Abstract
Octamer transcription factor 1 (OCT1) is an androgen receptor (AR)‐interacting partner and regulates the expression of target genes in prostate cancer cells. However, the function of OCT1 in castration‐resistant prostate cancer (CRPC) is not fully understood. In the present study, we used 22Rv1 cells as AR‐positive CRPC model cells to analyze the role of OCT1 in CRPC. We showed that OCT1 knockdown suppressed cell proliferation and migration of 22Rv1 cells. Using microarray analysis, we identified four AR and OCT1‐target genes, disks large‐associated protein 5 (DLGAP5), kinesin family member 15 (KIF15), non‐SMC condensin I complex subunit G (NCAPG), and NDC80 kinetochore complex component (NUF2) in 22Rv1 cells. We observed that knockdown of DLGAP5 and NUF2 suppresses growth and migration of 22Rv1 cells. Furthermore, immunohistochemical analysis showed that positive expression of DLGAP5 in prostate cancer specimens is related to poor cancer‐specific survival rates of patients. Notably, enhanced expression of DLGAP5 was observed in CRPC tissues of patients. Thus, our findings suggest that these four genes regulated by the AR/OCT1 complex could have an important role in CRPC progression.
Keywords: androgen receptor, CRPC, DLGAP5, OCT1, prostate cancer
Abbreviations
- ANLN
anillin actin binding protein
- AR
androgen receptor
- ARE
androgen responsive element
- CRPC
castration‐resistant prostate cancer
- DAB
3,3′‐diaminobenzidine
- DHT
dihydrotestosterone
- DLGAP5
disks large‐associated protein 5
- KIF15
kinesin family member 15
- NCAPG
non‐SMC condensin I complex subunit G
- NUF2
NDC80 kinetochore complex component
- OCT1
octamer transcription factor 1
1. INTRODUCTION
Androgen and AR signaling pathways play a key role in the development and progression of prostate cancer.1, 2 Although androgen deprivation therapy (ADT) has significant efficacy in treating advanced prostate cancer, most prostate cancers develop resistance and are termed CRPC.3 Despite a low testosterone environment, AR as well as its target genes such as prostate‐specific antigen (PSA) are still highly expressed in the majority of CRPC lesions.4
Transcription factors (TF) collaborating with AR are important for AR‐regulated gene expression. Usually, AR binds to the canonical ARE (AGAACAnnnTGTTCT) situated in the enhancer and promoter region5 of the target gene to activate gene expression. Interestingly, this canonical ARE exists in only 10% of AR‐binding regions, whereas 68% of the AR‐binding regions harbor non‐canonical ARE.6 In addition to AR, these non‐canonical ARE need three TF, GATA binding protein 2 (GATA2), forkhead box A1 (FOXA1), and OCT1 to activate gene expression.6 Thus, OCT1 forms a hierarchical network with GATA2 and FOXA1 in AR signaling pathways.6, 7 Previously, we reported that OCT1 promotes proliferation and migration in LNCaP cells.8 We showed that acyl‐CoA synthetase long‐chain family member 3 (ACSL3) was the most highly expressed gene regulated by AR and OCT1 in LNCaP cells. We also showed that ACSL3 activates intratumoral steroid genesis in LNCaP‐derived CRPC model cells.9, 10 These results raised the hypothesis that downstream target genes of the OCT1 and AR complex play important roles in prostate cancer progression; however, the role of OCT1 in CRPC is not fully understood.
In recent work, we investigated an OCT1 genome‐wide network in a CRPC model cell line, 22Rv1.11 We then observed that OCT1 binding is greater in CRPC model cells compared with AR‐dependent prostate cancer, LNCaP. ANLN was identified as one of the major OCT1‐target genes in CRPC, which is highly induced specifically in 22Rv1 cells and facilitates cell cycle progression.12 In the present study, we analyzed the role of OCT1 in CRPC cell growth and migration by analyzing its target genes in 22Rv1 cells. We found that OCT1 knockdown suppressed 22Rv1 cell proliferation and migration. Furthermore, we found four novel target genes for AR and OCT1, DLGAP5, KIF15, NCAPG, and NUF2 that are highly expressed in CRPC tissues as well as in 22Rv1 cells. Our functional analyses suggest that these new OCT1 signals could be involved in the development of CRPC.
2. MATERIALS AND METHODS
2.1. Cell culture and reagents
Prostate cancer cell lines, 22Rv1 and LNCaP, were purchased from ATCC. Short tandem repeat (STR) analysis was used to authenticate the genotype of these cell lines. These cells were grown in RPMI‐1640 medium supplemented with 10% FBS, 50 U/mL penicillin and 50 μg/mL streptomycin. To analyze androgen responsiveness, we treated cells with 10 nmol/L DHT (Wako). Before androgen treatment, cells were cultured in phenol red free RPMI‐1640 medium (Nacalai Tesque) with 2.5% charcoal stripped FBS for 72 hours.
2.2. Small interfering RNA
We transfected cells with siRNA using Lipofectamine RNAi MAX (Thermo Fisher Scientific) according to the manufacture's protocol. We purchased control siRNA and other siRNAs from Sigma Genosys Japan as described.11 We designed siRNAs targeting OCT1. Sequences were: siOCT1 #A, 5′‐GUGAAGGCUAGGUGAGAAGC‐3′, siOCT1 #B, 5′‐GUGCUAGAUAGGUUUAUAAGU‐3′. To knock down AR, KIF15, NUF2, NCAPG and DLGAP5, we used silencer select siRNAs (AR #A: s1539; AR #B: 1538; KIF15: s35246; NUF2: s37981; NCAPG: s34514; DLGAP5: s18914) from Thermo Fisher Scientific.
