Abstract
Purpose:
To predict microsatellite instability (MSI) status of colon cancer on preoperative CT imaging using radiomic analysis.
Methods:
This retrospective study involved radiomic analysis of preoperative CT imaging of patients who underwent resection of stage II–III colon cancer from 2004–2012. A radiologist blinded to MSI status manually segmented the tumor region on CT images. 254 intensity-based radiomic features were extracted from the tumor region. Three prediction models were developed with: 1) only clinical features, 2) only radiomic features, and 3) “combined” clinical and radiomic features. Patients were randomly separated into training (n=139) and test (n=59) sets. The model was constructed from training data only; the test set was reserved for validation only. Model performance was evaluated using AUC, sensitivity, specificity, PPV, and NPV.
Results:
Of the total 198 patients, 134 (68%) patients had microstatelite stable (MSS) tumors and 64 (32%) patients had MSI tumors. The combined model performed slightly better than the other models, predicting MSI with an AUC of 0.80 for the training set and 0.79 for the test set (specificity=96.8% and 92.5%, respectively), whereas the model with only clinical features achieved an AUC of 0.74 and the model with only radiomic features achieved an AUC of 0.76. The model with clinical features alone had the lowest specificity (70%) compared with the model with radiomic features alone (95%) and the combined model (92.5%).
Conclusions:
Preoperative prediction of MSI status via radiomic analysis of preoperative CT adds specificity to clinical assessment and could contribute to personalized treatment selection.
Keywords: Colon, colonic neoplasms, microsatellite repeats, microsatellite instability, immunotherapy
Introduction
Colon cancer is the third most common cancer and the second most common cause of cancer mortality in men and women combined worldwide; however, not all colon cancers are genetically similar. Recently, colon cancers with microsatellite instability (MSI – about 15%) and the remaining microsatellite stable (MSS) tumors have been highlighted in clinical practice to have unique clinical and pathologic characteristics and distinct responses to treatment. MSI colon cancers are caused by loss of DNA mismatch repair activity and have a more indolent course and a slightly better prognosis when compared to MSS cancers [1]. MSI colon tumors are typically observed in younger patients, frequently arise in the right colon, have increased lymphocytic infiltrate, are associated with Lynch Syndrome (3% of all colon cancers and 3% of MSI tumors), and respond favorably to immune check point blockade [2]. MSS cancers on the other hand are more likely to occur in older patients and in non-hereditary conditions, arise in the left colon [3], and are treated with standard-of-care 5-FU based chemotherapy [4].
MSI status is currently assessed preoperatively by colonoscopic biopsy requiring a team of anesthesiologists, gastroenterologists, and various nursing and support staff for the procedure which can be costly. This procedure is the standard of care but has two challenges. First, biopsy is an invasive test in which the DNA extracted from the sample may not meet the minimum quality/quantity criteria for the genetic assay, thus resulting in unknown MSI status [5]. Second, many patients with colon cancer are first diagnosed via CT imaging rather than colonoscopy when they become symptomatic (such as with pain or bloating) and seek medical attention. In particular, this may be seen in younger patients who are more likely to have MSI tumors, as they are too young to meet the current United States Preventive Services Task Force screening recommendation for colonoscopy, which is for patients over 50 years of age [6], or the new American Cancer Society guidelines of 45 years of age [7]. These patients then experience a treatment delay while waiting for either a colonoscopy or surgery to determine MSI status as these tumors are currently indistinguishable by a radiologist on routine CT imaging.
The approach to the treatment of many luminal gastrointestinal cancers (esophageal [8; 9], gastric [10], rectal [11]) is migrating towards a neoadjuvant paradigm. Correspondingly, preoperative chemotherapy for locally advanced operable primary colon cancer has been found to be feasible, safe, and capable of inducing significant downstaging according to preliminary results from the FOxTROT Collaborative group [12]. In this context, the early detection of MSI status could enable risk reduction in patients with MSI tumors who would benefit most from immunotherapy; at the same time, side effects from 5-FU based chemotherapy, which has limited value in MSI colon cancers, could be avoided, and treatment could be better individualized. One approach to enabling early detection is to leverage computational analyses of high resolution routine cross-sectional imaging using radiomics.
