Clin. Neurophysiol 116 (12), 2719–2733. [DOI] [PubMed] [Google Scholar]
- Heusser AC, Poeppel D, Ezzyat Y, Davachi L, 2016. Episodic sequence memory is supported by a theta-gamma phase code. Nat. Neurosci 19 (10), 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, McCandliss BD, Black JM, Gantman A, Zakerani N, Hulme C, Lyytinen H, Whitfield-Gabrieli S, Glover GH, Reiss AL, et al. , 2011. Neural systems predicting long-term outcome in dyslexia. Proc. Natl. Acad. Sci 108 (1), 361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoexter MQ, Miguel EC, Diniz JB, Shavitt RG, Busatto GF, Sato JR, 2013. Predicting obsessive-compulsive disorder severity combining neuroimaging and machine learning methods. J. Affect. Disord 150 (3), 1213–1216. [DOI] [PubMed] [Google Scholar]
- Honnorat N, Satterthwaite T, Gur R, Gur R, Davatzikos C, 2017. sgrasp: a graph-based method for the derivation of subject-specific functional parcellations of the brain. J. Neurosci. Methods 277, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S-H, Mullen TR, Jung T-P, Cauwenberghs G, 2016. Real-time adaptive EEG source separation using online recursive independent component analysis. IEEE Trans. Neural Syst. Rehabil. Eng. 24 (3), 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Liu AA, Lafon B, Friedman D, Dayan M, Wang X, Bikson M, Doyle WK, Devinsky O, Parra LC, 2017. Measurements and models of electric fields in the in vivo human brain during transcranial electric stimulation. Elife 6, e18834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh Y, Jung D, Seo T, Sun S, Kim SH, Rhim H, Chung S, Kim C-H, Kwon Y, Bikson M, et al. , 2018. Brain stimulation patterns emulating endogenous thalamocortical input to parvalbumin-expressing interneurons reduce nociception in mice. Brain Stimul. Basic Transl. Clin. Res. Neuromodulation [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illari PM, Williamson J, 2012. What is a mechanism? Thinking about mechanisms across the sciences. Eur. J. Philos. Sci 2 (1), 119–135. [Google Scholar]
- Insel TR, Cuthbert BN, 2015. Brain disorders? Precisely. Science 348 (6234), 499–500. [DOI] [PubMed] [Google Scholar]
- Jerbi K, Ossandon T, Hamame CM, Senova S, Dalal SS, Jung J, Minotti L, Bertrand O, Berthoz A, Kahane P, et al. , 2009. Task-related gamma-band dynamics from an intracerebral perspective: review and implications for surface EEG and MEG. Hum. Brain Mapp. 30 (6), 1758–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman R, 1959. On the general theory of control systems. IRE Trans. Autom. Control. 4 (3), 110–110. [Google Scholar]
- Kennel MB, 1997. Statistical test for dynamical nonstationarity in observed time-series data. Phys. Rev. E 56 (1), 316. [Google Scholar]
- Khambhati AN, Kahn AE, Costantini J, Ezzyat Y, Solomon EA, Gross RE, Jobst BC, Sheth SA, Zaghloul KA, Worrell G, et al. , 2018. Predictive control of electrophysiological network architecture using direct, single-node neurostimulation in humans. Biorxiv 292748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleim JA, Chan S, Pringle E, Schallert K, Procaccio V, Jimenez R, Cramer SC, 2006. Bdnf val66met polymorphism is associated with modified experience-dependent plasticity in human motor cortex. Nat. Neurosci 9 (6), 735. [DOI] [PubMed] [Google Scholar]
- Kong R, Li J, Orban C, Sabuncu MR, Liu H, Schaefer A, Sun N, Zuo X-N, Holmes AJ, Eickhoff SB, et al. , 2018. Spatial topography of individual-specific cortical networks predicts human cognition, personality, and emotion. Cereb. Cortex [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus D, Naros G, Bauer R, Khademi F, Le ao MT, Ziemann U, Gharabaghi A, 2016. Brain state-dependent transcranial magnetic closed-loop stimulation controlled by sensorimotor desynchronization induces robust increase of corticospinal excitability. Brain Stimul. 9 (3), 415–424. [DOI] [PubMed] [Google Scholar]
