Abstract
Purpose:
To develop a subspace learning method for the recently proposed subspace-based MRSI approach known as SPICE, and achieve ultrafast 1H-MRSI of the brain.
Theory and Methods:
A novel strategy is formulated to learn a low-dimensional subspace representation of MR spectra from specially acquired training data and use the learned subspace for general MRSI experiments. Specifically, the subspace learning problem is formulated as learning “empirical” distributions of molecule-specific spectral parameters (e.g., concentrations, lineshapes and frequency shifts) by integrating physics-based model and the training data. The learned spectral parameters and quantum mechanical simulation basis can then be combined to construct acquisition-specific subspace for spatiospectral encoding and processing. High-resolution MRSI acquisitions combining ultrashort-TE/short-TR excitation, sparse sampling, and the elimination of water suppression have been performed to evaluate the feasibility of the proposed method.
Results:
The accuracy of the learned subspace and the capability of the proposed method in producing high-resolution 3D 1H metabolite maps and high-quality spatially-resolved spectra (with a nominal resolution of ~2. 4×2.4×3 mm3 in 5 minutes) were demonstrated using phantom and in vivo studies. By eliminating water suppression, we are also able to extract valuable information from the water signals for data processing (B0 map, frequency drift, and coil sensitivity) as well as for mapping tissue susceptibility and relaxation parameters.
Conclusion:
The proposed method enables ultrafast 1H-MRSI of the brain using a learned subspace, eliminating the need of acquiring subject-dependent navigator data (known as ) in the original SPICE technique. It represents a new way to perform MRSI experiments and an important step towards practical applications of high-resolution MRSI.
Keywords: MR spectroscopic imaging, union-of-subspaces model, subspace learning, rapid spatiospectral encoding, no water suppression
INTRODUCTION
MRSI has been considered a potentially powerful in vivo molecular imaging modality, with the capability to detect and quantify various endogenous metabolites and neurotransmitters simultaneously (1–5). However, practical applications of MRSI have been hindered by several fundamental technical hurdles, including the inherently low SNR, high dimensionality of the spatiospectral imaging problem, the presence of strong nuisance signals (e.g., water/lipid signals in 1H-MRSI) and the effects of B0 field inhomogeneity (which can lead to signal loss and spectral distortion). Despite the significant efforts devoted to improve individual components of MRSI, including faster acquisitions (6–12), nuisance signal suppression (13–17), more sophisticated image reconstruction methods (18–27), and advanced instrumentation (28–33), the combination of speed, resolution, SNR and organ coverage offered by the state-of-the-art methods still remains rather limited.
Recently, SPICE (SPectroscopic Imaging by exploiting spatiospectral CorrElation) has emerged as a new approach for achieving fast, high-resolution MRSI by modeling high-dimensional spatiospectral functions of interest using low-dimensional subspace representations (34–36). The key components within the SPICE framework include the estimation of the subspace structure and the use of this subspace for rapid spatiospectral encoding and data processing. The current strategy to address the subspace estimation issue is to acquire a set of high-SNR, low-resolution navigator data (denoted as ) during each experiment, and extract a data-dependent subspace for subsequent processing (37–39). Such an acquisition often requires sophisticated experimental setup for water/lipid suppression and challenging data processing steps for correcting B0 inhomogeneity effects and removing residual nuisance signals due to the limited k-space coverage. Moreover, for 1H-MRSI data acquired without solvent suppression, removal of the 3 to 4 order-of-magnitude stronger nuisance signals requires much more accurate water and lipid signal subspaces, which are very difficult to obtain from only low-resolution (39).
We present in this work a new approach to improve SPICE by formulating a subspace learning strategy that combines physics-based spectral models and the acquisition of spectral training data. Specifically, we recognize that a general in vivo spectrum is inherently a combination of signals originating from a small number of molecules, each of which has a well-characterized resonance structure and experiment/molecule-dependent variations captured by a few parameters (40). Leveraging this well-established spectral prior, we use quantum mechanical simulations to predict the resonance structures of individual metabolites and acquire specially designed training data to learn the distributions of the molecule-dependent parameters. The combination of these two ingredients allows us to efficiently capture the physiologically meaningful spectral variations, from which low-dimensional subspaces can be determined and used for general MRSI experiments. As a result, the acquisition of experiment-dependent data (37,38,41) is no longer necessary, further reducing the imaging time. A rapid acquisition strategy that synergistically integrates ultrashort-TE, short-TR excitation, sparse spatiospectral encoding and the elimination of water/lipid suppression is then deployed for the actual 3D 1H-MRSI experiments. This allows us to generate high-resolution data in a short period with high SNR efficiency and self-calibration capability. A union-of-subspaces model based processing strategy with the learned subspace is developed to separate the different signal components with large dynamic range differences, i.e., the unsuppressed water/lipid signals, the metabolite signal of interest, and the macromolecule baseline, and to obtain high-SNR metabolite reconstruction from the noisy data.
Phantom and in vivo experiments have been carried out to evaluate the proposed method, and demonstrated the accuracy of the learned subspace in representing general in vivo spectral variations and its utility to achieve ultrafast high-resolution MRSI of the brain. Using the learned subspace, 3D MRSI with a large brain coverage and millimeter-level resolution (e.g., a nominal 2.5 mm in-plane and 3 mm through-plane resolution) without any water/lipid suppression in just a few minutes (i.e., 5 to 8 minutes) can be achieved. The detailed subspace learning, data acquisition and processing methods as well as the experimental results are described in the subsequent sections.
THEORY
SPICE: Subspace-Based MRSI
SPICE is an emerging approach to high-resolution MRSI that is characterized by the use of the following union-of-subspaces (UoSS) model to drive both data acquisition and image reconstruction (37–39)
| [1] |
It assumes that different signal components in the spatiospectral function of interest ρ(r, f), i.e., metabolites (m), water (w), lipids (f) and macromolecules (MM), reside in individual low-dimensional subspaces, each spanned by the basis functions , , and , respectively, and with Lm, Lw, Lf, LMM ≪ Nf (Nf denotes the number of spectral samples to achieve the desired spectral resolution). This model dramatically reduces the number of degrees-of-freedom by transforming the original problem of recovering the very-high-dimensional ρ(r, f) into the estimation of the spatial co-efficients and the bases (with x being m, w, f, or MM), thereby enabling a better tradeoff between speed, resolution and SNR. For example, consider a discretized spatiospectral function with 100×100 voxels each having a 512-point spectrum, the model in Eq. [1] with an order of 16 reduces the number of unknowns from 100×100×512 to 100×100×16+16×512 (a factor of about 30). However, directly estimating both and from noisy, high-resolution MRSI data is not practical (37), thus special acquisition and reconstruction strategies need to be designed to solve these problems.