2.3. Cell proliferation assay
22Rv1 cells were seeded at 500 cells/well and LNCaP cells were seeded at 1000 cells/well in 96‐well plates and cultured in RPMI‐1640. MTS assay was carried out using CellTiter 96 AQueous One (Promega) according to the manufacturer's instructions. Assays were carried out in five wells, and data are presented as mean and SD.
2.4. Migration assay
Migration assay was done by using a cell culture insert with 8.0‐μm pore size polyester filters (BD Biosciences). Before the assay, the lower surface of the filter was immersed for 30 minutes in 10 μg/mL fibronectin (Sigma) diluted with PBS. Then, we added RPMI‐1640 medium with 10% FBS in the lower chamber and RPMI‐1640 medium with 10% FBS containing cells (5 × 103) in the upper chamber. After 48 hours of incubation, we analyzed cells on the lower side of the inserts. We fixed cells in methanol for 30 minutes. Fixed cells were washed with PBS and stained with Giemsa solution (Muto Pure Chemicals) for 30 seconds. Cells that migrated to the lower surface were counted in five randomly selected fields under a microscope at a magnification of ×200.
2.5. Western blot analysis
Whole‐cell lysates were prepared using lysis buffer (50 mmol/L Tris‐HCl [pH 8.0], 150 mmol/L NaCl, 1% NP‐40, protease inhibitor cocktail [Nacalai Tesque]). Protein concentration was determined by BCA assay (Pierce). Protein (50 μg) was loaded on 8% SDS‐PAGE, separated by electrophoresis, and electroblotted onto Immobilon‐P Transfer Membranes (Millipore). Membranes were incubated with primary antibodies overnight and were then incubated with secondary antibodies. Antibody‐antigen complexes were detected using Pierce ECL Plus Western Blotting Substrate (Life Technologies). The following antibodies were used in this study: anti‐OCT1 antibody (ab15112; Abcam), anti‐AR‐V7 (ab198394; Abcam), anti‐AR‐antibody (H‐280; Santa Cruz Biotechnology), anti‐DLGAP5 (E‐7; Santa Cruz Biotechnology), anti‐β‐actin antibody (Sigma), anti‐phospho‐AR (Ser81) (04‐078; Millipore).
2.6. Quantitative reverse transcription polymerase chain reaction
Total RNA was obtained using ISOGEN reagent (Nippon Gene). First‐strand cDNAs were generated using the PrimeScript RT reagent kit (Takara) according to the manufacturer's protocol. Expression level of genes was measured by qRT‐PCR using the KAPA SYBR green mix (Sigma Genosys) on StepOne Real‐Time PCR System (Thermo Fisher Scientific). The following primer sequences were used in the present study: GAPDH forward: 5′‐GGTGGTCTCCTCTGACTTCAACA‐3′, reverse: 5′‐GTGGTCGTTGAGGGCAATG‐3′; KIF15 forward: 5′‐CCTTGGAGGTAATGCCAAAA ‐3′, reverse: 5′‐GCTCACATTTCCTTGGGTGT‐3′; NUF2 forward: 5′‐TCCAAATCCAAAGCCTGAAG ‐3′, reverse: 5′‐GTCATTCACCCGGCAGATAG‐3′; NACPG forward: 5′‐TGGGAAGTATGCCAGAAATG ‐3′, reverse: 5′‐TAACCACTGGGCATTCATCA‐3′; DLGAP5 forward: 5′‐GCCAAGGGCAATGAAAACTA‐3′, reverse: 5′‐TCTTTGGCCTTTGACCTTGT‐3′.
2.7. Microarray
Procedures of the microarray experiment in 22Rv1 cells have been described.12 Using the data, we determined OCT1‐regulated genes that had OCT1‐binding regions within 3 kilobases (kb) from transcription start sites in 22Rv1 cells.12 In the present study, we selected genes that were more expressed in 22Rv1 cells than in LNCaP cells, induced by DHT (DHT/vehicle >2.0‐fold) and upregulated in metastatic CRPC tissues. To analyze the expression level of OCT1 target genes in CRPC tissues, we used datasets registered in Oncomine ( https://www.oncomine.org/). To further evaluate the mRNA expression level in CRPC tissues, we downloaded the data of two cohorts (GSE GSE35988, GSE3325) including CRPC tissues from the Gene Expression Omnibus (GEO) at NCBI.
2.8. Immunohistochemistry
Immunohistochemical analysis was carried out using the streptavidin‐biotin amplification method previously described.8, 11 Briefly, primary antibody (1:200 dilution) against ANLN actin binding protein (ab211872; Abcam) and DLGAP5 was applied and followed by Histofine Simple Stain MAX‐PO (Nichirei). The antigen‐antibody complex was visualized with DAB solution (1 mmol/L DAB, 50 mmol/L Tris‐HCl buffer [pH 7.6], and 0.006% H2O2). Immunoreactivity (IR) score was calculated by the staining intensity (score 0, none; score 1, weak; score 2, moderate; score 3, strong) of positively stained cells and the proportion of positive cells to total cells (under 200× magnification) (score 0, none; score 1, <1/100; score 2, 1/100 to 1/10; score 3, 1/10 to 1/3; score 4, 1/3 to 2/3; score 5, >2/3).8, 11 Immunostained slides were evaluated blind to clinical information by two urological pathologists and specimens were defined as positive when the score was 5 or greater.