- Langley P, Laird JE, Rogers S, 2009. Cognitive architectures: research issues and challenges. Cogn. Syst. Res 10 (2), 141–160. [Google Scholar]
- Langs G, Wang D, Golland P, Mueller S, Pan R, Sabuncu MR, Sun W, Li K, Liu H, 2016. Identifying shared brain networks in individuals by decoupling functional and anatomical variability. Cereb. Cortex 26 (10), 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen M-Y, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NU, et al. , 2015. Functional system and areal organization of a highly sampled individual human brain. Neuron 87 (3), 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezak MD, Howieson DB, Loring DW, Fischer JS, 2004. Neuropsychological Assessment. Oxford University Press, USA. [Google Scholar]
- Liang H, Wang H, 2017. Structure-function network mapping and its assessment via persistent homology. PLoS Comput. Biol. 13 (1), e1005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Alonso V, Cheeran B, Río-Rodríguez D, Fernández-del Olmo M, 2014. Inter-individual variability in response to non-invasive brain stimulation paradigms. Brain Stimul. 7 (3), 372–380. [DOI] [PubMed] [Google Scholar]
- Luber BM, Davis S, Bernhardt E, Neacsiu A, Kwapil L, Lisanby SH, Strauman TJ, 2017. Using neuroimaging to individualize tms treatment for depression: toward a new paradigm for imaging-guided intervention. NeuroImage 148, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CJ, Breeden A, Gordon EM, Cherry JB, Turkeltaub PE, Vaidya CJ, 2018. Precision inhibitory stimulation of individual-specific cortical hubs disrupts information processing in humans. Biorxiv 254417. [DOI] [PubMed] [Google Scholar]
- Maier-Hein KH, Neher PF, Houde J-C, Côté M-A, Garyfallidis E, Zhong J, Chamberland M, Yeh F-C, Lin Y-C, Ji Q, et al. , 2017. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun 8 (1), 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeig S, Debener S, Onton J, Delorme A, 2004. Mining event-related brain dynamics. Trends Cogn. Sci. 8 (5), 204–210. [DOI] [PubMed] [Google Scholar]
- Makeig S, Onton J, 2011. ERP features and EEG dynamics: an ICA perspective In: Luck S, Kappenman E (Eds.), Oxford Handbook of Event-related Potential Components. pp. 51–88. [Google Scholar]
- Malaguti A, Rossini D, Lucca A, Magri L, Lorenzi C, Pirovano A, Colombo C, Smeraldi E, Zanardi R, 2011. Role of comt, 5-ht1a, and sert genetic polymorphisms on antidepressant response to transcranial magnetic stimulation. Depress. Anxiety 28 (7), 568–573. [DOI] [PubMed] [Google Scholar]
- Malioutov D, Cetin M, Willsky AS, 2005. A sparse signal reconstruction perspective for source localization with sensor arrays. IEEE Trans. Signal Process. 53 (8), 3010–3022. [Google Scholar]
- Månsson KN, Frick A, Boraxbekk C-J, Marquand A, Williams S, Carlbring P, Andersson G, Furmark T, 2015. Predicting long-term outcome of internet-delivered cognitive behavior therapy for social anxiety disorder using fmri and support vector machine learning. Transl. Psychiatry 5 (3), e530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, et al. , 2001. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos. Trans. R. Soc. Lond. B Biol. Sci. 356 (1412), 1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaglia JD, Harvey DY, White N, Kelkar A, Zimmerman J, Bassett DS, Hamilton RH, 2018. Network controllability in the inferior frontal gyrus relates to controlled language variability and susceptibility to tms. J. Neurosci 17–0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaglia JD, Huang W, Karuza EA, Kelkar A, Thompson-Schill SL, Ribeiro A, Bassett DS, 2018. Functional alignment with anatomical networks is associated with cognitive flexibility. Nat. Hum. Behav 2 (2), 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaglia JD, Lynall M-E, Bassett DS, 2015. Cognitive network neuroscience. J. Cogn. Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaglia JD, Ramanathan DM, Venkatesan UM, Hillary FG, 2011. The challenge of non-ergodicity in network neuroscience. Netw. Comput. Neural Syst. 22 (1-4), 148–153. [DOI] [PubMed] [Google Scholar]