A critical step for the subspace imaging framework specified by Eq. [1] is the determination of {ϕlx(·)} (also referred to as the subspace below). With predetermined subspaces, only the spatial coefficients need to be determined for high-resolution spatiospectral reconstruction. This further reduces the degrees-of-freedom and also effectively incorporates molecule spectral structure constraints, allowing for better separation of the different signal components with large dynamic ranges, e.g., the much stronger water/lipid signals and the weak metabolite signals (37, 38). In previously proposed SPICE-based methods, this was addressed by acquiring high-SNR, low-spatial-resolution and high-spectral-resolution navigator data () during each experiment, from which the subspaces for the metabolites and water/lipids were estimated (e.g., through SVD analysis) and used for subsequent processing (37). However, this strategy typically requires sophisticated experimental setup, adds extra scan time to each experiment, and imposes additional challenges for data processing due to the limited k-space coverage.
Subspace-Based MRSI with Learned Spectral Bases
We propose in this work a new strategy to learn the subspace from a set of specially acquired training data instead of from experiment-dependent . The proposed method is motivated by recognizing the significant amount of prior knowledge available for the spectral variations of individual molecules of interest (especially in brain tissues). Specifically, a voxel spectrum is inherently a combination of signals originating from a small number of molecules, each of which is characterized by a well-defined resonance structure φm(t) (also known as the metabolite basis in the spectral quantification literature) with m being the molecule index and some parameters θm that specify the spectral variations. With these two ingredients, the most commonly used model to represent an experimentally acquired FID assumes and can be written as
| [2] |
where cm denotes the metabolite concentrations, and δfm denote the subject/tissue-dependent linshape and frequency shifts for each molecule, respectively. The Fourier transform of the resonance structure φm(t) contains relative frequencies, amplitudes and phases of different spectral components for a single molecule. The additional modulation function h(t) absorbs other system imperfection related components (e.g., B0 field inhomogeneity or eddy currents induced lineshape distortions).
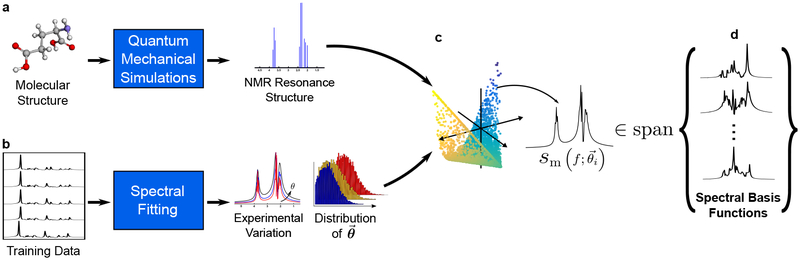
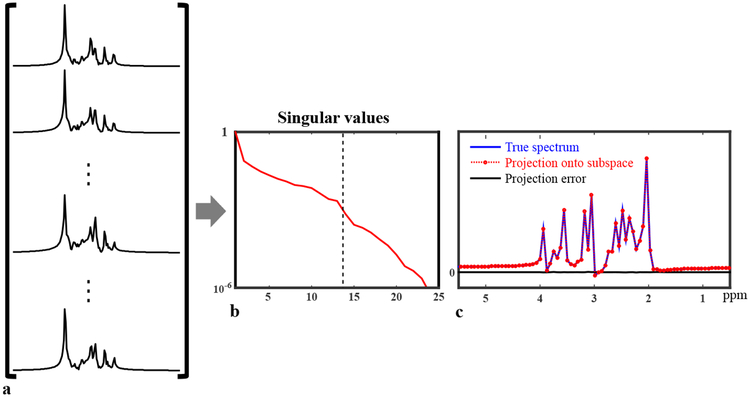
Accordingly, all the FIDs governed by the parametric Eq. [2] can be considered as points residing on a nonlinear low-dimensional manifold embedded in the high-dimensional vector space (42,43). This manifold can be learned given a sufficiently large number of data samples. But with special constraints on individual components, a low-dimensional subspace approximation can also be estimated. More specifically, we assume that a) φm(t) can be predicted by molecular structures and quantum mechanical (QM) simulations (as illustrated in Fig. 1a) and this feature remains stable across different tissue types; b) the specific values of and δfm can vary for different voxels and subjects, but their distributions under typical physiological conditions should be similar and thus can be learned from training data (Fig. 1b); and c) a significant portion of experiment-to-experiment variations in the spectra originates from h(t), e.g., caused by B0 inhomogeneity variations and eddy currents, and h(t) can be estimated from unsuppressed water spectroscopic signals and separated from the molecule and tissue dependent spectral variations. With these assumptions and specially designed training data to capture representative distributions of the spectral parameters (i.e., cm, and δfm), we can learn a linear subspace to accurately approximate the low-dimensional manifold that captures the physiologically meaningful spectral variations (Fig. 1c), and use this learned subspace for general high-resolution MRSI experiments (Fig. 1d). To illustrate that spectral functions generated using parameters from a certain distribution can be approximated by a subspace, we simulated 10,000 metabolite spectra with uniform randomly distributed cm, Gaussian distributed (with mean and standard deviations from literature values) and δfm (mean 0 and standard deviation 3 Hz)1. As shown in Fig. 2, the Casorati matrix formed by these spectra (a) has rapidly decaying singular values, implying they reside in a low-dimensional subspace, which is also supported by comparing a newly generated spectrum and its projection onto a 14-dimensional subspace from the current 10,000 (c). The projection error is negligible.
Figure 1:
An illustration of the proposed subspace learning strategy: (a) Resonance structures for individual metabolites are obtained by QM simulations; (b) High-SNR training data are acquired and fitted to sample the distributions of spectral parameters (i.e., cm, , δfm); (c) Integration of these two generates sample spectra that reside on a low-dimensional manifold. Assuming that the parameters for physiologically meaningful spectra came from an underlying distribution independent of subjects, this manifold can be accurately approximated by a low-dimensional subspace that can be learned from training data (d) and used for general MRSI experiments.
Figure 2:
Illustration of subspace approximation for spectra with parameters generated from specific distributions: 10000 metabolite spectra were synthesized using Eq. [2] with randomly distributed {cm}, {}and {δfm}, and arranged into a Casorati matrix (a), which has very rapidly decaying singular values as shown in (b), indicating that these spectra can be well approximated by a low-dimensional subspace. The dash line indicates where the rank truncation error falls below 1e−3 (rank 14). A new spectra (not among the existing 10000) was generated and projected onto a 14-dimensional subspace (c); the projection error is negligible, further validating the subspace model.
While learning the metabolite signal subspace requires special high-SNR data, the water and lipid subspaces can be directly determined from the high-resolution MRSI data if acquired without water/lipid suppression pulses, as these two components have excellent SNR. The key issue for this step is how to handle spatiospectral encodings with more flexible sampling grids, e.g., the sparse sampling scheme used in (39,41). To this end, we can perform an initial spatiotemporal reconstruction leveraging both parallel imaging and regularization methods to interpolate the undersampled k-space data (as described in (41)) and apply the following multi-peak exponential model fitting to this initial reconstruction (ignoring the metabolites since they are negligible compared to the unsuppressed water/lipids)
| [3] |
The ρw(r) and ρf(r) functions capture the linear coefficients, φf(t) the resonance structure for subcutaneous lipids (44), and the apparent relaxation constants, and Δf(r) the B0 field inhomogeneity. The fitted water and lipid signals can then be synthesized (at the desired temporal grids) and arranged into Casorati matrices from which their individual bases can be extracted using SVD. Note that the B0 information estimated in this step can also be used for inhomogeneity correction during the subsequent metabolite reconstruction. For data acquired with short or ultrashort-TE, the macromolecule (MM) baseline signal needs to be taken into account during reconstruction. To this end, we estimated the MM subspace from previously obtained metabolite-nulled CSI data as described in (45) and incorporate it into the processing.
