Abstract
Clear cell renal cell carcinoma (ccRCC) is one of the most common malignant tumors in the urinary system. Surgical intervention is the preferred treatment for ccRCC, but targeted biological therapy is required for postoperative recurrent or metastatic ccRCC. Autophagy is an intracellular degradation system for misfolded/aggregated proteins and dysfunctional organelles. Defective autophagy is associated with many diseases. Mul1 is a mitochondrion‐associated E3 ubiquitin ligase and involved in the regulation of divergent pathophysiological processes such as mitochondrial dynamics, and thus affects the development of various diseases including cancers. Whether Mul1 regulates ccRCC development and what is the mechanism remain unclear. Histochemical staining and immunoblotting were used to analyze the levels of Mul1 protein in human renal tissues. Statistical analysis of information associated with tissue microarray and The Cancer Genome Atlas (TCGA) database was conducted to show the relationship between Mul1 expression and clinical features and survival of ccRCC patients. Impact of Mul1 on rates of cell growth and migration and autophagy flux were tested in cultured cancer cells. Herein we show that Mul1 promoted autophagy flux to facilitate the degradation of P62‐associated protein aggresomes and adipose differentiation‐related protein (ADFP)‐associated lipid droplets and suppressed the growth and migration of ccRCC cells. Levels of Mul1 protein and mRNA were significantly reduced so that autophagy flux was likely blocked in ccRCC tissues, which is potentially correlated with enhancement of malignancy of ccRCC and impairment of patient survival. Therefore, Mul1 may promote autophagy to suppress the development of ccRCC.
Keywords: autophagy, clear cell renal cell carcinoma, Mul1 protein, P62 protein, perilipin‐2
1. INTRODUCTION
Clear cell renal cell carcinoma (ccRCC) is one of the most common urinary tract malignant tumors with high morbidity and mortality. In the past two decades, the incidence of ccRCC worldwide has doubled, and the death toll is rising at a rate of 1% per year.1 Development of ccRCC is difficult to predict, although epidemiological studies have found that tobacco consumption, obesity, hypertension and chronic nephropathy can induce ccRCC.1, 2, 3, 4, 5 Localized ccRCC can be treated with partial, radical nephrectomy or thermal ablation.6, 7, 8 However, approximately 30% of patients with localized ccRCC who underwent surgical intervention developed further metastatic ccRCC (mccRCC) and required systemic treatment.7 Although many targeted drugs and biologicals for ccRCC are currently in the research and development stage, their efficacy remains to be explored. Hence, the identification of new and effective biomarkers is essential for a better understanding of the biological progression of ccRCC and to improve the diagnosis and prognosis of this cancer in clinical practice.
Autophagy is an intracellular degradation system that captures damaged or non‐functional proteins and organelles and transports them to lysosomes for degradation.9 It plays an important homeostatic role in controlling the quality and quantity of proteins and organelles. Autophagy processes include cargo sequestration, transport to lysosomes, cargo degradation and recycling of degraded products, and the function of each different stage may be regulated differentially.10 LC3 functions as a core protein in autophagy and is mainly used for the recognition and recruitment of autophagy cargo. It elongates and seals the cargo and transports it to the autophagosomes.11 At present, there are conflicting reports about the roles of autophagy in the development of cancer. Some studies related to breast cancer, colon cancer, lung cancer, prostate cancer and pancreatic cancer have shown that autophagy mainly promotes cancer development,12, 13, 14, 15, 16 whereas others related to liver cancer and ccRCC have shown the opposite results in that autophagy suppresses cancer development.17, 18, 19 Our previous study found that microtubule‐associated protein family 1 (MAP1S) can promote autophagy clearance of lipid droplets and reduce DNA double‐strand breaks and genome instability, consequently suppressing the development of ccRCC and promoting patient survival.19 Similarly, other groups have shown that autophagy can suppress ccRCC development.20, 21, 22 Therefore, whether autophagy promotes or inhibits cancer development or whether autophagy has different regulatory mechanisms for cancer development in different tissues requires further investigation.9, 23
Mitochondrial E3 ubiquitin ligase 1 (Mul1), a multifunctional mitochondrial membrane‐associated protein located on the outer membrane of the mitochondrion with two transmembrane domains and one Ring‐finger domain,24 regulates different biological processes such as mitochondrial dynamics, cell growth, apoptosis and mitophagy (autophagy degradation of mitochondria), through ubiquitination and SUMOylation.25, 26, 27 Mitophagy and mitochondrial dynamics cause frequent changes in both quantity and morphology of cells and organelles.28 The imbalance of homeostasis is clearly associated with diseases such as Parkinson's disease, viral infection and carcinomas.29, 30, 31, 32, 33 It was reported that Mul1 mediates the ubiquitination of Akt and HSPA5 to inhibit the development of head and neck cancer.32, 33 Herein, we show that Mul1 promotes autophagy flux and suppresses ccRCC development with cell culture models and its loss of expression in ccRCC tissues from patients may lead to the blockade of autophagy flux, development of ccRCC and impairment of survival.