- Menichetti G, Remondini D, Panzarasa P, Mondragón RJ, Bianconi G, 2014. Weighted multiplex networks. PLoS One 9 (6), e97857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill RD, Ito T, Cole MW, 2017. From connectome to cognition: the search for mechanism in human functional brain networks. NeuroImage [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar PC, 2004. A manifesto on psychology as idiographic science: bringing the person back into scientific psychology, this time forever. Measurement 2 (4), 201–218. [Google Scholar]
- Morbidi F, Garulli A, Prattichizzo D, Rizzo C, Manganotti P, Rossi S, 2007. Off-line removal of tms-induced artifacts on human electroencephalography by Kalman filter. J. Neurosci. Methods 162 (1-2), 293–302. [DOI] [PubMed] [Google Scholar]
- Muldoon SF, Bassett DS, 2016. Network and multilayer network approaches to understanding human brain dynamics. Philos. Sci 83 (5), 710–720. [Google Scholar]
- Muldoon SF, Pasqualetti F, Gu S, Cieslak M, Grafton ST, Vettel JM, Bassett DS, 2016. Stimulation-based control of dynamic brain networks. PLoS Comput. Biol. 12 (9), e1005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen T, Kothe C, Chi YM, Ojeda A, Kerth T, Makeig S, Cauwenberghs G, Jung T-P, 2013. Real-time modeling and 3d visualization of source dynamics and connectivity using wearable EEG. In: Engineering in Medicine and Biology Society (EMBC), 2013 35th Annual International Conference of the IEEE pp. 2184–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TR, Kothe CA, Chi YM, Ojeda A, Kerth T, Makeig S, Jung T-P, Cauwenberghs G, 2015. Real-time neuroimaging and cognitive monitoring using wearable dry eeg. IEEE Trans. Biomed. Eng. 62 (11), 2553–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrabadi NM, 2007. Pattern recognition and machine learning. J. Electron. Imaging 16 (4), 049901. [Google Scholar]
- Ngandu T, Lehtisalo J, Solomon A, Levälahti E, Ahtiluoto S, Antikainen R, Bäckman L, Hänninen T, Jula A, Laatikainen T, et al. , 2015. A 2year multidomain intervention of diet, exercise, cognitive training, and vascular risk monitoring versus control to prevent cognitive decline in at-risk elderly people (finger): a randomised controlled trial. Lancet 385 (9984), 2255–2263. [DOI] [PubMed] [Google Scholar]
- Nouretdinov I, Costafreda SG, Gammerman A, Chervonenkis A, Vovk V, Vapnik V, Fu CH, 2011. Machine learning classification with confidence: application of transductive conformal predictors to mri-based diagnostic and prognostic markers in depression. NeuroImage 56 (2), 809–813. [DOI] [PubMed] [Google Scholar]
- Nummenmaa A, McNab JA, Savadjiev P, Okada Y, Hämäläinen MS, Wang R, Wald LL, Pascual-Leone A, Wedeen VJ, Raij T, 2014. Targeting of white matter tracts with transcranial magnetic stimulation. Brain Stimul. 7 (1), 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz A, Fox MD, Craddock RC, Colcombe S, Milham MP, 2016. An integrated framework for targeting functional networks via transcranial magnetic stimulation. NeuroImage 127, 86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz A, Legon W, Rowlands A, Bickel WK, Paulus W, Tyler WJ, 2013. Physiological observations validate finite element models for estimating subject-specific electric field distributions induced by transcranial magnetic stimulation of the human motor cortex. NeuroImage 81, 253–264. [DOI] [PubMed] [Google Scholar]