Spatiospectral Reconstruction Using Learned Subspaces
With the learned signal subspaces, reconstruction from high-resolution MRSI data acquired in an experiment without can be obtained. The unsuppressed water/lipid signals are first removed from the data using the estimated water/lipid bases and a UoSS-based least-squares fitting method adapted from (39). After water/lipids removal, the residual water sidebands were further removed using the method described in (46). The final metabolite reconstruction was subsequently obtained from the water/lipid-removed data. More specifically, we first solve the following explicit subspace constrained reconstruction problem (37)
| [4] |
where Φm and ΦMM are matrix representations of the learned metabolite and macromolecule (MM) bases, respectively, Cx represent the corresponding spatial coefficients, B captures the B0 field inhomogeneity induced frequency shifts and lineshape modulation functions derived from the water FIDs, F denotes the Fourier transform operator, the vector dres contains the (k,t)-space data coupled with a sampling operator Ω and Dw contains edge weightings calculated from the companion water images. The regularization parameters λm and λMM were chosen based on the discrepancy principle with noise variance estimated from the background. With and , an initial spatiotemporal reconstruction can be synthesized as or . These estimates can be used as the final reconstruction but they can have structured errors due to modeling bias. To reduce this bias, an additional refitting step can be performed. For example, we use here the initial estimates ( and ) from the first step as references for a second reconstruction, i.e.,
| [5] |
will be the final metabolite spatiotemporal reconstruction. This additional step helps to reduce potential model bias and improve data consistency by imposing a softer constraint, which is further discussed below and demonstrated with supporting results.
METHODS
Data Acquisition
Data were acquired from both a physical phantom and healthy volunteers to evaluate the performance of the proposed method. All experiments were carried out on a Prisma 3T system (Siemens Healthineers, Erlangen, Germany) using a 20-channel head coil. The in vivo study was approved by the local Institutional Review Board and carried out with participants’ consent. The metabolite phantom was constructed using a glass water jar with a customized screw-on polycarbonate lid. Nine glass vials with three different diameters were glued into the phantom and filled with brain metabolite solutions at physiological concentrations (more details on the metabolite phantom are provided in the appendix). Since the molecular composition of the phantom is stable, training data was acquired only once using a long FID-CSI sequence with 2.3 ms TE, 60×60 matrix size, 200×200 mm2 FOV, and 10 mm slice thickness. The spectral bandwidth (BW) was 2000 Hz with 1024 FID samples. The subspace estimated from this data was used for all the future experiments. For in vivo experiments, a low-resolution, 3D FID-EPSI sequence was implemented to acquire the training data for subspace learning, with TR/TE = 310/2.5 ms, a resolution of approximately 1 cm3, 1500 Hz spectral bandwidth, 80 Hz weak water suppression and outer volume suppression (OVS). The total acquisition time is approximately three minutes with three signal averages. An accompanying nonwater-suppressed data with matched resolution was also acquired in each training scan (additional 1 min). Data were acquired from 5 different healthy volunteers to demonstrate the feasibility of the proposed method, while more training data can be obtained in the future for a denser sampling of the spectral parameter space for improved subspace learning.
With the subspaces learned from training data, high-resolution MRSI acquisitions can be performed in a general experiment session (without the need of acquiring ). Specifically, we used a rapid acquisition strategy that integrates the following features (41): 1) an FID pulse-acquire excitation scheme that achieved ultrashort TEs (less than 3 ms); 2) an EPSI-like gradient waveform that can produce (k,t)-space trajectories with an extended k-space coverage along the frequency encoding direction (e.g., kx) and sparse spectral sampling for rapid spatiospectral encoding after each excitation; 3) a small number of spectral encodings acquired each TR, which combined with the FID excitation achieved very short TRs (less than 200 ms) with optimized SNR efficiency (11); 4) a variable-density k-space sparse sampling strategy that extends the coverage along the phase encoding directions (e.g., ky and kz) for high resolution while keeping a center k-space region fully sampled to maintain a sufficient SNR for metabolite reconstructions; and 5) the elimination of solvent suppression modules, and inclusion of a set of navigators (including FIDs acquired at the central k-space, linear projections and orbital projections) to track system instability and subject motion with minimal perturbation of the steady state. These features work synergistically within the subspace framework, resulting in an unprecedented ultrashort-TE, very-short-TR, rapid acquisition scheme that can produce ultrahigh-resolution MRSI data in a short period. For phantom scans, high-resolution 3D MRSI data were acquired using the following parameters: TR/TE = 280/2.5 ms, matrix size = 80×80×24, FOV = 210×210×72 mm3 (a nominal voxel size of 2.6×2.6×3 mm3), number of echoes = 120, readout dwell time = 6 μs, echospacing=1.4 ms,2 and the navigators acquired every 50 TRs (14 s). In vivo data were acquired with TR/TE = 210/2.5 ms, matrix size = 96×110×24, FOV = 230×230×72 mm3 (a nominal voxel size of 2.4×2.1×3 mm3), number of echoes = 110, readout dwell time = 6 μs, and echospacing = 1.76 ms. The total acquisition time was about 5 minutes with a factor of two undersampling along ky. The center 32 ky encodings were fully sampled.
Subspace Estimation Implementation
The residual nuisance water and lipid signals were first removed from each water-suppressed training data using the method described in Refs. (37, 39) followed by B0 inhomogeneity corrected reconstruction using additionally acquired high-resolution field maps (37). The correction was also applied to the accompanying nonwater-suppressed data, from which voxel-wise lineshape distortion h(t) was extracted. To this end, we first performed a single-Lorentzian-peak fit to the unsuppressed data and used the initial fit as a reference signal to determine the residual temporal modulation function h(t) through a generalized-series model refitting (45). The estimated h(t) was incorporated into Eq. [2] for a subsequent spectral fitting of the water-suppressed data to obtain estimates of cm, and δfm, using the QUEST quantification method (47). As each data set contains many voxels, the estimated spectral parameters can be treated as samples from their underlying distributions (referred to as the empirical distributions). Combining these sampled parameter values and metabolite basis φm(t) generated for the specific imaging acquisition (FID, ultrashort-TE excitation in this case) through QM simulations using the NMR-SCOPE package (48), a collection of spectra can be generated and arranged into a Casorati matrix (a total of 5000 spectra were obtained), from which a subspace can be constructed using SVD. Note that alternative strategies can be used for this step, although we took a simplified approach for the purpose of feasibility demonstration in this work. For example, more sophisticated parameter distributions can be assumed and explicitly learned from the training data so that an even larger collection of training spectra can be generated by sampling from these distributions, which may lead to improved subspace accuracy and generalization capability.