2.9. Data accession
ChIP‐seq data and microarray data used in the present study were deposited in GEO repository (accession numbers: GSE123517, GSE123565, and GSE62492).
2.10. Statistical analysis
Cell proliferation, cell migration assays, and xenograft experiments were evaluated using Student's t test, Mann‐Whitney U test and ANOVA with Dunnett's multiple comparisons test. Cancer‐specific survival curves were obtained by the Kaplan‐Meier method and verified by the log‐rank (Mantel‐Cox) test. Statistical assessments were implemented in GraphPad Prism for Mac 6.0 (GraphPad Software, Inc.) and JMP 9.0 software (SAS Institute Japan, Inc.) and P‐values <0.05 were considered statistically significant.
3. RESULTS
3.1. Octamer transcription factor 1 promotes proliferation and migration in 22Rv1 cells
First, we analyzed the difference between the hormone‐naïve prostate cancer LNCaP and CRPC model 22Rv1 cells. Endogenous expression levels of AR and an AR‐variant, AR‐V7, were elevated in 22Rv1 cells compared with LNCaP cells, suggesting the upregulated AR signals in 22Rv1 cells in both the presence and absence of androgen (Figure 1A). By MTS assay, we examined the response of each cell line to a hormone treatment and hormone depletion (Figure 1B). Interestingly, significant enhanced cell growth in the absence of androgen was observed in 22Rv1 cells compared with LNCaP. Therefore, we used 22Rv1 cells to analyze the role of OCT1 in CRPC as a model.
Figure 1.

Knockdown of octamer transcription factor 1 (OCT1) reduces proliferation and migration of 22Rv1 cells. A, Comparison of androgen receptor (AR) and AR‐V7 expression in castration‐resistant prostate cancer (CRPC) model cells, 22Rv1, with hormone naïve prostate cancer cells, LNCaP. Cells were treated with 10 nmol/L dihydrotestosterone (DHT) or vehicle for 24 h. Western blot analysis for AR and AR‐V7 expression was carried out and β‐actin was used as a loading control. B, Difference of androgen‐independent and ‐dependent cell growth in 22Rv1 cells from LNCaP cells. Cells were treated with 10 nmol/L DHT or vehicle. MTS assay was done at the indicated time points (N = 4). Results are presented as mean and SD. **P < 0.01, ***P < 0.001. C, Validation of siRNA targeting for OCT1. 22Rv1 cells were transfected with 10 nmol/L siControl or 10 nmol/L siOCT1 (#A and #B) for 48 h and then treated with 10 nmol/L DHT or vehicle for 24 h. Western blot analysis for OCT1 expression was carried out and β‐actin was used as a loading control. D, Knockdown of OCT1 inhibits CRPC cell proliferation. 22Rv1 cells were treated with 10 nmol/L siControl or 10 nmol/L siOCT1 (#A and #B). MTS assay was carried out at the indicated time points (N = 6). Results are presented as mean and SD. *P < 0.05, ***P < 0.001. E, Knockdown of OCT1 inhibits CRPC cell migration. 22Rv1 cells were treated with siControl or siOCT1 #A or #B. After 48 h incubation, cell migration assay was done (N = 5). Results are presented as mean and SD. ***P < 0.001. Left panels show representative views of migrated cells (siControl and siOCT1 #A and #B) in 22Rv1 cells
To investigate the effects of OCT1 silencing on CRPC cell proliferation and migration, we generated a series of two siRNAs targeting OCT1 (siOCT1 #A and #B). Transfection of 10 nmol/L siOCT1 #A and #B to LNCaP and 22Rv1 cells significantly reduced endogenous OCT1 expression (Figure 1C and Figure S1A). First, by using MTS assay, we observed that transfection of siOCT1 #A and #B repressed DHT‐treated LNCaP cell proliferation as expected from our past report8 (Figure S1B). Then, MTS assay was carried out using 22Rv1 cells treated with these siRNAs. Transfection of both siOCT1 #A and #B significantly inhibited 22Rv1 cell proliferation compared to siControl after 4 days (Figure 1D). In terms of 22Rv1 cell‐migration ability, numbers of migrated cells treated with both siOCT1#A and #B for 48 hours were significantly decreased compared to those with siControl (Figure 1E).
3.2. Identification of candidate OCT1‐target genes involved in prostate cancer progression
To identify OCT1‐responsive genes in 22Rv1 cells, we previously carried out microarray analysis combined with our previous ChIP‐seq data and identified 911 putative OCT1‐regulated genes with OCT1 binding in the promoter region.12 Microarray data showed that the expression levels of 35 genes, including 33 androgen‐responsive genes, in 22Rv1 cells were higher than in LNCaP cells (Figure 2A). In addition, 10 of 33 genes were highly expressed in metastatic CRPC tissues rather than in localized prostate cancer according to the Oncomine dataset which is a cancer microarray database and web‐based data‐mining platform (Figure 2B).13, 14 Interestingly, five of 10 genes, DLGAP5, KIF15, NCAPG, NUF2, and anillin actin binding protein (ANLN), were associated with cell cycle, particularly mitosis (M)‐phase. Cell cycle (M‐phase) is the major molecular hallmark in AR‐mediated development of CRPC15 and OCT1‐target signals in 22Rv1 cells.12 In addition to ANLN,12 we observed significantly decreased expression of the other four genes by treating cells with siOCT1 #A and #B (Figure 2C). Therefore, in the present study, we further analyzed the functions of these four novel OCT1‐target genes. ChIP‐seq data indicated that OCT1 and AR binding were colocated in the promoter or enhancer regions of these four genes (Figure 3A). In line with the results of AR‐ChIP‐seq, we observed that the mRNA expression of these four genes was upregulated by androgen treatment and repressed by AR inhibition (Figure 3B), suggesting cooperative regulation by AR and OCT1 to increase transcription.