- Ozen S, Sirota A, Belluscio MA, Anastassiou CA, Stark E, Koch C, Buzsáki G, 2010. Transcranial electric stimulation entrains cortical neuronal populations in rats. J. Neurosci 30 (34), 11476–11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Gandal MJ, Geschwind DH, 2015. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet 16 (8), 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Leone A, Walsh V, Rothwell J, 2000. Transcranial magnetic stimulation in cognitive neuroscience-virtual lesion, chronometry, and functional connectivity. Curr. Opin. Neurobiol 10 (2), 232–237. [DOI] [PubMed] [Google Scholar]
- Peterchev AV, Murphy DL, Goetz SM, 2015. Quiet transcranial magnetic stimulation: status and future directions. In: Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE pp. 226–229. [DOI] [PubMed] [Google Scholar]
- Petersen SE, Sporns O, 2015. Brain networks and cognitive architectures. Neuron 88 (1), 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfurtscheller G, Da Silva FL, 1999. Event-related EEG/MEG synchronization and desynchronization: basic principles. Clin. Neurophysiol 110 (11), 1842–1857. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen M-Y, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, et al. , 2015. Long-term neural and physiological phenotyping of a single human. Nat. Commun 6, 8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov T, Jensen O, Schoffelen J-M, 2018. Dorsal and ventral cortices are coupled by cross-frequency interactions during working memory. NeuroImage [DOI] [PubMed] [Google Scholar]
- Posner MI, 1980. Orienting of attention. Q. J. Exp. Psychol 32 (1), 3–25. [DOI] [PubMed] [Google Scholar]
- Posner MI, Rothbart MK, 2007. Research on attention networks as a model for the integration of psychological science. Annu. Rev. Psychol 58, 1–23. [DOI] [PubMed] [Google Scholar]
- Raco V, Bauer R, Tharsan S, Gharabaghi A, 2016. Combining tms and tacs for closed-loop phase-dependent modulation of corticospinal excitability: a feasibility study. Front. Cell. Neurosci 10, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Verstrepen KJ, 2007. Timescales of genetic and epigenetic inheritance. Cell 128 (4), 655–668. [DOI] [PubMed] [Google Scholar]
- Rogasch NC, Fitzgerald PB, 2013. Assessing cortical network properties using TMS-EEG. Hum. Brain Mapp. 34 (7), 1652–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogasch NC, Thomson RH, Farzan F, Fitzgibbon BM, Bailey NW, Hernandez-Pavon JC, Daskalakis ZJ, Fitzgerald PB, 2014. Removing artefacts from TMS-EEG recordings using independent component analysis: importance for assessing prefrontal and motor cortex network properties. NeuroImage 101, 425–439. [DOI] [PubMed] [Google Scholar]
- Rogasch NC, Thomson RH, Farzan F, Fitzgibbon BM, Bailey NW, Hernandez-Pavon JC, Daskalakis ZJ, Fitzgerald PB, 2014. Removing artefacts from TMS-EEG recordings using independent component analysis: importance for assessing prefrontal and motor cortex network properties. NeuroImage 101, 425–439. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O, 2010. Complex network measures of brain connectivity: uses and interpretations. NeuroImage 52 (3), 1059–1069. [DOI] [PubMed] [Google Scholar]
- Saalmann YB, Pinsk MA, Wang L, Li X, Kastner S, 2012. The pulvinar regulates information transmission between cortical areas based on attention demands. Science 337 (6095), 753–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack AT, Cohen Kadosh R, Schuhmann T, Moerel M, Walsh V, Goebel R, 2009. Optimizing functional accuracy of TMS in cognitive studies: a comparison of methods. J. Cogn. Neurosci 21 (2), 207–221. [DOI] [PubMed] [Google Scholar]
- Sack AT, Kohler A, Bestmann S, Linden DE, Dechent P, Goebel R, Baudewig J, 2007. Imaging the brain activity changes underlying impaired visuospatial judgments: simultaneous fmri, tms, and behavioral studies. Cereb. Cortex 17 (12), 2841–2852. [DOI] [PubMed] [Google Scholar]