Radiomics is a burgeoning field and has shown promising results in distinguishing a complete response from residual tumor after total neoadjuvant treatment in rectal cancer [13], estimating disease free survival in early stage lung cancer [14], assessing tumor recurrence risk in breast cancer [15], and assessing Gleason score in prostate cancer [16]. Given the ubiquity of CT in the preoperative evaluation and TNM staging of colon cancer and the potential gap in knowledge of a tumor’s MSI status, we hypothesized that radiomic features extracted from portal venous phase CT could identify MSI colon cancers. Therefore, the purpose of this study was to design a prediction model based on radiomic features extracted from preoperative CT to non-invasively identify MSI colon cancers.
Methods
Patient Selection
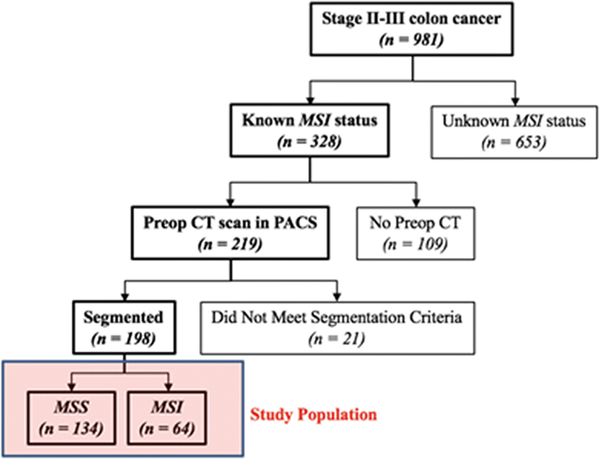
In this Health Insurance Portability and Accountability Act-approved retrospective study, a waiver of informed patient consent was obtained from our institutional review board. A surgical database was searched for consecutive patients that underwent resection of stage II–III colon cancer between January 2004 and December 2012. Patients with 90-day postoperative deaths and another stage IV malignancy were excluded. Thus, the search yielded 981 patients. Patients with known mismatch repair proteins expression status determined using immunohistochemistry (IHC) were considered eligible for our study (n = 328). Among these patients, those with a preoperative portal-venous phase CT scan of the abdomen and pelvis within 8 weeks of surgery available on the institutional picture archiving and communication system (Centricity PACS, GE Healthcare) were included (n = 219). Finally, patients with CT images of poor quality or radiographically occult tumor were excluded (n = 21). Therefore, the final cohort consisted of 198 patients with stage II or III colon cancer with IHC-proven MSI status. During the study period, IHC testing was routinely performed on specimens from every patient ≤ 50 years old undergoing a colectomy for colon cancer at our institution and was selectively performed in patients > 50 years old who had a family history of colorectal cancer and/or clinical features suspicious for MSI. Data regarding demographics and clinicopathologic features were collected by review of the electronic medical record.
Clinical and Pathological Variables
Within the Department of Pathology, after standard gross and histopathologic assessment was performed, IHC for mismatch repair proteins expression was performed using the standard streptavidin-biotin-peroxidase procedure. Primary MoAbs against MLH1 (clone G168–728, diluted 1:250 (PharMingen®)), MSH2 (clone FE11, diluted 1:50 (Oncogene Research Products®)), MSH6 (clone GRBP.P1/2.D4, diluted 1:200 (Serotec Inc®)), and PMS2 (clone A16–4, diluted 1:200 (BD PharMingen®)) were used. Tumors deficient in MLH1, MSH2, MSH6, or PMS2 proteins were used as external controls. Tumors that showed a total absence of nuclear staining while their adjacent benign tissue showed nuclear staining were scored negatively for the expression of that protein and were deemed MSI [17].
CT Acquisition
Patients included in the study were scanned as part of routine clinical care. As a result, scans were acquired at our center as well as outside institutions with images uploaded to PACS. CT acquisition was performed using a variety of CT manufacturers including: Elscint (2), GE (121), Philips (16), Siemens (44), Toshiba (14), and unknown (1). Mean acquisition parameters were: 120 kVp (range: 100–140 kVp), exposure time 751 ms (range: 500–1782 ms), and tube current 333 mA (range: 100–752 mA). Images were reconstructed at a slice thickness varying from 1–7.5 mm (mean: 4.6 mm) with a reconstruction diameter range from 274–500 (mean: 400). All contrast agents (for example: Omnipaque, Isovue, Optiray, and Ultravist), contrast rates, and dosages were acceptable.