Figure 2.
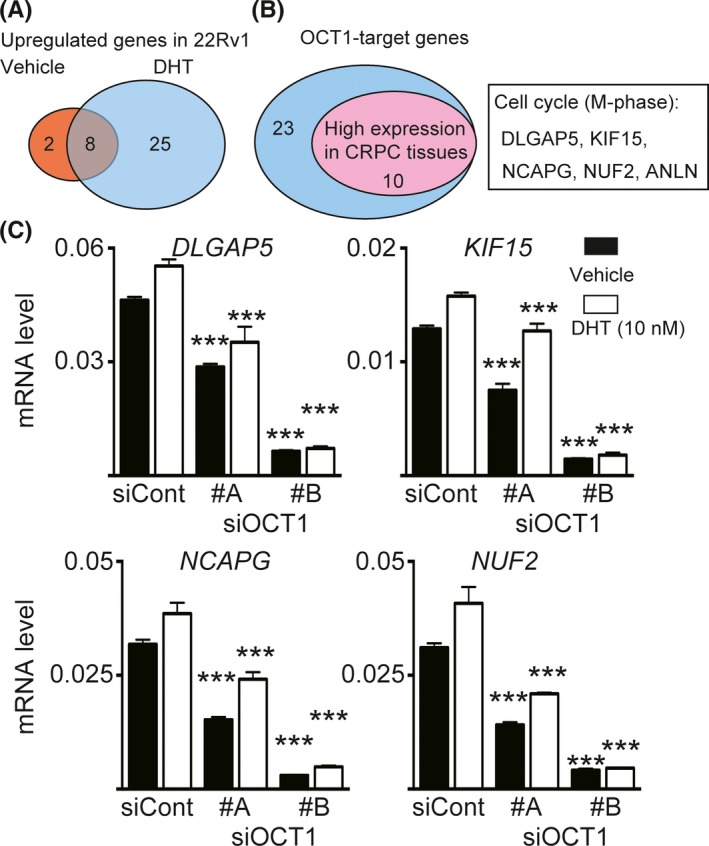
Identification of octamer transcription factor 1 (OCT1)‐target genes associated with castration‐resistant prostate cancer (CRPC). A, Candidate OCT1‐target genes in 22Rv1 cells. Among OCT1‐regulated genes in the presence of dihydrotestosterone (DHT),12 genes that satisfied the following three conditions were extracted: (i) upregulated in 22Rv1 cells compared with LNCaP cells in the absence of DHT; (ii) induced by DHT treatment in 22Rv1 cells; and (iii) high expression level was observed in 22Rv1 cells. Similarly, 10 genes were also extracted in the absence of DHT. B, Identification of OCT1‐target genes highly expressed in CRPC tissues. Using the Oncomine datasets (Varambally et al33 and Grasso et al32), we selected 10 candidate genes highly expressed in metastatic CRPC tissues. C, Validation of OCT1 regulation. 22Rv1 cells were treated with 10 nmol/L siControl or siOCT1 (#A and #B) for 48 h and then treated with 10 nmol/L DHT or vehicle for 24 h. We then measured mRNA expression levels of four candidate genes by qRT‐PCR. Results are presented as mean and SD (N = 3). ***P < 0.001, compared with siControl in the same condition. ANLN, anillin actin binding protein; DLGAP5, disks large‐associated protein 5; KIF15, kinesin family member 15; NCAPG, non‐SMC condensin I complex subunit G; NUF2, NDC80 kinetochore complex component
Figure 3.

Transcriptional regulatory mechanism of four novel octamer transcription factor 1 (OCT1)‐target genes. A, ChIP‐seq analysis of OCT1 and androgen receptor (AR) binding in the enhancer/promoter regions of disks large‐associated protein 5 (DLGAP5), kinesin family member 15 (KIF15), non‐SMC condensin I complex subunit G (NCAPG), and NDC80 kinetochore complex component (NUF2) in 22Rv1 cells are shown. Arrows indicate the direction of each gene. B, Four OCT1‐target genes were regulated by AR. 22Rv1 cells were treated with 5 nmol/L siControl (siCont.) or siAR (#A and #B) for 48 h and then treated with 10 nmol/L dihydrotestosterone (DHT) or vehicle for 24 h. mRNA expression level of each gene was measured by qRT‐PCR. Results are presented as mean and SD (N = 3). *P < 0.05, **P < 0.01, ***P < 0.001, compared with cells treated with siControl
3.3. Effects of knockdown of OCT1‐target genes on 22Rv1 cell proliferation and migration
Next, we evaluated the efficacy of knockdown of these OCT1‐target genes for inhibiting CRPC cell growth. We used siRNAs specifically targeting these OCT1 signals, and then confirmed substantial reduction of endogenous expression in 22Rv1 cells treated with these siRNAs compared with siControl (Figure 4A). MTS assay and cell migration assay were carried out using 22Rv1 cells treated with specific siRNA for each gene (siDLGAP5, siKIF15, siNCAPG, and siNUF2). Cell proliferation was significantly repressed in 22Rv1 cells treated with siDLGAP5 and siNUF2 compared with siControl (Figure 4B). Cell migration assays showed that the number of invaded cells was significantly decreased in 22Rv1 cells treated with siDLGAP5, siKIF15 and siNUF2 compared with siControl (Figure 4C).