- Schiff SJ, 2012. Neural Control Engineering: The Emerging Intersection Between Control Theory and Neuroscience. MIT Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilberg L, Schuhmann T, Sack AT, 2017. Interindividual variability and intraindividual reliability of intermittent theta burst stimulation-induced neuroplasticity mechanisms in the healthy brain. J. Cogn. Neurosci 29 (6), 1022–1032. [DOI] [PubMed] [Google Scholar]
- Schirrmeister RT, Springenberg JT, Fiederer LDJ, Glasstetter M, Eggensperger K, Tangermann M, Hutter F, Burgard W, Ball T, 2017. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 38 (11), 5391–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CE, Leach JB, 2003. Neural tissue engineering: strategies for repair and regeneration. Annu. Rev. Biomed. Eng 5 (1), 293–347. [DOI] [PubMed] [Google Scholar]
- Schultz AP, Chhatwal JP, Huijbers W, Hedden T, Van Dijk KR, McLaren DG, Ward AM, Wigman S, Sperling RA, 2014. Template based rotation: a method for functional connectivity analysis with a priori templates. NeuroImage 102, 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutter D, 2009. Antidepressant efficacy of high-frequency transcranial magnetic stimulation over the left dorsolateral prefrontal cortex in double-blind sham-controlled designs: a meta-analysis. Psychol. Med 39 (1), 65–75. [DOI] [PubMed] [Google Scholar]
- Scott J, 2017. Social Network Analysis. Sage. [Google Scholar]
- Seghier ML, Price CJ, 2018. Interpreting and utilising intersubject variability in brain function. Trends Cogn. Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi H, Takeuchi S, Kadota H, Kohno Y, Nakajima Y, 2011. TMS-induced artifacts on eeg can be reduced by rearrangement of the electrode’s lead wire before recording. Trends Cogn. Sci. 122 (5), 984–990. [DOI] [PubMed] [Google Scholar]
- Silvanto J, Muggleton NG, Cowey A, Walsh V, 2007. Neural adaptation reveals state-dependent effects of transcranial magnetic stimulation. Eur. J. Neurosci 25 (6), 1874–1881. [DOI] [PubMed] [Google Scholar]
- Silvanto J, Pascual-Leone A, 2008. State-dependency of transcranial magnetic stimulation. Brain Topogr. 21 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotema CW, Dirk Blom J, Hoek HW, Sommer IE, 2010. Should we expand the toolbox of psychiatric treatment methods to include repetitive transcranial magnetic stimulation (rtms)? A meta-analysis of the efficacy of RTMS in psychiatric disorders. J. Clin. Psychiatry 71 (7), 873. [DOI] [PubMed] [Google Scholar]
- Smith SM, 2004. Overview of fmri analysis. Br. J. Radiol 77 (suppl_2), S167–S175. [DOI] [PubMed] [Google Scholar]
- Snowball A, Tachtsidis I, Popescu T, Thompson J, Delazer M, Zamarian L, Zhu T, Kadosh RC, 2013. Long-term enhancement of brain function and cognition using cognitive training and brain stimulation. Curr. Biol 23 (11), 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M, Kamm T, Tergau F, Ulm G, Paulus W, 2002. Repetitive paired-pulse transcranial magnetic stimulation affects corticospinal excitability and finger tapping in Parkinson’s disease. Clin. Neurophysiol 113 (6), 944–950. [DOI] [PubMed] [Google Scholar]
- Sompolinsky H, 2014. Computational neuroscience: beyond the local circuit. Curr. Opin. Neurobiol 25, xiii–xviii. [DOI] [PubMed] [Google Scholar]
- Spaak E, de Lange FP, Jensen O, 2014. Local entrainment of alpha oscillations by visual stimuli causes cyclic modulation of perception. J. Neurosci 34 (10), 3536–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R, 2005. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1 (4), e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Schlagenhauf F, Huys QJ, Raman S, Aponte EA, Brodersen KH, Rigoux L, Moran RJ, Daunizeau J, Dolan RJ, et al. , 2017. Computational neuroimaging strategies for single patient predictions. NeuroImage 145, 180–199. [DOI] [PubMed] [Google Scholar]