RESULTS
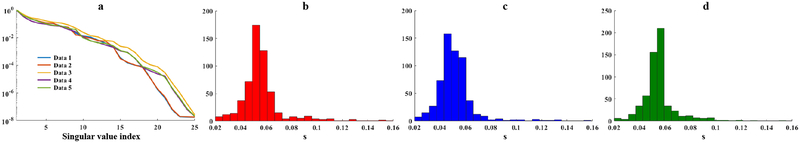
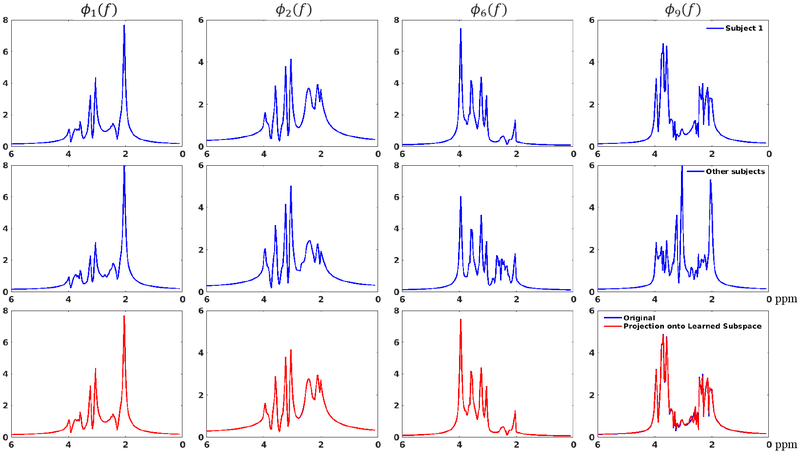
Figures 3 and 4 show the subspace learning results from the in vivo training data. Figure 3a demonstrates the rapidly decaying singular values for the Casorati matrices formed by fitted spectra from all five training data, which implies that all the spectral functions reside in a low-dimensional subspace. Similar histograms for the estimated spectral parameters (i.e., ) are shown for three subjects (Figs. 3b–d), implying an underlying consistent subspace structure across different subjects (The histograms for all five subjects are shown in the Supporting Information Figure S1). The accuracy of the subspace learned from all the training data is evaluated in Fig. 4, which compares the spectral basis estimated from one test data set to the subspace learned from other training data using a projection test. As shown by the projection results, the learned subspace can represent the test spectral basis accurately even though individual spectra may vary. A projection error eproj, as defined below, was also calculated for a quantitative comparison:
| [6] |
where V1 denotes the basis estimated from the first training data, V0 the learned basis from the other four and a projection operator that projects V1 onto the subspace spanned by V0. Similar tests have been performed for basis estimated from each individual training data and yielded similarly small errors, shown in Supporting Information Figure S2. These data support the feasibility of using training data to learn a subject-independent subspace that can be used for general MRSI experiments.
Figure 3:
Subspace learning from in vivo data: (a) Singular value decays for the Casorati matrices formed by the fitted spatiospectral functions from all five training data (different volunteers); the rapid decays demonstrate the accuracy of low-dimensional subspace representation; (b–d) Histograms of the estimated parameters for three data sets. Similar distributions can be observed, supporting the concept of using training data to determine a subject-independent distribution from which a subspace can be constructed.
Figure 4:
Evaluation of the learned subspace: The first row shows the spectral basis estimated from the first training data set (the 1st, 2nd, 6th and 9th bases are shown in different columns, respectively); The second row shows the basis learned from the remaining four data; The third row compares the basis in the first row (blue) and their projections onto the subspace spanned by the basis in the second row (red). As can be seen, while individual spectra may vary, the projections match the original spectral basis very well, implying an accurate representation using the learned subspace. The project error was around 3%.
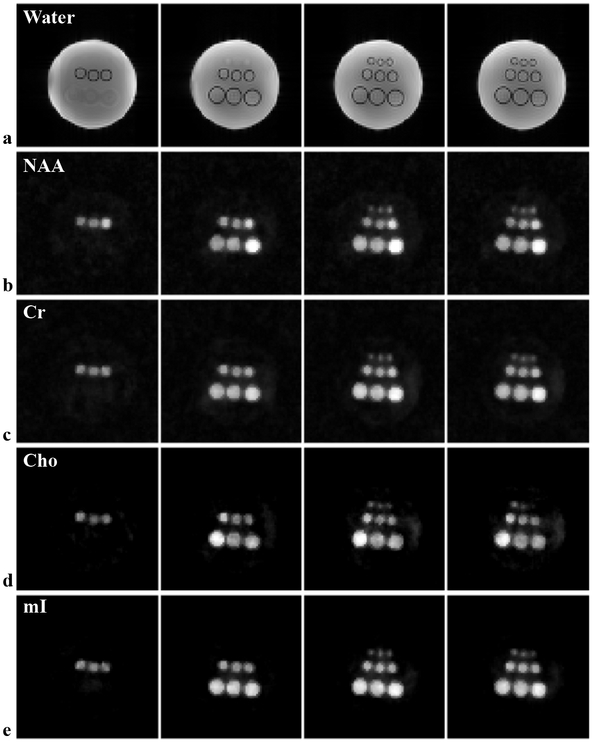
Figure 5 shows a set of representative results obtained by the proposed method from an 8 min phantom scan. As no water suppression is applied, structural images can be reconstructed from the unsuppressed water signals (Fig. 5a), which can be used for subsequent processing (e.g., providing spatial support or edge information). Reconstructed spatial maps of different metabolites (Figs. 5b–e), i.e., NAA, Cr, Cho, and myoinositol (mI), are shown to demonstrate the performance in terms of resolution and SNR. The concentration differences between the vials can be clearly visualized in these metabolite maps. High-quality spatially-resolved spectra (corresponding to a nominal voxel size of 2.6 2.6×3 mm3) were produced (Supporting Information Figure S3). A further quantitative comparison of metabolite ratios between different vials can be found in the Supporting Information Figure S4.
Figure 5:
High-resolution, 3D 1H-MRSI results from the metabolite phantom using an 8 min scan: (a) Images reconstructed from the unsuppressed water signals illustrating the structural arrangement of the phantom (note that the lengths of vials are different for different rows thus the changing features across slices); (b–e) Reconstructed metabolite maps, i.e., NAA (b), Cr (c), Cho (d) and mI (e) for the corresponding slices in (a). As can be seen, the proposed method produced high-resolution, high-SNR reconstructions, allowing visualization of even the smallest vials as well as resolving the concentration differences.