Radiomic Analysis
CT scans were transferred from PACS to a local workstation for advanced image analysis. A radiologist with expertise in body imaging manually segmented the tumor region on all CT slices using ITK-SNAP software; segmentation was then secondarily reviewed and edited by a second body imaging radiologist. Both radiologists were blinded to MSI status. A total of 254 well-established intensity-based radiomic features were extracted from the tumor region using grey-level co-occurrence matrices (GLCMs) [18], run-length matrices (RLMs) [19], local binary patterns (LBPs) [20–22], fractal dimension (FD) [23; 24], intensity histogram (IH) and angle co-occurrence matrices (ACMs) [25; 26]. These features included 19 features from GLCM, ACM1, and ACM2, respectively; 11 from RLM, 5 from IH; 127 from LBP; and 54 from FD [27; 28]. Each feature was extracted from each CT slice and averaged to obtain a single value for each patient. These features quantify heterogeneity in CT enhancement patterns. GLCM features, for example, encode the spatial distribution of two neighboring pixels located at a distance and direction, whereas RLM features represent the coarseness of an image via counting the number of consecutive pixels. FD features capture the self-similarity present within the image. ACMs describe the orientation patterns present in the tumor region. Software created in MATLAB R2015a (The MathWorks, Inc.) was used to extract the radiomic features. These radiomic features have been previously described in the literature [25].
Statistical Analysis of Clinicopathologic Variables
The significance of associations between MSI and clinicopathologic variables was assessed using the Fisher’s exact or Chi square test for categorical variables, and the Student’s t-test or Mann-Whitney test, as appropriate by the type of distribution, for continuous variables. A p-value ≤ 0.05 was considered significant. Statistical analyses of clinicopathologic variables were performed using statistical software (Statistical Package for the Social Sciences, Inc., SPSS software version 25).
Prediction Model Building
Three MSI prediction models were designed using our available preoperative data: (1) prediction model using clinical variables, (2) prediction model using radiomic features extracted from the tumor region in CT, and (3) prediction model using a combination of clinical variables and radiomic features. The first prediction model included clinical variables significantly associated with MSI on univariate statistical analysis. The second prediction model included radiomic features that were significant on the Wilcoxon rank-sum test. In this prediction model, highly correlated radiomic features (i.e., features with correlation coefficient > 0.9) were removed from the feature set to reduce the number of variables. Finally, in the third prediction model, clinical variables were combined with radiomic features. A random forest classifier was utilized to create all three MSI prediction models. Random forest is considered as one of the most accurate general-purpose machine learning techniques that handles a large number of features without overfitting [29]. The patient cohort was randomly divided into training and test sets to ensure that the prediction models were evaluated on independent data: 70% of the data was used to design the prediction model, while the remaining 30% was used to evaluate the performance of the model. The performance of the models was evaluated using the following: AUC under the receiver operating characteristic curve, sensitivity, specificity, PPV, and NPV. The random forrest models were designed in MATLAB.
Results
Patients
A total of 198 patients met the inclusion criteria and were included in the analysis. Figure 1 shows the patient selection. CT scans performed at outside institutions were reviewed by a radiologist for adequate contrast enhancement and overall image quality, and 21 patients were excluded due to poor image quality. Reasons for this exclusion included: no or poorly timed (arterial instead of portal venous phase) contrast (n = 7), slice spacing issues resulting in an inability to download series for transfer to segmentation software (n = 7), unclear or occult tumor (n = 2), breathing artifact resulting in missing or partially duplicated tumor (n = 2), and 1 patient had indistinct borders due to the tumor acting as a lead point for intussusception.
Fig. 1.
Flowchart of the study population. MSS: microsatellite stability, MSI: microsatellite instability
Among the 198 patients, 152 had CT scans performed at outside institutions and 46 had scans performed at our institution; 134 (68%) patients had MSS tumors and 64 (32%) patients had MSI tumors. Table 1 lists clinicopathologic variables stratified by MSI status. MSI status was significantly associated with age, primary tumor location (left or right side), node positivity, number of positive nodes, stage, differentiation, mucinous adenocarcinoma, tumor budding, tumor infiltrating lymphocytes, vascular invasion, and KRAS mutant status on univariate analysis.