2. MATERIALS AND METHODS
2.1. Patients and tissue samples
Preliminary screening of levels of Mul1 protein by immunohistochemical analysis and immunoblot analysis with an antibody against Mul1 (ab84067) from Abcam were conducted on renal tissues from 11 patients and three patients diagnosed with ccRCC in the Fifth Affiliated Hospital, Guangzhou Medical University, respectively. Confirmation of levels of Mul1 protein by immunohistochemical analysis with the same antibody was conducted on a tissue microarray (TMA) from Xi'an Alenabio Co., Ltd (Cat No: PR803c) including 100 ccRCC tissues and 50 normal renal tissues with clinical information as summarized in Table 1. Further analyzing the relationship of levels of Mul1 mRNA with clinical features and survival times was conducted on information in a TCGA dataset including tumor tissues from 534 ccRCC patients with clinical features as summarized in Table 2. All procedures carried out in studies involving human patients were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. The present study is a retrospective study in accordance with ethics review regulations and was reviewed and approved by the Ethics Committee, The Fifth Affiliated Hospital, Guangzhou Medical University (Guangzhou, China).
Table 1.
Correlation of Mul1 expression with clinicopathological characteristics in patients with ccRCC (TMA)
| Clinical features | TMA | ||
|---|---|---|---|
| Case | Mean ± SD | P‐value | |
| Tissue | |||
| Cancer | 100 | 0.91 ± 0.75 | .000** |
| Normal | 50 | 2.78 ± 0.91 | |
| Age (y) | |||
| <60 | 84 | 1.45 ± 1.14 | .352 |
| ≥60 | 66 | 1.64 ± 1.26 | |
| Gender | |||
| Male | 102 | 1.56 ± 1.17 | .705 |
| Female | 48 | 1.48 ± 1.25 | |
| Pathological grade | |||
| G1 | 66 | 0.88 ± 0.75 | .941 |
| G2‐G3 | 30 | 0.87 ± 0.73 | |
| Clinical stage | |||
| I | 62 | 1.16 ± 0.73 | .000** |
| II | 38 | 0.50 ± 0.60 | |
| Tumor invasion | |||
| T1 | 62 | 1.16 ± 0.73 | .000** |
| T2 | 38 | 0.50 ± 0.60 | |
| Lymph node metastasis | |||
| N0 | 100 | 0.91 ± 0.75 | — |
| N1 | 0 | — | |
| Distant metastasis | |||
| M0 | 100 | 0.91 ± 0.75 | — |
| M1 | 0 | — | |
Abbreviation: ccRCC, clear cell renal cell carcinoma; Mul1, mitochondrial E3 ubiquitin ligase 1; TMA, tissue microarray.
*P < .05; **P < .01.
"—" mean:data cannot be analyzed.
Table 2.