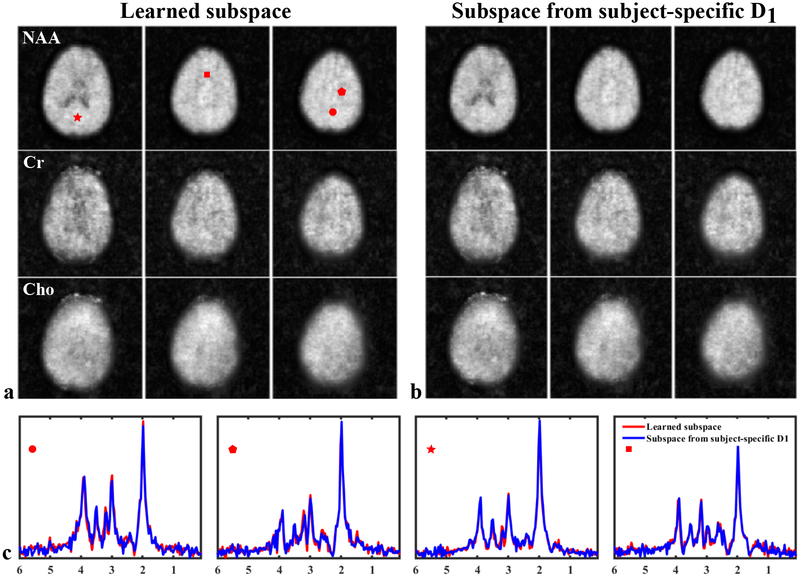
A set of 3D brain 1H-MRSI results from our ultrafast acquisition obtained using the subspace learned from training data of the other four volunteers are shown in Figure 6, with a comparison to the results obtained using the subspace estimated from a acquired for this particular subject. The metabolite maps of NAA, Cr and Cho for different slices across the imaging volume are shown (different rows in Fig. 6a and b), along with representative spatially-resolved spectra from the two different reconstructions (Fig. 6c, encoded with different colors). High-SNR metabolite reconstructions can be produced from only a 5 min scan. The spatially-resolved spectra from selected voxels also exhibit high SNR and clearly resolved metabolite peaks with negligible water/lipid residuals and good separation from the macromolecule baseline. Furthermore, the reconstructions produced using the learned subspace and subspace from subject-specific are fairly consistent, demonstrated by both visual inspection and a relative error. Such an error comparison for all 5 high-resolution acquisitions each associated with an experiment-specific can be found in the Supporting Information Table S1. We have also evaluated the reconstruction residuals with and without using the refitting step in Eq. [5] to demonstrate the improved data consistency and reduced modeling bias offered by this additional step. As shown in the Supporting Information Figure S5, the residuals from the second step are more similar to random noise with reduced structured errors observed in the residuals from the direct subspace fitting.
Figure 6:
Spatiospectral reconstructions from a 5 min, 3D brain 1H-MRSI acquisition. The metabolite reconstructions were produced using the learned subspace (a) and the subspace estimated from a single subject-specific (b), respectively. The maps of NAA, Cr, and Cho are shown in different rows. The plots in (c) compare the reconstructed spatially-resolved spectra at selected voxels, locations indicated by the red symbols in (a). As can be seen, the two spatiospectral reconstructions are very similar to each other; but the learned subspace can achieve this result without acquiring subject-specific . The relative errors for the NAA, Cr and Cho maps are 2.4%, 6.4%, and 8.0%, respectively.
Figure 7 shows another set of ultrafast 1H-MRSI results from a new data set not among the previous 5 acquisitions. Again, high-resolution, high-SNR metabolite maps can be reconstructed from the data using the learned subspace (Fig. 7a–d). Moreover, as no water suppression was used, the companion unsuppressed water spectroscopic signals in our data offer unique self-calibrating capabilities and richer information than conventional spectroscopic acquisitions. For example, structural information can be obtained from water reconstruction (Fig. 7e) and tissue susceptibility (χ) maps from the phase variations (41). Other useful information that can be extracted from the water FIDs is shown in the Supporting Information Figure S6.
Figure 7:
Ultrafast, 3D 1H-MRSI of the brain from another 5 min scan without water suppression using the learned subspace: (a-d) Reconstructed metabolite maps of NAA, Cr, Cho and mI for different slices across the 3D volume; (e) anatomical images with contrast reconstructed from the unsuppressed water signals; (f) tissue quantitative susceptibility maps (QSM) obtained from the phase variations encoded in the water spectroscopic signals.
DISCUSSION
We have presented a new strategy for learning a signal subspace that can capture molecule-specific spectral variations from specially designed training data, and using this learned subspace for MRSI data acquisition and reconstruction, removing the need of acquiring experiment/subject-dependent data as described in the original SPICE method (37). With the proposed method, rapid, high-resolution, volumetric MRSI without solvent suppression has been demonstrated using both phantom and in vivo experiments. An impressive combination of speed, resolution and SNR (approximately 3 mm isotropic resolution in a few minutes) was achieved. Note that while ultrahigh-resolution 2D MRSI results have recently been produced on ultrahigh-field scanners (e.g., 7 and 9.4 T scanners) (33, 49), the subspace learning approach and acquisition strategy can readily be extended to these systems and effectively take advantage of the improved sensitivity for better performance. It is also worth noting that the dimensionality reduction (which leads to noise reduction) offered by low-dimensional subspace models is the main factor contributing to the significant SNR improvement, while the spatial regularization offers additional improvement by taking advantage of anatomical prior information from water images.
The proposed method effectively integrated physics-based modeling and experimental data for constructing a low-dimensional subspace representation of spectral functions. While a subspace can also be constructed by directly taking an SVD of the collection of measured spectra from the training data, it is important to note that this scheme represents a distinct approach to subspace learning from the proposed method. Specifically, learning the subspace directly from the measured spectra can be framed as learning their distribution by treating each spectral function as a point in a high-dimensional space, which requires a large amount of training data. By introducing the physical model (e.g., the QM simulated basis), we translated the subspace learning problem to learning the distributions of a small number of spectral parameters, which theoretically should require significantly less data. Furthermore, as the QM basis is acquisition dependent (e.g., SE versus FID excitation) while the spectral parameters are not, our proposed strategy allows us to use acquisitions optimized for the molecules of interest to learn their parameter distributions and construct acquisition-specific subspace for general high-resolution MRSI experiments, and is thus much more flexible. There are also other practical benefits of our strategy, such as separation of residual nuisance signals and other signal distortions from the molecular signals of interest by incorporating the physics constraints.
We have simplified 1H-MRSI experiments by eliminating the need for sophisticated water and lipid suppression modules. Besides allowing for more flexible experimental designs by 1) reducing the burden or complexity in optimizing the RF pulses and gradients for saturation, and 2) alleviating the SAR issue especially when translating the proposed acquisition to higher fields, it also allows us to retain all the valuable information from the water protons. The reference information extracted from the unsuppressed water signals offers improved consistency to the MRSI data compared to the conventional approaches where the MRSI data and water referencing data are acquired separately. For spatiospectral encoding, the scheme shown in this paper is just one of the many options to implement the subspace-based acquisition strategy with differential emphasis on spatial and spectral dimensions. We chose the echo-planar-based trajectories due to its high efficiency in traversing the (k,t)-space, ease of implementation, lower susceptibility to trajectory errors (due to gradient imperfection), and reduced computational burdens in data processing. But other non-Cartesian trajectories such as radial (50), spiral (8) and concentric rings (51) that offer higher efficiency in covering the (k,t)-space with potential further acceleration and/or extended k-space coverage can be considered.