Table 1:
Clinicopathological features of patients who underwent surgery for stage II–III colon cancer stratified by MSI statusa
| Characteristics | MSS (n = 134) | MSI (n = 64) | p value |
|---|---|---|---|
| Median age, years (range) | 51.5 (27–85) | 62.0 (29–86) | 0.007 |
| Gender | 0.84 | ||
| Female | 67 (50.0) | 33 (51.6) | |
| Male | 67 (50.0) | 31 (48.4) | |
| Primary tumor location | < 0.05 | ||
| Right | 63 (47.0) | 58 (90.6) | |
| Left | 71 (53.0) | 6 (9.4) | |
| T stage | 0.55 | ||
| T1 | 3 (2.2) | 1 (1.6) | |
| T2 | 10 (7.5) | 2 (3.1) | |
| T3 | 100 (74.6) | 53 (82.8) | |
| T4 | 21 (15.7) | 8 (12.5) | |
| Node-positive primary | |||
| No | 62 (46.3) | 46 (71.9) | <0.05 |
| Yes | 72 (53.7) | 18 (28.1) | <0.05 |
| N1 | 54 (40.3) | 12 (18.8) | |
| N2 | 18 (13.4) | 6 (9.4) | |
| Median number of positive nodes (range) | 1.0 (0–10) | 0.0 (0–8) | < 0.05 |
| Median total number of nodes (range) | 23.5 (11–119) | 28.0 (6–117) | 0.18 |
| CRC stage | < 0.05 | ||
| II | 62 (46.3) | 46 (71.9) | |
| III | 72 (53.7) | 18 (28.1) | |
| Tumor differentiation | < 0.05 | ||
| Poor | 26 (19.4) | 29 (45.3) | |
| Moderate | 108 (80.6) | 35 (54.7) | |
| Well | 0 (0.0) | 0 (0.0) | |
| Mucinous adenocarcinoma | < 0.05 | ||
| Yes | 37 (27.6) | 32 (50.0) | |
| No | 97 (72.4) | 32 (50.0) | |
| Tumor budding | < 0.05 | ||
| Yes | 40 (29.9) | 7 (10.9) | |
| No | 29 (21.6) | 15 (23.4) | |
| Unknown | 65 (48.5) | 42 (65.6) | |
| Tumor infiltrating lymphocytes | < 0.05 | ||
| Yes | 16 (11.9) | 22 (34.4) | |
| No | 60 (44.8) | 11 (17.2) | |
| Unknown | 58 (43.3) | 31 (48.4) | |
| Perineural invasion | 0.098 | ||
| Yes | 45 (33.6) | 14 (21.9) | |
| No | 86 (64.2) | 50 (78.1) | |
| Unknown | 3 (2.2) | 0 (0.0) | |
| Vascular invasion | < 0.05 | ||
| Yes | 78 (58.2) | 24 (37.5) | |
| No | 56 (41.8) | 40 (62.5) | |
| Positive margin | 0.15 | ||
| Yes | 0 (0.0) | 1 (1.6) | |
| No | 134 (100.0) | 63 (98.4) | |
| KRAS mutant | < 0.05 | ||
| Yes | 6 (4.5) | 4 (6.3) | |
| No | 11 (8.2) | 13 (20.3) | |
| Unknown | 117 (87.3) | 47 (73.4) | |
| BRAF mutant | 0.17 | ||
| Yes | 5 (3.7) | 5 (7.8) | |
| No | 14 (10.4) | 11 (17.2) | |
| Unknown | 115 (85.8) | 48 (75.0) | |
| HNPCCb | - | ||
| Yes | - | 15 (23.4) | |
| Sporadic | - | 16 (25.0) | |
| Unknown | - | 33 (51.6) |
Values in the table are numbers of patients (percentages) unless otherwise indicated
Confirmed after V600E BRAF mutation and/or MLH1 hypermethylation promoter assays MSI, Microsatellite instability; MSS, Microsatellite stability; CRC, Colorectal cancer; HNPCC, Hereditary nonpolyposis colorectal cancer
Multivariate preoperative MSI prediction models
Of the 198 patients, 139 were included in the training set and 59 were reserved strictly for tesing the performance of the prediction models. The first model included two clinical variables significant on univariate analysis and commonly cited in the literature (age and location of tumor [right or left side]) and achieved an AUC of 0.74 with the test data.
The second model included radiomic features. In total, 93 of 254 (37%) radiomic features showed a significant association with MSI. After removing correlated features, 40 radiomic features were selected for inclusion in the second prediction model, which achieved an AUC of 0.76 with the test data.
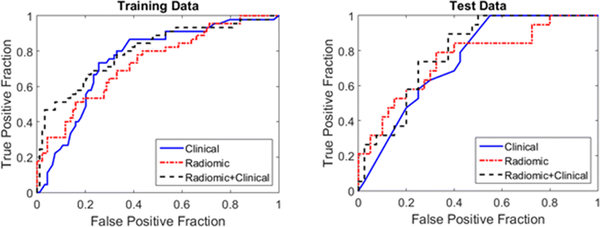
The third model combined both clinical variables and radiomic features into a single prediction model. This model achieved an AUC of 0.79 with the test data. The receiver operator curves for both training and test data are demonstrated in Figure 2.