Figure 4.

Functional analyses of four octamer transcription factor 1 (OCT1)‐target genes. A, Validation of siRNAs targeting each OCT1‐target gene. 22Rv1 cells were transfected with 5 nmol/L siControl or siRNA targeting four OCT1‐target genes identified in the present study (siDLGAP5, siKIF15, siNCAPG and siNUF2) for 48 h. We then measured mRNA expression levels of each gene by qRT‐PCR. Results are presented as mean and SD (N = 3). ***P < 0.001, compared with siControl. B, Role of four OCT1‐target genes in castration‐resistant prostate cancer (CRPC) cell proliferation. After treatment with 5 nmol/L siControl or siRNA targeting OCT1‐target genes, MTS assay was carried out at the indicated time points (N = 5). Results are presented as mean and SD. *P < 0.05, ***P < 0.001 vs siControl. C, Role of four OCT1‐target genes in CRPC cell migration. 22Rv1 cells were treated with siControl or siRNA targeting OCT1‐target genes. After 48 h incubation, cell migration assay was done (N = 5). Results are presented as mean and SD. ***P < 0.001. D, DLGAP5 was upregulated in 22Rv1 cells compared with LNCaP cells and induced by androgen. Western blot analysis for DLGAP5 and OCT1 expression was carried out and β‐actin was used as a loading control. E, 22Rv1 cells were treated with 10 nmol/L DHT or vehicle for 24 h. mRNA expression level of DLGAP5 was measured by qRT‐PCR. Results are presented as mean and SD (N = 3). **P < 0.01, ***P < 0.001, compared with LNCaP cells. F, ChIP‐seq analysis of OCT1 and AR binding in the enhancer/promoter regions of DLGAP5 in 22Rv1 and LNCaP cells is shown. Arrows indicate the direction of each gene. G, Involvement of OCT1 and DLGAP5 in AR activity in 22Rv1 cells. 22Rv1 cells were treated with siControl or siDLGAP5 (5 nmol/L). After 48 h incubation, cells were treated with 10 nmol/L DHT or vehicle for 24 h. Cell lysates were extracted for western blot analysis. Western blot analysis for indicated protein expressions were carried out and β‐actin was used as a loading control. DLGAP5, disks large‐associated protein 5; KIF15, kinesin family member 15; NCAPG, non‐SMC condensin I complex subunit G; NUF2, NDC80 kinetochore complex component
Next, we investigated how the downstream signaling pathways such as DLGAP5 are triggered in CRPC. DLGAP5 as well as other OCT1‐target genes were increased in 22Rv1 cells compared with LNCaP cells (Figure 4D, E and Figure S2A). ChIP‐seq analysis around the DLGAP locus showed multiple AR‐binding regions (Figure 3A). However, AR and OCT1 binding at the putative promoter region was obviously observed in 22Rv1 cells compared with LNCaP cells (Figure 4F), suggesting that recruitment of these transcription factors is a key event for inducing DLGAP5 in 22Rv1 cells. Moreover, we examined the role of OCT1 and DLGAP5 in AR expression and AR activity by western blot analysis (Figure 4G). We then observed decreased AR phosphorylation level and AR expression in response to DHT treatment by silencing DLGAP5 and OCT1. Therefore, these findings suggest that these signals can also be important for AR activity.
3.4. Clinical significance of DLGAP5 expression in prostate cancer
We found that DLGAP5 and NUF2 affect both migration and proliferation of 22Rv1 cells. As the relationship between the expression level of DLGAP5 in prostate cancer tissues and clinical characteristics has not been fully determined, we investigated the clinical significance of DLGAP5 expression in prostate cancer tissues. First, we confirmed that DLGAP5 as well as other three genes were highly expressed in metastatic CRPC tissues compared with localized prostate cancer by using data in the Oncomine database (Figure 5A and Figure S2B). Moreover, we conducted immunohistochemistry (IHC) analysis using specimens of prostate tissues obtained from 95 hormone therapy naïve prostate cancer patients by radical prostatectomy (Table 1) and CRPC tissues from six patients by transurethral resection of the prostate (TURP). In IHC analysis using DLGAP5 antibody (Figure S3A,B), we evaluated DLGAP5 expression by IR score and five was defined as the cut‐off value. Thus, the foci were classified as positive IR when IR ≧5 (Figure 5B). We observed a small number (N = 4) of DLGAP5‐positive cases in hormone naïve prostate cancer specimens. Interestingly, positive IR of DLGAP5 was significantly associated with poor prognosis of patients after the operation (Figure 5C). However, analysis of clinical background of these four patients showed no significant parameters (Table 1). Furthermore, we observed an increased number of cancer cells expressing DLGAP5 in CRPC tissues. In total, we detected positive IR in four out of six (67%) CRPC patients (Figure 5D). Furthermore, high OCT1 expression was observed in all CRPC tissues (Figure S4A,B). To measure the activation status of OCT1 in CRPC tissues, we evaluated the expression level of the OCT1‐major target, ANLN, by IHC analysis (Figure S4C). We then observed positive ANLN expression in four out of six cases in which DLGAP5 expression level was also high, in line with the increased OCT1 activity in these cases (Figure S4D). Thus, these findings supported that high OCT1 expression and activity induces DLGAP5 expression specifically in CRPC.
Figure 5.