Correlation of Mul1 expression with clinicopathological characteristics in patients with ccRCC (TCGA)
| Clinical features | TCGA | |||
|---|---|---|---|---|
| Case | Low, n (%) | High, n (%) | P‐value | |
| Tissue | ||||
| Cancer | 534 | 267 (50) | 267 (50) | — |
| Normal | — | — | ||
| Age (y) | ||||
| <60 | 246 | 144 | 102 | .114 |
| ≥60 | 287 | 152 | 135 | |
| Gender | ||||
| Male | 345 | 215 | 130 | .000** |
| Female | 188 | 81 | 107 | |
| Pathological grade | ||||
| G1‐G2 | 242 | 115 | 127 | .000** |
| G3‐G4 | 283 | 179 | 104 | |
| Clinical stage | ||||
| I‐II | 324 | 165 | 159 | .005* |
| III‐IV | 206 | 129 | 77 | |
| Tumor invasion | ||||
| T1‐T2 | 342 | 178 | 164 | .019* |
| T3‐T4 | 191 | 118 | 73 | |
| Lymph node metastasis | ||||
| N0 | 239 | 134 | 105 | .409 |
| N1 | 16 | 10 | 6 | |
| Distant metastasis | ||||
| M0 | 421 | 230 | 191 | .065 |
| M1 | 79 | 51 | 28 | |
Abbreviation: ccRCC, clear cell renal cell carcinoma; Mul1, mitochondrial E3 ubiquitin ligase 1; TCGA, The Cancer Genome Atlas.
*P < .05; **P < .01.
"—" mean:data cannot be analyzed.
2.2. Generation of stable cell lines
Cell lines HK2 (CC4008), 769‐P (CC1504) and ACHN (CC1505) were purchased from Cellcook, Guangzhou. 769‐P cells were transfected with control siRNA and Mul1‐specific siRNAs Mul1‐342, Mul1‐486 and Mul1‐1080 from Shanghai Genepharma or control lentiviral expression vector, and the vector with Mul1 was inserted between cut sites of restriction enzyme EcoRI and BamHI from Guangzhou HYY to select stable cell lines with the expression of Mul1 suppressed or elevated as confirmed by both quantitative RT‐PCR and immunoblot analyses.
2.3. Assays of cell growth rates and migration
Cell growth rates were carried out through the CCK‐8 proliferation assay. The same number of cells were seeded and cultured in normal medium. After being cultured for 4 hours, 24 hours, 48 hours and 72 hours, cells were treated with 20 μL CCK‐8 solution for 2 hours and subjected to absorbance measurement at 450 nm by microplate reader. Cell migration rates were analyzed by wound‐healing assay. Cells grown to full confluence on culture plates were treated with 4 μg/mL mitomycin (Cat#: 10107409001) from Roche for 2 hours and scratched. Cells were continuously cultured and observed under a microscope to measure distances of cell migration at specific time points.
2.4. Assays of the impact of Mul1 on autophagy
Stable cell lines with suppression or elevation of Mul1 were cultured to full confluence and treated with 10 μmol/L Bafilomycin A1 (S1413) from Selleck for 6 hours. Cells were collected and lysed to prepare cell lysates. Cell lysates with the same amount of total protein as measured by the bicinchoninic acid (BCA) method were separated on SDS‐PAGE and separated proteins were transferred to membranes to conduct immunoblot with Mul1 antibody as described above. LC3 antibody (ab192890) was from Abcam, and P62 antibody (#88588) was from CST. Levels of adipose differentiation‐related protein (ADFP) in renal tissues from ccRCC patients from our hospital were analyzed by immunohistochemical staining with ADFP‐specific antibody (cat# ab181452) from Abcam.
2.5. Assigment of immunoreactivity scores and statistical analyses
To quantify the levels of Mul1 protein in renal tissues from ccRCC patients as shown by immunohistochemical staining, five random fields captured under microscope with a magnitude of 400‐fold were selected. Percentage of positively stained cells to total cells was calculated and scored with the standard: <5% (0 points), 6%‐25% (1 point), 26%‐50% (2 points), 51%‐75% (3 points), and >75% (4 points). Staining intensity was visually scored and stratified according to the following criteria: no staining (0 points), mild (1 point), moderate (2 points), and strong (3 points). The final immunoreactivity score of each case was calculated by adding the two scores based on the immunostaining percentage and the immunostaining intensity.