Fig. 2.
Model performance comparison for the training and test data
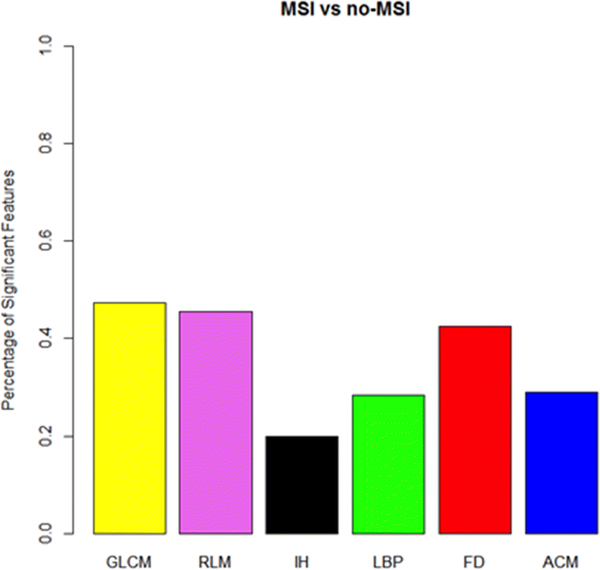
Model performance was compared between the models using sensitivity, specificity, NPV, and PPV with the training and test data (Table 2). The operating point for these performance metrics was determined using Youden’s index. Radiomic features from all feature categories showed significant performance, with GLCM showing the largest percentage and demonstrating increased homogeneity (Figure 3), followed by RLM and FD (Figure 4).
Table 2:
MSI prediction model results for training and test data
| Training | Test | |||||
|---|---|---|---|---|---|---|
| Clinical | Radiomic | Clinical + radiomic | Clinical | Radiomic | Clinical + radiomic | |
| AUC | 0.75 | 0.74 | 0.8 | 0.74 | 0.76 | 0.79 |
| Sensitivity | 68.9 | 28.9 | 44.4 | 63.2 | 31.6 | 31.6 |
| Specificity | 74.5 | 95.8 | 96.8 | 70 | 95 | 92.5 |
| PPV | 56.4 | 76.5 | 87 | 50 | 75 | 66.7 |
| NPV | 83.3 | 73.8 | 78.5 | 80 | 74.5 | 74 |
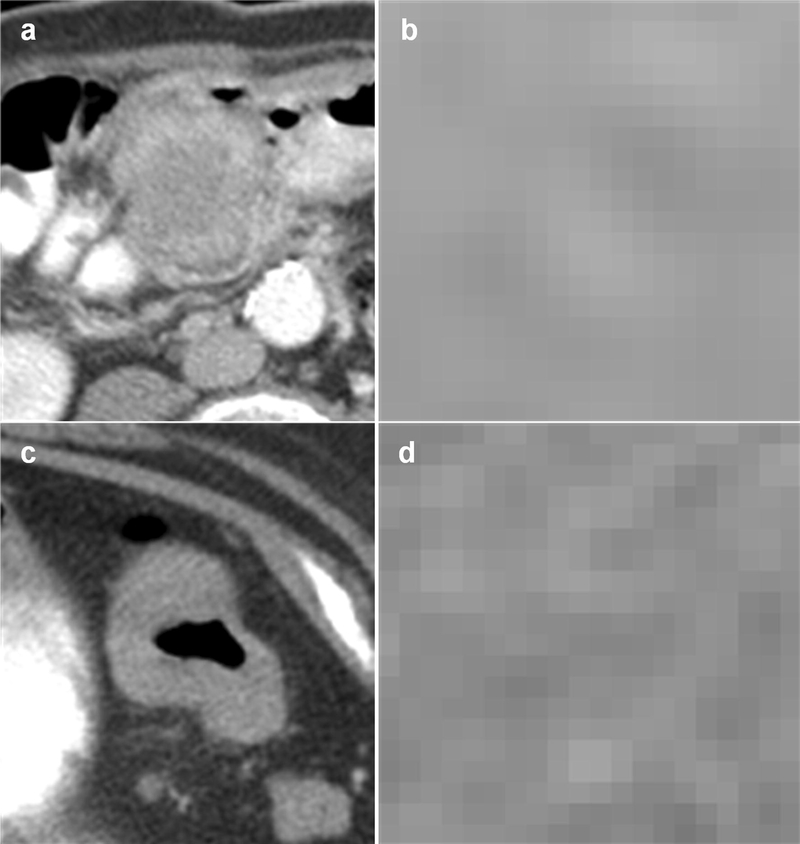
Fig. 3.