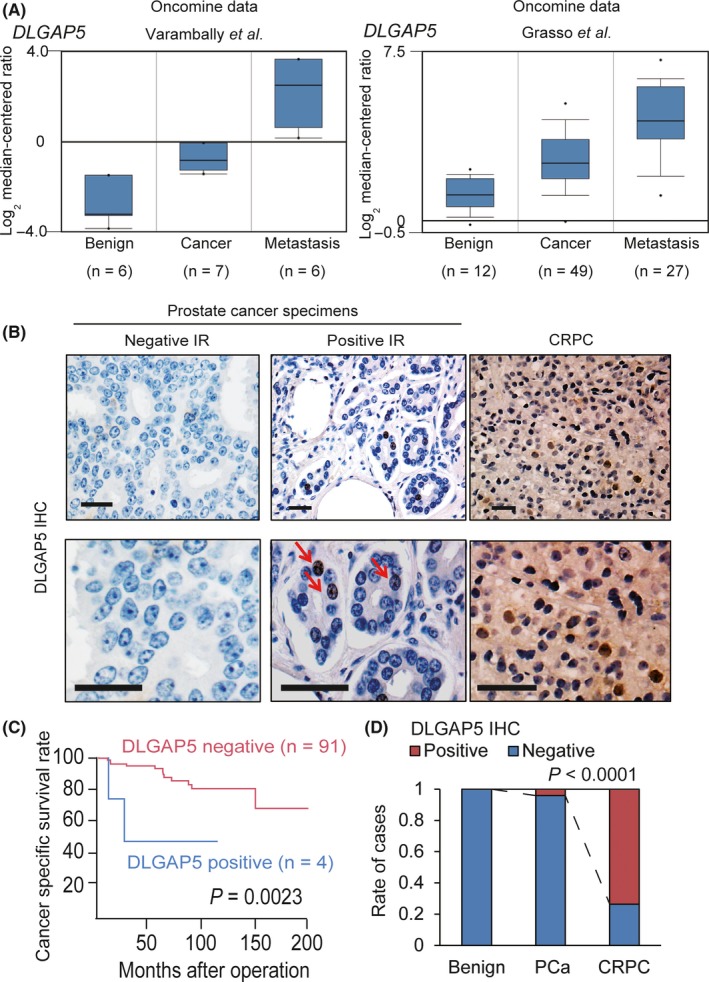
Disks large‐associated protein 5 (DLGAP5) expression in prostate cancer tissues. A, mRNA expression level of DLGAP5 in castration‐resistant prostate cancer (CRPC) tissues. DLGAP5 expression in CRPC tissues was analyzed by using data in the two Oncomine datasets (Varambally et al33 and Grasso et al32). B, Representative images of immunohistochemistry (IHC) of DLGAP5 in prostate cancer tissues. Representative images of negative and positive immunoreactive (IR) cases of prostate cancer specimens and CRPC tissues are shown. (Arrows, positive cells; scale bar, 50 μm). C, Positive expression of DLGAP5 is associated with poor prognosis of prostate cancer patients. Cancer‐specific survival of prostate cancer patients is shown (n = 95). Survival curve was obtained by Kaplan‐Meier method and P‐value was determined by log‐rank (Mantel‐Cox) test. D, Rate of cases in which positive IR was detected by DLGAP5 IHC in benign, prostate cancer (PCa), and CRPC tissues. Chi‐squared test was done to calculate P‐value
Table 1.
Relationship between DLGAP5 immunoreactivity and clinicopathological findings in human prostate cancer (n = 95)
| DLGAP5 immunoreactivity | |||
|---|---|---|---|
| Negative (n = 91) | Positive (n = 4) | P‐value | |
| Age (y) | 67.9 ± 5.9 | 70.2 ± 3.8 | .45 |
| Gleason score | |||
| 5‐7 | 62 | 2 | .46 |
| 8‐10 | 29 | 2 | |
| Pathological T stage | |||
| 2 | 42 | 1 | .60 |
| 3a | 28 | 1 | |
| 3b | 19 | 1 | |
| 4 | 2 | 1 | |
| Pathological N stage | |||
| N0 | 73 | 3 | .80 |
| N1 | 18 | 1 | |
Immunoreactivity (IR) score (0‐8) was obtained as the sum of the proportion and the intensity of IR. Proportion (0, none; 1, <1/100; 2, 1/100 to 1/10; 3, 1/10 to 1/3; 4, 1/3 to 2/3; and 5, >2/3), Intensity (0, none; 1, weak; 2, moderate; and 3, strong). IR scores of 0‐4 and 5, 6 were defined as negative and positive IR, respectively.
Abbreviation: DLGAP5, disks large‐associated protein 5.
4. DISCUSSION
Our previous study showed that high expression of OCT1 in prostate cancer tissues was correlated with poor prognosis of patients,8 indicating that OCT1 has a critical role in prostate cancer progression. However, the molecular function of OCT1 in CRPC cell proliferation has not been fully investigated. In the present study, we focused on target genes of OCT1 in CRPC model cells, 22Rv1.16 By western blot analysis and MTS assay, we observed the expression of AR‐V7, a hallmark of CRPC, and androgen‐independent cell growth in 22Rv1 cells. Consistent with our results, ADT is not sufficiently effective for 22Rv1 cells on account of AR hypersensitivity and expression of AR variants such as AR‐V7 unlike hormone‐naïve LNCaP cells in the previous report.17 Therefore, we used 22Rv1 cells as model cells to investigate the role of OCT1 in CRPC.