- Stiso J, Khambhati AN, Menara T, Kahn AE, Stein JM, Das SR, Gorniak R, Tracy J, Litt B, Davis KA, et al. , 2018. White Matter Network Architecture Guides Direct Electrical Stimulation Through Optimal State Transitions. arXiv preprint arXiv: 1805.01260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney KT, Ward TE, McLoone SF, 2012. Artifact removal in physiological signals-practices and possibilities. IEEE Trans. Inf. Technol. Biomed. 16 (3), 488–500. [DOI] [PubMed] [Google Scholar]
- Thielscher A, Antunes A, Saturnino GB, 2015. Field modeling for transcranial magnetic stimulation: a useful tool to understand the physiological effects of TMS?. In: Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE pp. 222–225. [DOI] [PubMed] [Google Scholar]
- Thomas Yeo B, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, et al. , 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol 106 (3), 1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thut G, Miniussi C, 2009. New insights into rhythmic brain activity from TMS-EEG studies. Trends Cogn. Sci. 13 (4), 182–189. [DOI] [PubMed] [Google Scholar]
- Thut G, Veniero D, Romei V, Miniussi C, Schyns P, Gross J, 2011. Rhythmic TMS causes local entrainment of natural oscillatory signatures. Curr. Biol 21 (14), 1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T, Aganj I, Ge T, Polimeni JR, Fischl B, 2017. Functional density and edge maps: characterizing functional architecture in individuals and improving cross-subject registration. NeuroImage 158, 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero-Cabré A, Amengual J, Stengel C, Pascual-Leone A, Coubard OA, 2017. Transcranial magnetic stimulation in basic and clinical neuroscience: a comprehensive review of fundamental principles and novel insights. Neurosci. Biobehav. Rev [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, de Reus MA, Feldman Barrett L, Scholtens LH, Coopmans FM, Schmidt R, Preuss TM, Rilling JK, Li L, 2015. Comparison of diffusion tractography and tract-tracing measures of connectivity strength in rhesus macaque connectome. Hum. Brain Mapp. 36 (8), 3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Maas HL, Molenaar PC, 1992. Stagewise cognitive development: an application of catastrophe theory. Psychol. Rev 99 (3), 395. [DOI] [PubMed] [Google Scholar]
- Varoquaux G, Thirion B, 2014. How machine learning is shaping cognitive neuroimaging. GigaScience 3 (1), 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, Langs G, Pan R, Qian T, Li K, et al. , 2015. Parcellating cortical functional networks in individuals. Nat. Neurosci 18 (12), 1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang X, Scheich H, 1996. LTD and LTP induced by transcranial magnetic stimulation in auditory cortex. Neuroreport 7 (2), 521–525. [DOI] [PubMed] [Google Scholar]
- Whelan R, Garavan H, 2014. When optimism hurts: inflated predictions in psychiatric neuroimaging. Biol. Psychiatry 75 (9), 746–748. [DOI] [PubMed] [Google Scholar]
- Wu W, Keller CJ, Rogasch NC, Longwell P, Shpigel E, Rolle CE, Etkin A, 2018. Artist: a fully automated artifact rejection algorithm for single-pulse TMS-EEG data. Hum. Brain Mapp. 39 (4), 1607–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Opitz A, Craddock RC, Wright MJ, Zuo X-N, Milham MP, 2016. Assessing variations in areal organization for the intrinsic brain: from fingerprints to reliability. Cereb. Cortex 26 (11), 4192–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Westfall J, 2017. Choosing prediction over explanation in psychology: lessons from machine learning. Perspect. Psychol. Sci 12 (6), 1100–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangen A, Roth Y, Voller B, Hallett M, 2005. Transcranial magnetic stimulation of deep brain regions: evidence for efficacy of the h-coil. Clin. Neurophysiol 116 (4), 775–779. [DOI] [PubMed] [Google Scholar]
- Zrenner C, Belardinelli P, Desideri D, Ziemann U, 2017. Phase of brain oscillations determines the direction of induced plasticity in real-time EEG-triggered TMS. Brain Stimul. 2 (10), 429. [Google Scholar]