Other mathematical structures can be incorporated into our subspace imaging framework to further improve reconstruction. For instance, exploiting the Hankel low-rank structures induced by the linear predictability properties in spectroscopy data may offer additional benefits (35, 52, 53). More advanced strategies to impose spatial prior information can also lead to potential improvements in SNR, and/or further reduction of artifacts (e.g., residual nuisance signals and spectral distortion) (54). The current subspace learning strategy assumes consistent subspace structures across different subjects (even when individual voxel spectra vary). This can be violated for specific patient populations where spectral parameters fall far outside of the normal physiological ranges, or there are novel spectral components due to a certain pathology. In these cases, additional training data need to be acquired to establish pathology-specific metabolite subspaces. This issue is currently being investigated and will be addressed in subsequent publications in the context of specific applications.
CONCLUSION
We have successfully developed a subspace learning method for subspace MRSI (i.e., SPICE) and demonstrated its feasibility in achieving rapid, high-resolution 1H-MRSI of the brain with large organ coverage and without water/lipid suppression. With the proposed method, the experiment-dependent navigator data for subspace estimation is no longer needed, which represents a new way of performing MRSI experiments. The new ultrafast MRSI capability also offers richer information than conventional methods. Such a capability, we believe, is an important step towards practical applications of high-resolution MRSI, and with further optimization and development will open up new opportunities for in vivo metabolic studies in various neuroscience and clinical investigations.
Supplementary Material
Supporting Information Figure S1: Histograms of the estimated values from all five training data. Note that values less than 20 ms or larger than 200 ms were filtered out, thus the different numbers in each bin for different data.
Supporting Information Figure S2: A “leave-one-out” comparison of the subspace estimated from a single training data and the subspace learned from the remaining four. The spectral basis functions from individual training data and their projections onto the subspace learned from other four are shown in different rows. The projection errors are 4.5%, 4.1%, 3.5% and 5.8% for data 2, 3, 4 and 5, respectively. The same result for data 1 has been shown in the main text.
Supporting Information Figure S3: Representative spatially-resolved voxel spectra from different vials in the phantom. The nominal voxel size is 2.6×2.6×3 mm3 and the locations are marked by the numbers in the water image shown on the left.
Supporting Information Figure S4: The concentration ratios among different vials for different metabolites in the phantom, using vial #3 as the reference. As can be seen, the ratios between vials for the same metabolite produced by the proposed method (Proposed) and the original SPICE method using data-dependent (Original SPICE) are very similar, both consistent to the true ratios. There is still room for improvement, e.g., reducing estimation bias, for the lower concentration metabolites.
Supporting Information Figure S5: Reconstruction residuals (FIDs) for two selected voxels (shown in different rows). The left column shows the residuals for the reconstruction produced without using the second reconstruction step; the right column shows the residuals for the reconstruction produced using the second step. Structured errors can be observed in the first column which are noticeably reduced in the right column, demonstrating the utility of the second step in improving data consistency and reducing model bias.
Supporting Information Figure S6: An illustration of various kinds of information that can be extracted from the companion water spectroscopic signal generated by our acquisition: (a) structural images (different echoes will have different mixed T1 and contrasts); (b) B0 maps; (c) maps and (d) tissue susceptibility maps. System instability, such as frequency (e) and phase drifts (f) can be tracked and corrected for using the interleaved navigators.
Supporting Information Figure S7: Variance analysis for the subspace constrained reconstruction: (a) Co-variance matrix for an individual 256-point FID that underwent the subspace fitting (dimensionality of the subspace was 10). The unit for each element is σ2, which is the variance of the Fourier reconstruction; (b) Variances for individual FID points, corresponding to the diagonal elements of the covariance matrix in (a). Significant noise variance reduction can be observed.
Supporting Information Table S1: Comparison of reconstructions using the learned subspace and the subspace estimated from subject/data-specific . Relative errors between the metabolite maps from the two different reconstructions were calculated and compared in the table below. The results for 5 different datasets were obtained in a leave-one-out fashion.
ACKNOWLEDGMENTS
This work was supported in part by the following research grants: NIH-R21-EB021013–01. The authors would like to acknowledge Beckman Institute for Advanced Science and Technology and the Biomedical Imaging Center for their supports in research facilities and data collection.
APPENDIX
Phantom Design
The metabolite phantom is a cylindrical glass jar fully filled with sodium chloride doped water and contains nine vials glued in three rows to the cap. Each row of vials had the same diameter and were filled with solutions of NAA, Cr, glycerophosphocholine (Cho), myo-inositol (mI), glutamate (Glu) and γ-aminobutyric acid (GABA) at physiological concentrations (5). More specifically, each row of vials were designed to have three different concentrations: (C1) 15 mM NAA, 12 mM Cr, 3 mM Cho, 12 mM mI, 8 mM Glu, 2 mM GABA; (C2) 10 mM NAA, 10 mM Cr, 3 mM Cho, 10 mM mI, 8 mI Glu, 2 mI GABA; (C3) 8 mM NAA, 8 mM Cr, 6 mM Cho, 8 mM mI, 10 mM Glu, 1 mM GABA. Accordingly, each column of vials with the same concentration will have three different sizes. The pH value for all the solutions was adjusted to approximately 7.2 using concentrated NaOH and HCL. A customized screw-on cap was made with polycarbonate and used to seal that glass jar.
Variance Analysis
To further demonstrate the SNR improvement offered by the subspace imaging approach, we provide here a theoretical variance analysis for subspace-based reconstruction against standard Fourier reconstruction. To simplify the mathematical expressions without loss of generality, we ignore B0 field inhomogeneity and sparse sampling here. As a result, we can express the standard Fourier reconstruction as
| [7] |
where ρ is an T × N matrix denoting the true spatiospectral function with T being the number of FID points (e.g., 256) and N the number of voxels, and n denotes the noise matrix (assumed to be i.i.d. Gaussian). Accordingly, the covariance matrix of each voxel FID (i.e., each column in , treated as a random vector) is , where IT denotes a T × T identity matrix. With the subspace model ρ = VC where and denote the subspace (with orthogonal basis) and the spatial coefficients, respectively, the least-squares coefficient estimate can be expressed as (VHV = IL)
| [8] |
The covariance matrix of each column in (corresponding to each voxel) is then (V is a deterministic matrix)
| [9] |
Since each voxel FID in the subspace-based reconstruction, ρs,n, in this case is simply
| [10] |
its covariance matrix can be written as
| [11] |
which we can use to quantify the variance changes/reduction achieved by the subspace constraint. Specifically, using a rank-10 subspace estimated from the training data as described in the main text, we calculated the entire covariance matrix in Eq. [11], shown in Supporting Information Figure S7. Each element in the matrix has a unit of σ2 and the diagonal elements represent the variances for each FID points. As can be seen, the noise in the reconstruction became correlated but the variances were significantly reduced. Figure S7b also shows the diagonal elements of VVH which further demonstrates the variance reduction at individual FID points.
Footnotes
Note that these distributions just serve the illustration purpose and are not the actual ones we learned from training data to capture the physiologically meaningful subspace.
The echospacing is defined as the interval between two echoes acquired with the same gradient polarity.