Examples of MSI positive and negative tumors classified correctly by our prediction model. (a) shows the cross-sectional axial image of an MSI tumor and (b) shows the tumor at the pixel level. (c) shows the cross-sectional axial image of an MSS tumor and (d) shows the tumor at the pixel level. Homogeneity is measured in MSI tumors which tend to be more indolent than MSS tumors. This is consistent with radiomics of other cancers indicating tumor heterogeneity is a marker for disease aggressiveness
Fig. 4.
Percentage of significant radiomic features in the training data by radiomic feature category
Discussion
We demonstrate that CT radiomic analysis can identify MSI colon cancers on initial pretreatment portal venous phase CT. We achieved similar results with the test and training sets using all three prediction models. Our combination model of radiomic features and clinical features had the best discriminatory ability in both the training cohort (AUC 0.80) and test cohort (AUC 0.79), higher than that of the model with radiomic features alone and the model with clinical features alone (test cohort: AUC of 0.76 and 0.74, respectively). The model with clinical features alone had the lowest specificity (70%) compared with the model with radiomic features alone (95%) and the combined model (92.5%). While our sensitivity for the combined model was low (test 31.6%) compared to the clinical features alone (test 63.2%), it is most likely due to the low sensitivity of the radiomic features (test 31.6%) which is lowering our sensitivity in the combined cohort. At the pixel level, increased homogeneity in the MSI tumors was observed, consistent with radiomics investigations of other cancers where increased tumor heterogeneity on imaging is a biomarker for disease aggressiveness [30].
Our results are of clinical value to medical and surgical oncologists. Since MSI and MSS tumors have different treatment strategies, the ability to predict MSI on preoperative CT scans routinely performed for detection and initial TNM staging of colon cancer may be useful to noninvasively stratify neoadjuvant treatment for colon cancer patients. Currently, MSI and MSS colon cancers are indistinguishable to radiologists on routine CT imaging. However, as standard of care moves from an adjuvant (postoperative) chemotherapy paradigm to a neoadjuvant (preoperative) paradigm for the treatment of colon cancer in higher risk tumors, the radiomics-based prediction model could be clinically impactful to differentiate MSI tumors early on. A model such as ours could serve as an imaging biomarker for MSI and could individualize initial treatment and reduce the risk to patients from unnecessary side effects due to the traditional 5-FU based chemotherapy in the neoadjuvant setting for MSI colon cancers. While not an adequate screening test due to the low sensitivity, our high specificity suggests patients who test positive for MSI could potentially go straight to neoadjuvant immunotherapy, but those who do not will still need to rely on other methods of MSI detection. Additionally, a validated imaging biomarker could decrease the time interval between diagnosis and treatment, and in the future, potentially act as a virtual biopsy.
An important consideration for all patients undergoing tissue biopsy, not just for colon cancer, is the rate of sampling error due to intratumoral heterogeneity or insufficient sample size [31; 32] as well as challenges to practitioners because MSI testing may not be routinely available—all of which may lead to an unknown MSI status. The availability of radiomics assessment of MSI status could aid in diagnosis, prognosis, and selection of optimal treatment for patients on initial CT evaluation. Our results demonstrate the potential of quantitative image analysis to detect MSI with high specificity in colon cancers on routine portal venous CT scan.
While a growing body of radiomics investigations in a wide range of cancers are being published, to our knowledge, our study is the first of its kind on CT radiomics for the prediction of MSI colon cancers. Our results indicate and further support the potential contribution of radiomics to aid in the development of precision medicine for patients with cancer.
This study has several limitations. First, to maximize our sample size, we included CT scans performed at outside institutions which were acquired under varying protocols. Given the potential differences in reproducibility in radiomic features with varying scanners and imaging protocols [33], further studies are needed to see if radiomic features for colon cancers are reproducible under different contrast-enhanced CT conditions. Nevertheless, the predictive potential of our radiomic features despite variation in CT imaging parameters is encouraging because colon cancer, once detected, typically does not undergo further advanced imaging such as magnetic resonance imaging (MRI) because portal venous phase CT remains the standard of care for TNM staging [34]. A sentinel paper on the field of radiomics discussed the intent of conducting radiomics analysis with standard of care images [35]. Our results using varying CT protocols, therefore, may be broadly applicable due to the ubiquity of CT imaging. Another limitation is that as a single-center retrospective analysis, our results need further validation in a prospective multicenter study to evaluate the reproducibility of our prediction models [36]. A third limitation is that manual segmentation may be a source of observer variability. In the future, automated segmentation may be possible which would help remove some of this variability.