Although we previously identified ACSL3 as the major OCT1‐target gene promoting tumor growth,9 the expression levels of TF collaborating with AR and their target genes are quite different between hormone‐naïve prostate cancer and CRPC.18 Therefore, we hypothesized that overexpressed OCT1 may induce other target genes in CRPC in addition to ACSL3. Global OCT1 downstream signals were analyzed by integrating microarray and ChIP‐seq analyses in 22Rv1 cells. We then identified that cell cycle‐related genes such as ANLN were predominantly enriched among OCT1‐regulated genes.12 In the present study, we identified four novel OCT1‐target genes which are related to the cell cycle and highly expressed in CRPC tissues of patients. Although DLGAP5 and NUF2 promoted both cell proliferation and migration, KIF15 promoted cell migration of 22Rv1 cells. Interestingly, these three genes act by associating with microtubules during cell division. High expression of cell division‐related genes is involved in CRPC development.15, 19 Cell division‐associated gene UBE2C is a representative target of AR‐V7, the short variant of AR induced in CRPC.19 Therefore, these findings suggest that OCT1 could function with AR and AR‐V7 for promoting cell division in CRPC.
KIF15 is a motor protein that acts on cell division and belongs to the kinesin superfamily.20 It works on chromosome division by moving on microtubules.21 Previous reports indicate that KIF15 acts on tumor proliferation through the MEK‐ERK pathway and its expression levels in pancreatic cancer and breast cancer tissues are positively correlated with poor prognosis.22, 23 NUF2 belongs to the NDC80 complex involving NDC80, spindle pole body component 24 (SPC24), and spindle pole body component 25 (SPC25) and functions as a kinetochore protein.24 NUF2 is reported as one of the cancer testis antigens which is ectopically secreted by cancer, and the expression levels of NUF2 is elevated in prostate cancer tissues.25 NCAPG functions as condensin I complex cooperating with structural maintenance of chromosomes protein 2 (SMC2), structural maintenance of chromosomes protein 4 (SMC4), chromosome‐associated protein D 2 (CAP‐D2), and chromosome‐associated protein H (CAP‐H). Condensin I complex aggregates chromosomes in cell division.26 NCAPG has been reported to promote hepatocellular cancer cell proliferation and migration.27
DLGAP5 promotes the formation of microtubules by functioning as a kinetochore protein.28 Together with 30 other cell cycle progression genes, the expression level of DLGAP5 is positively related to the prostate cancer recurrence rate.29 Although further studies are necessary in order to elucidate the mechanisms of these genes on cellular migration, some studies suggested that the effects on microtubules in cell division are quite similar to the effects in cell migration.30, 31 In our IHC study, there were a small number of cases with localized prostate cancer with positive IR of DLGAP5. Notably, this positive IR is significantly associated with poor cancer‐specific survival of patients. By analyzing the relationship between DLGAP5 IR and clinicopathological findings, the ratio of elevated Gleason score (≧8) was relatively high in DLGAP5‐positive cases (50%) compared with ‐negative cases (32%), although no significant clinical parameters were found. Further investigation using a larger cohort will be required to determine the clinical background of cases with DLGAP5 high expression. We also showed enhanced expression of DLGAP5 in CRPC tissues. In line with our results, high mRNA expression level of DLGAP5 was observed in the CRPC samples of two cohorts, Grasso et al,32 and Varambally et al.33 These results suggest that expression of DLGAP5 is characteristic of CRPC tissues. Alternatively, according to a recent report, there might be a nest of treatment‐resistant cells from early‐stage prostate cancer.34 The prostate gland consists of basal and luminal epithelial cells as well as neuroendocrine cells that are well known to be resistant to chemotherapy.34 Furthermore, most of the luminal cells, the main components of the prostate gland, are sensitive to castration. However, a small number of luminal cells can remain and evolve into CRPC.35, 36 These findings suggest that high expression of DLGAP5 may be associated with such treatment‐resistant cells in prostate cancer. Meanwhile, we suggest that further clinical study by using more CRPC tissues is important to explore the usefulness of DLGAP expression as a promising biomarker or therapeutic target in CRPC.
In conclusion, we here identified four new OCT1‐target genes specifically expressed in CRPC. Our functional assays showed the role of these genes in CRPC cell growth and migration. In addition, we found that positive expression of DLGAP5 in prostate cancer tissues correlated with poor prognosis of patients. Interestingly, high expression of DLGAP5 was frequently observed in CRPC tissues. Our results provide an insight into the critical role of OCT1 in prostate cancer progression to CRPC.
DISCLOSURE
Authors declare no conflicts of interest for this article.
Supporting information
ACKNOWLEDGMENTS
This study was supported by grants of P‐CREATE (S.I., JP18cm0106144) from AMED, Japan; by grants (D.O., 19H03793, K.T., 17H04334) from JSPS, Japan; by grants from Takeda Science Foundation (S.I. and K.T.).
Yamamoto S, Takayama K‐I, Obinata D, et al. Identification of new octamer transcription factor 1‐target genes upregulated in castration‐resistant prostate cancer. Cancer Sci. 2019;110:3476–3485. 10.1111/cas.14183
Shinichiro Yamamoto and Ken‐ichi Takayama contributed equally to this work.