References
- 1.Lauterbur PC, Kramer DM, House WV, Chen CN, Zeugmatographic high resolution nuclear magnetic resonance spectroscopy: Images of chemical inhomogeneity within macroscopic objects. J Amer Chem Soc 1975;97:6866 – 6868. [Google Scholar]
- 2.Brown TR, Kincaid BM, Ugurbil K, NMR chemical shift imaging in three dimensions. Proc Natl Acad Sci 1982;79:3523 – 3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maudsley AA, Hilal SK, Perman WH, Simon HE, Spatially resolved high resolution spectroscopy by “four-dimensional” NMR. J Magn Reson 1983;51:147 – 152. [Google Scholar]
- 4.Luyten PR, Marien AJ, Heindel W, van Gerwen PH, Herholz K, den Hollander JA, Friedmann G, Heiss WD, Metabolic imaging of patients with intracranial tumors: H-1 MR spectroscopic imaging and PET. Radiology 1990;176:791 – 799. [DOI] [PubMed] [Google Scholar]
- 5.de Graaf RA, In Vivo NMR Spectroscopy: Principles and Techniques. Hoboken, NJ: John Wiley and Sons, 2007. [Google Scholar]
- 6.Mansfield P, Spatial mapping of the chemical shift in NMR. Magn Reson Med 1984;1:370 – 386. [DOI] [PubMed] [Google Scholar]
- 7.Posse S, Tedeschi G, Risinger R, Ogg R, Le Bihan D, High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn Reson Med 1995;33:34 – 40. [DOI] [PubMed] [Google Scholar]
- 8.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM, Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn Reson Med 1998;39:889 – 898. [DOI] [PubMed] [Google Scholar]
- 9.Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C, Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI). Magn Reson Med 2009;61:548 – 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schirda CV, Tanase C, Boada FE, Rosette spectroscopic imaging: Optimal parameters for alias-free, high sensitivity spectroscopic imaging. J Magn Reson Imag 2009;29:1375 – 1385. [DOI] [PubMed] [Google Scholar]
- 11.Pohmann R, von Kienlin M, Haase A, Theoretical evaluation and comparison of fast chemical shift imaging methods. J Magn Reson 1997;129:145 – 160. [DOI] [PubMed] [Google Scholar]
- 12.Andronesi OC, Gagoski BA, Sorensen AG, Neurologic 3D MR spectroscopic imaging with low-power adiabatic pulses and fast spiral acquisition. Radiology 2012;262:647 – 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haase A, Frahm J, Hanicke W, Matthaei D, 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol 1985;30:341. [DOI] [PubMed] [Google Scholar]
- 14.Ogg RJ, Kingsley RB, Taylor JS, WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson 1994;104:1 – 10. [DOI] [PubMed] [Google Scholar]
- 15.Duyn JH, Gillen J, Sobering G, van Zijl PC, Moonen CT, Multisection proton MR spectroscopic imaging of the brain. Radiology 1993;188:277 – 282. [DOI] [PubMed] [Google Scholar]
- 16.Barkhuysen H, de Beer R, van Ormondt D, Improved algorithm for noniterative time-domain model fitting to exponentially damped magnetic resonance signals. J Magn Reson 1987;73:553 – 557. [Google Scholar]
- 17.Haupt CI, Schuff N, Weiner MW, Maudsley AA, Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med 1996;35:678 – 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Levin DN, Lauterbur PC, Spraggins T, SLIM: Spectral localization by imaging. Magn Reson Med 1988;8:314 – 322. [DOI] [PubMed] [Google Scholar]
- 19.Liang ZP, Lauterbur PC, A generalized series approach to MR spectroscopic imaging. IEEE Trans Med Imag 1991;10:132 – 137. [DOI] [PubMed] [Google Scholar]
- 20.Haldar JP, Hernando D, Song SK, Liang ZP, Anatomically constrained reconstruction from noisy data. Magn Reson Med 2008;59:810 – 818. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Gabr RE, Schar M, Weiss RG, Bottomley PA, Magnetic resonance spectroscopy with linear algebraic modeling (SLAM) for higher speed and sensitivity. J Magn Reson 2012;218:66 – 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eslami R, Jacob M, Robust reconstruction of MRSI data using a sparse spectral model and high resolution MRI priors. IEEE Trans Med Imag 2010;29:1297 – 1309. [DOI] [PubMed] [Google Scholar]
- 23.Kasten J, Klauser A, Lazeyras F, Van De Ville D, Magnetic resonance spectroscopic imaging at super-resolution: Overview and perspectives. J Magn Reson 2016;263:193 – 208. [DOI] [PubMed] [Google Scholar]
- 24.Cao P, Wu EX, Accelerating phase-encoded proton MR spectroscopic imaging by compressed sensing. J Magn Reson Imag 2015;41:487 – 495. [DOI] [PubMed] [Google Scholar]
- 25.Chatnuntawech I, Gagoski B, Bilgic B, Cauley SF, Setsompop K, Adalsteinsson E, Accelerated 1H MRSI using randomly undersampled spiral-based k-space trajectories. Magn Reson Med 2014;74:13 – 24. [DOI] [PubMed] [Google Scholar]
- 26.Wilson NE, Iqbal Z, Burns BL, Keller M, Thomas MA, Accelerated five-dimensional echo planar J-resolved spectroscopic imaging: Implementation and pilot validation in human brain. Magn Reson Med 2016;75:42 – 51. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya I, Jacob M, Compartmentalized low-rank recovery for high-resolution lipid unsuppressed MRSI. Mag Reson Med 2017;78:1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dydak U, Weiger M, Pruessmann KP, Meier D, Boesiger P, Sensitivity-encoded spectroscopic imaging. Magn Reson Med 2001;46:713 – 722. [DOI] [PubMed] [Google Scholar]
- 29.Lin FH, Tsai SY, Otazo R, Caprihan A, Wald LL, Belliveau JW, Posse S, Sensitivity-encoded (SENSE) proton echo-planar spectroscopic imaging (PEPSI) in the human brain. Magn Reson Med 2007;57:249 – 257. [DOI] [PubMed] [Google Scholar]
- 30.Tsai SY, Otazo R, Posse S, Lin YR, Chung HW, Wald LL, Wiggins GC, Lin FH, Accelerated proton echo planar spectroscopic imaging (PEPSI) using GRAPPA with a 32-channel phased-array coil. Magn Reson Med 2008;59:989 – 998. [DOI] [PubMed] [Google Scholar]
- 31.Henning A, Fuchs A, Murdoch JB, Boesiger P, Slice-selective FID acquisition, localized by outer volume suppression (FIDLOVS) for 1H-MRSI of the human brain at 7T with minimal signal loss. NMR Biomed 2009;22:683 – 696. [DOI] [PubMed] [Google Scholar]
- 32.Bogner W, Gruber S, Trattnig S, Chmelik M, High-resolution mapping of human brain metabolites by free induction decay 1H MRSI at 7T. NMR Biomed 2012;25:873 – 882. [DOI] [PubMed] [Google Scholar]
- 33.Chadzynski GL, Bause J, Shajan G, Pohmann R, Scheffler K, Ehses P, Fast and efficient free induction decay MR spectroscopic imaging of the human brain at 9.4 Tesla Magn Reson Med 2016;Doi: 10.1002/mrm.26539. [DOI] [PubMed] [Google Scholar]
- 34.Liang ZP, Spatiotemporal imaging with partially separable functions In: Proc IEEE Int Symp Biomed Imag, Arlington, VA, USA, 2007; pp. 988 – 991. [Google Scholar]