Finally, two interesting elements of our patient data set were (1) the high number of MSI tumors at our tertiary care cancer center and (2) the average age of MSI patients was actually older than the MSS counterpart (62 years in MSI cohort compared to 51.5 years in MSS). Review of the literature cites an incidence of MSI in the population of about 15%. In our study, 32% of patients had MSI, which may be partly due to the referral pattern of our large tertiary referral center. While this enriched population may have influenced our results, our patients were accrued consecutively and thus are not necessarily a source of bias. Regarding the older age of MSI patients (62 years) which is contrary to the typically younger MSI CRC population frequently cited in the literature, it is only the patients with MSI CRC due to Lynch syndrome who tend to be younger compared to those with sporadic MSI CRC who tend to be older due to loss of MLH1 expression which increases with age [3]. Our results, therefore, might reflect the balance in the two different MSI populations as sporadic MSI CRC is more common (12%) than Lynch-associated MSI CRC (3%).
In conclusion, our results using a radiomics prediction model to identify MSI colon cancers has important clinical implications for the future of precision medicine. In this work, we show that a radiomics prediction model could enable a diagnosis to be established that influences appropriate preoperative treatment (traditional 5-FU based chemotherapy versus immunotherapy) and this could be highly useful in the coming era of neoadjuvant chemotherapy for colon cancer.
Acknowledgements
The authors thank Joanne Chin, MFA, for her editorial support of this article. This study has received funding by NIH/NCI P30 CA008748 Cancer Center Support Grant and the Colorectal Cancer Research Center CC50367 at Memorial Sloan Kettering.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest: The authors declare that they have no conflict of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent: Written informed consent was waived by the Institutional Review Board.
References
- 1.Cox VL, Saeed Bamashmos AA, Foo WC et al. (2018) Lynch Syndrome: Genomics Update and Imaging Review. Radiographics 38:483–499 [DOI] [PubMed] [Google Scholar]
- 2.Le DT, Uram JN, Wang H et al. (2015) PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372:2509–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138:2073–2087.e2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andre T, Boni C, Mounedji-Boudiaf L et al. (2004) Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med 350:2343–2351 [DOI] [PubMed] [Google Scholar]
- 5.Zauber NP, Sabbath-Solitare M, Marotta S, Perera LP, Bishop DT (2006) Adequacy of colonoscopic biopsy specimens for molecular analysis: a comparative study with colectomy tissue. Diagn Mol Pathol 15:162–168 [DOI] [PubMed] [Google Scholar]
- 6.Bibbins-Domingo K, Grossman DC, Curry SJ et al. (2016) Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. Jama 315:2564–2575 [DOI] [PubMed] [Google Scholar]
- 7.American Cancer Society American Cancer Society Guideline for Colorectal Cancer Screening. Available via https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/acs-recommendations.html. Accessed November 1, 2018
- 8.van Heijl M, van Lanschot JJ, Koppert LB et al. (2008) Neoadjuvant chemoradiation followed by surgery versus surgery alone for patients with adenocarcinoma or squamous cell carcinoma of the esophagus (CROSS). BMC Surg 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurogochi T, Honda M, Yamashita K et al. (2018) Safety and efficacy of preoperative chemotherapy followed by esophagectomy versus upfront surgery for resectable esophageal squamous cell carcinoma. Surg Today. 10.1007/s00595-018-1718-8 [DOI] [PubMed] [Google Scholar]
- 10.Hosoda K, Azuma M, Katada C et al. (2018) A phase II study of neoadjuvant chemotherapy with docetaxel, cisplatin, and S-1, followed by gastrectomy with D2 lymph node dissection for high-risk advanced gastric cancer: results of the KDOG1001 trial. Gastric Cancer. 10.1007/s10120-018-0884-0 [DOI] [PubMed] [Google Scholar]
- 11.Cercek A, Roxburgh CSD, Strombom P et al. (2018) Adoption of Total Neoadjuvant Therapy for Locally Advanced Rectal Cancer. JAMA Oncol 4:e180071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foxtrot Collaborative G (2012) Feasibility of preoperative chemotherapy for locally advanced, operable colon cancer: the pilot phase of a randomised controlled trial. Lancet Oncol 13:1152–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvat N, Veeraraghavan H, Khan M et al. (2018) MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology 287:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Liu Z, He L et al. (2016) Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 281:947–957 [DOI] [PubMed] [Google Scholar]