REFERENCES
- 1. Chen CD, Welsbie DS, Tran C, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33‐39. [DOI] [PubMed] [Google Scholar]
- 2. Takayama K, Inoue S. Transcriptional network of androgen receptor in prostate cancer progression. Int J Urol. 2013;20(8):756‐768. [DOI] [PubMed] [Google Scholar]
- 3. Scher HI, Halabi S, Tannock I, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26(7):1148‐1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Obinata D, Takayama K, Takahashi S, Inoue S. Crosstalk of the androgen receptor with transcriptional collaborators: potential therapeutic targets for castration‐resistant prostate cancer. Cancers (Basel) 2017;9(3):E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835‐839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor‐dependent prostate cancer growth. Mol Cell. 2007;27(3):380‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prefontaine GG, Walther R, Giffin W, Lemieux ME, Pope L, Hache RJ. Selective binding of steroid hormone receptors to octamer transcription factors determines transcriptional synergism at the mouse mammary tumor virus promoter. J Biol Chem. 1999;274(38):26713‐26719. [DOI] [PubMed] [Google Scholar]
- 8. Obinata D, Takayama K, Urano T, et al. Oct1 regulates cell growth of LNCaP cells and is a prognostic factor for prostate cancer. Int J Cancer. 2012;130(5):1021‐1028. [DOI] [PubMed] [Google Scholar]
- 9. Obinata D, Takayama K, Fujiwara K, et al. Targeting Oct1 genomic function inhibits androgen receptor signaling and castration‐resistant prostate cancer growth. Oncogene. 2016;35(49):6350‐6358. [DOI] [PubMed] [Google Scholar]
- 10. Migita T, Takayama KI, Urano T, et al. ACSL3 promotes intratumoral steroidogenesis in prostate cancer cells. Cancer Sci. 2017;108(10):2011‐2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Obinata D, Takada S, Takayama K, et al. Abhydrolase domain containing 2, an androgen target gene, promotes prostate cancer cell proliferation and migration. Eur J Cancer. 2016;57:39‐49. [DOI] [PubMed] [Google Scholar]
- 12. Takayama KI, Suzuki Y, Yamamoto S, Obinata D, Takahashi S, Inoue S. Integrative genomic analysis of OCT1 reveals coordinated regulation of androgen receptor in advanced prostate cancer. Endocrinology. 2019;160(2):463‐472. [DOI] [PubMed] [Google Scholar]
- 13. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data‐mining platform. Neoplasia. 2004;6(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rhodes DR, Kalyana‐Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen‐independent prostate cancer. Cell. 2009;138(2):245‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sramkoski RM, Pretlow TG II, Giaconia JM, et al. A new human prostate carcinoma cell line, 22Rv1. Vitro Cell Dev Biol Anim. 1999;35(7):403‐409. [DOI] [PubMed] [Google Scholar]
- 17. Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701‐711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma NL, Massie CE, Ramos‐Montoya A, et al. The androgen receptor induces a distinct transcriptional program in castration‐resistant prostate cancer in man. Cancer Cell. 2013;23(1):35‐47. [DOI] [PubMed] [Google Scholar]
- 19. Hu R, Lu C, Mostaghel EA, et al. Distinct transcriptional programs mediated by the ligand‐dependent full‐length androgen receptor and its splice variants in castration‐resistant prostate cancer. Cancer Res. 2012;72(14):3457‐3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu M, Nadar VC, Kozielski F, Kozlowska M, Yu W, Baas PW. Kinesin‐12, a mitotic microtubule‐associated motor protein, impacts axonal growth, navigation, and branching. J Neurosci. 2010;30(44):14896‐14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez‐Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19(20):1703‐1711. [DOI] [PubMed] [Google Scholar]
- 22. Wang J, Guo X, Xie C, Jiang J. KIF15 promotes pancreatic cancer proliferation via the MEK‐ERK signalling pathway. Br J Cancer. 2017;117(2):245‐255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zou JX, Duan Z, Wang J, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12(4):539‐549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tooley J, Stukenberg PT. The Ndc80 complex: integrating the kinetochore's many movements. Chromosome Res. 2011;19(3):377‐391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shiraishi T, Terada N, Zeng Y, et al. Cancer/Testis Antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy. J Transl Med. 2011;9:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115(1):109‐121. [DOI] [PubMed] [Google Scholar]
- 27. Zhang Q, Su R, Shan C, Gao C, Wu P. Non‐SMC condensin I complex, subunit G (NCAPG) is a novel mitotic gene required for hepatocellular cancer cell proliferation and migration. Oncol Res. 2018;26(2):269‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casanova CM, Rybina S, Yokoyama H, Karsenti E, Mattaj IW. Hepatoma up‐regulated protein is required for chromatin‐induced microtubule assembly independently of TPX2. Mol Biol Cell. 2008;19(11):4900‐4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12(3):245‐255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valiron O, Caudron N, Job D. Microtubule dynamics. Cell Mol Life Sci. 2001;58(14):2069‐2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng S, Song Y, Shen M, et al. Microtubule‐binding protein FOR20 promotes microtubule depolymerization and cell migration. Cell Discov. 2017;3:17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration‐resistant prostate cancer. Nature. 2012;487(7406):239‐243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8(5):393‐406. [DOI] [PubMed] [Google Scholar]
- 34. Shen MM, Abate‐Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24(18):1967‐2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rane JK, Droop AP, Pellacani D, et al. Conserved two‐step regulatory mechanism of human epithelial differentiation. Stem Cell Reports. 2014;2(2):180‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barros‐Silva JD, Linn DE, Steiner I, et al. Single‐cell analysis identifies LY6D as a marker linking castration‐resistant prostate luminal cells to prostate progenitors and cancer. Cell Rep. 2018;25(12):3504‐3518.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