- 35.Nguyen HM, Peng X, Do MN, Liang ZP, Denoising MR spectroscopic imaging data with low-rank approximations. IEEE Trans Biomed Eng 2013;60:78 – 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam F, Liang ZP, A subspace approach to high–resolution spectroscopic imaging. Magn Reson Med 2014;71:1349 – 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam F, Ma C, Clifford B, Johnson CL, Liang ZP, High-resolution 1H-MRSI of the brain using SPICE: Data acquisition and image reconstruction. Magn Reson Med 2016;76:1059 – 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma C, Lam F, Ning Q, Johnson CL, Liang ZP, High-resolution 1H-MRSI of the brain using short-TE SPICE. Magn Reson Med 2017;77:467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma C, Lam F, Johnson CL, Liang ZP, Removal of nuisance signals from limited and sparse 1H MRSI data using a union-of-subspaces model. Magn Reson Med 2016;75:488 – 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Lam F, Clifford B, Liang Z, A subspace approach to spectral quantification for MR spectroscopic imaging. IEEE Trans Biomed Eng 2017;64:2486 – 2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng X, Lam F, Li Y, Clifford B, Liang ZP, Simultaneous QSM and metabolic imaging of the brain using SPICE. Magn Reson Med 2018;79:13 – 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peyré G, Manifold models for signals and images. Comput Vis Image Underst 2009;113:249 – 260. [Google Scholar]
- 43.Nakarmi U, Wang Y, Lyu J, Liang D, Ying L, A kernel-based low-rank (KLR) model for low-dimensional manifold recovery in highly accelerated dynamic MRI. IEEE Trans Med Imaging 2017; 36:2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren J, Dimitrov I, Sherry AD, Malloy CR, Composition of adipose tissue and marrow fat in humans by 1H NMR at 7 Tesla. J Lipid Res 2008;49:2055 – 2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam F, Li Y, Clifford B, Liang Z, Macromolecule mapping of the brain using ultrashort-TE acquisition and reference-based metabolite removal. Magn Reson Med 2018;79:2460 – 2469. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Lam F, Guo R, Clifford B, Peng X, Liang Z, Removal of water sidebands from 1H-MRSI data acquired without water suppression In: Proc Intl Soc Mag Reson Med, Paris, 2018; p. 4016. [Google Scholar]
- 47.Ratiney H, Sdika M, Coenradie Y, Cavassila S, van Ormondt D, Graveron-Demilly D, Time-domain semi-parametric estimation based on a metabolite basis set. NMR Biomed 2005;18:1 – 13. [DOI] [PubMed] [Google Scholar]
- 48.Stefan D, Cesare FD, Andrasescu A, Popa E, Lazariev A, Vescovo E, Strbak O, Williams S, Starcuk Z, Cabanas M, van Ormondt D, Graveron-Demilly D, Quantitation of magnetic resonance spectroscopy signals: the jMRUI software package. Meas Sci Technol 2009;20:104035. [Google Scholar]
- 49.Hangel G, Strasser B, Povazan M, Heckova E, Hingerl L, Boubela R, Gruber S, Trattnig S, Bogner W, Ultra-high resolution brain metabolite mapping at 7T by short-TR Hadamard-encoded FID-MRSI. NeuroImage 2016;168:199 – 210. [DOI] [PubMed] [Google Scholar]
- 50.Silva AC, Barbier EL, Lowe IJ, Koretsky AP, Radial echo-planar imaging. J Magn Reson 1998;135:242 – 247. [DOI] [PubMed] [Google Scholar]
- 51.Chiew M, Jiang W, Burns B, Larson P, Steel A, Jezzard P, Albert Thomas M, Emir UE, Density-weighted concentric rings k-space trajectory for 1H magnetic resonance spectroscopic imaging at 7 T. NMR Biomed 2018;31:e3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ying J, Lu H, Wei Q, Cai J, Guo D, Wu J, Chen Z, Qu X, Hankel matrix nuclear norm regularized tensor completion for N-dimensional exponential signals. IEEE Trans Signal Process 2017;65:3702 – 3717. [Google Scholar]
- 53.Bhattacharya I, Jacob M, Denoising and deinterleaving of EPSI data using structured low-rank matrix recovery In: IEEE 15th Intl Symp Biomed Imaging, 2018; pp. 679 – 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klauser A, Courvoisier S, Kasten J, Kocher M, Guerquin-Kern M, Van De Ville D, Lazeyras F, Fast high-resolution brain metabolite mapping on a clinical 3T MRI by accelerated 1H-FID-MRSI and low-rank constrained reconstruction Magn Reson Med 2018; 10.1002/mrm.27623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1: Histograms of the estimated values from all five training data. Note that values less than 20 ms or larger than 200 ms were filtered out, thus the different numbers in each bin for different data.
Supporting Information Figure S2: A “leave-one-out” comparison of the subspace estimated from a single training data and the subspace learned from the remaining four. The spectral basis functions from individual training data and their projections onto the subspace learned from other four are shown in different rows. The projection errors are 4.5%, 4.1%, 3.5% and 5.8% for data 2, 3, 4 and 5, respectively. The same result for data 1 has been shown in the main text.
Supporting Information Figure S3: Representative spatially-resolved voxel spectra from different vials in the phantom. The nominal voxel size is 2.6×2.6×3 mm3 and the locations are marked by the numbers in the water image shown on the left.
Supporting Information Figure S4: The concentration ratios among different vials for different metabolites in the phantom, using vial #3 as the reference. As can be seen, the ratios between vials for the same metabolite produced by the proposed method (Proposed) and the original SPICE method using data-dependent (Original SPICE) are very similar, both consistent to the true ratios. There is still room for improvement, e.g., reducing estimation bias, for the lower concentration metabolites.
Supporting Information Figure S5: Reconstruction residuals (FIDs) for two selected voxels (shown in different rows). The left column shows the residuals for the reconstruction produced without using the second reconstruction step; the right column shows the residuals for the reconstruction produced using the second step. Structured errors can be observed in the first column which are noticeably reduced in the right column, demonstrating the utility of the second step in improving data consistency and reducing model bias.
Supporting Information Figure S6: An illustration of various kinds of information that can be extracted from the companion water spectroscopic signal generated by our acquisition: (a) structural images (different echoes will have different mixed T1 and contrasts); (b) B0 maps; (c) maps and (d) tissue susceptibility maps. System instability, such as frequency (e) and phase drifts (f) can be tracked and corrected for using the interleaved navigators.
Supporting Information Figure S7: Variance analysis for the subspace constrained reconstruction: (a) Co-variance matrix for an individual 256-point FID that underwent the subspace fitting (dimensionality of the subspace was 10). The unit for each element is σ2, which is the variance of the Fourier reconstruction; (b) Variances for individual FID points, corresponding to the diagonal elements of the covariance matrix in (a). Significant noise variance reduction can be observed.
Supporting Information Table S1: Comparison of reconstructions using the learned subspace and the subspace estimated from subject/data-specific . Relative errors between the metabolite maps from the two different reconstructions were calculated and compared in the table below. The results for 5 different datasets were obtained in a leave-one-out fashion.