- 15.Li H, Zhu Y, Burnside ES et al. (2016) MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology 281:382–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wibmer A, Hricak H, Gondo T et al. (2015) Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol 25:2840–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Middha S, Zhang L, Nafa K et al. (2017) Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO Precision Oncology. 10.1200/po.17.00084:1-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haralick RM, Shanmugam K, Dinstein I (1973) Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics SMC-3:610–621 [Google Scholar]
- 19.Tang X (1998) Texture information in run-length matrices. IEEE Transactions on Image Processing 7:1602–1609 [DOI] [PubMed] [Google Scholar]
- 20.Ojala T, Pietikäinen M, Harwood D (1996) A comparative study of texture measures with classification based on featured distributions. Pattern Recognition 29:51–59 [Google Scholar]
- 21.Pietikäinen M, Hadid A, Zhao G, Ahonen T (2011) Local binary patterns for still imagesComputer vision using local binary patterns Computational Imaging and Vision, vol 40 Springer, London, pp 13–47 [Google Scholar]
- 22.Mehta R, Egiazarian K (2013) Rotated Local Binary Pattern (RLBP): Rotation invariant texture descriptorInternational Conference on Pattern Recognition Applications and Methods. Institute of Electrical and Electronics Engineers IEEE, Barcelona, Spain, pp 497–502 [Google Scholar]
- 23.Al-Kadi OS, Watson D (2008) Texture Analysis of Aggressive and Nonaggressive Lung Tumor CE CT Images. IEEE Transactions on Biomedical Engineering 55:1822–1830 [DOI] [PubMed] [Google Scholar]
- 24.Costa AF, Humpire-Mamani G, Traina AJM (2012) An Efficient Algorithm for Fractal Analysis of Textures2012 25th SIBGRAPI Conference on Graphics, Patterns and Images, Ouro Preto, Brazil, pp 39–46 [Google Scholar]
- 25.Chakraborty J, Rangayyan RM, Banik S, Mukhopadhyay S, Desautels JEL (2012) Statistical measures of orientation of texture for the detection of architectural distortion in prior mammograms of interval-cancer. Journal of Electronic Imaging 21:033010 [Google Scholar]
- 26.Chakraborty J, Rangayyan RM, Banik S, Mukhopadhyay S, Desautels JL (2012) Detection of architectural distortion in prior mammograms using statistical measures of orientation of textureMedical Imaging 2012: Computer-Aided Diagnosis. International Society for Optics and Photonics, pp 831521 [Google Scholar]
- 27.Attiyeh MA, Chakraborty J, Gazit L et al. (2018) Preoperative risk prediction for intraductal papillary mucinous neoplasms by quantitative CT image analysis. HPB (Oxford). 10.1016/j.hpb.2018.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakraborty J, Langdon-Embry L, Cunanan KM et al. (2017) Preliminary study of tumor heterogeneity in imaging predicts two year survival in pancreatic cancer patients. PLoS One 12:e0188022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biau G (2012) Analysis of a Random Forests Model. Journal of Machine Learning Research 12:1063–1095 [Google Scholar]
- 30.Sala E, Mema E, Himoto Y et al. (2017) Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol 72:3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng J, Chakraborty J, Chapman WC et al. (2017) Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma Using Quantitative Image Analysis. J Am Coll Surg 225:778–788.e771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gado A, Ebeid B, Abdelmohsen A, Axon A (2011) Improving the Yield of Histological Sampling in Patients With Suspected Colorectal Cancer During Colonoscopy by Introducing a Colonoscopy Quality Assurance Program. Gastroenterology Res 4:157–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrin T, Midya A, Yamashita R et al. (2018) Short-term reproducibility of radiomic features in liver parenchyma and liver malignancies on contrast-enhanced CT imaging. Abdom Radiol (NY). 10.1007/s00261-018-1600-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Comprehensive Cancer Network (NCCN) (2018) NCCN Clinical Practice Guidelines in Oncology (NCCN guidelines): Colon Cancer. Version 4.2018 Available via https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf. Accessed December 7, 2018 [DOI] [PubMed]
- 35.Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images Are More than Pictures, They Are Data. Radiology 278:563–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Figueiras R, Baleato-Gonzalez S, Padhani AR et al. (2018) Advanced Imaging Techniques in Evaluation of Colorectal Cancer. Radiographics 38:740–765 [DOI] [PubMed] [Google Scholar]