SPSS 22.0 software was used for statistical analysis. Survival was analyzed using the Kaplan‐Meier method. Univariate analysis comparisons and multivariate survival comparisons were carried out using Cox proportional hazard regression models. Pearson's chi‐squared tests (TCGA database) and Student's t tests (TMA database) were used to analyze the association of Mul1 expression with ccRCC clinicopathological characteristics. Differences were assigned as statistically significant when P < .05.
3. RESULTS
3.1. Expression of Mul1 protein is reduced in human ccRCC tissues
To probe the role of Mul1 protein in the development of ccRCC, we conducted preliminary screening of the levels of Mul1 protein in ccRCC tissues and their respective adjacent normal renal tissues from 11 patients collected in our hospital by immunohistochemical staining. It was found that the levels of Mul1 protein were high in the distal convoluted tubules and the proximal convoluted tubules of normal renal tissues but near negative in ccRCC tissue from the same patients (Figure 1A‐D). The lower levels of Mul1 expression in ccRCC tissues compared to their respective normal renal tissues were further confirmed with tissues from three patients by immunoblot analysis (Figure 1E,F). To further verify the reduced expression of Mul1 in ccRCC tissues, we examined the levels of Mul1 in a commercially available TMA which contained 100 ccRCC and 50 adjacent non‐cancerous tissues by immunohistochemical staining. Levels of Mul1 protein reflected by immunoreactivity score (IRS) were significantly lower in ccRCC tissues than in normal tissues (Figure 1G‐L, Table 1). Therefore, the expression of Mul1 protein is dramatically reduced in ccRCC tissues.
Figure 1.
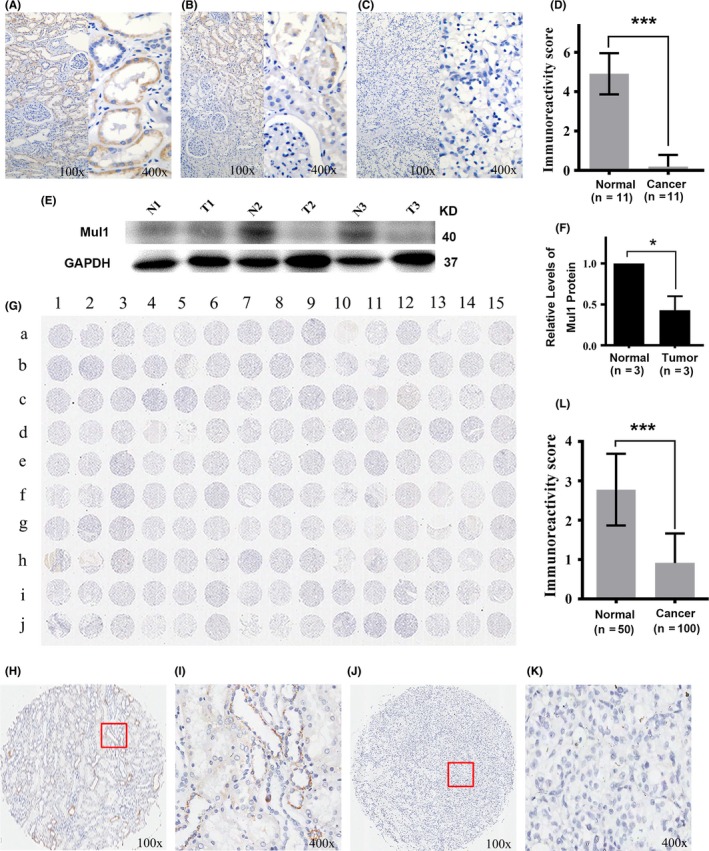
Expression of mitochondrial E3 ubiquitin ligase 1 (Mul1) protein in clear cell renal cell carcinoma (ccRCC) tissues and their adjacent normal tissues. (A‐C) Representative images showing adjacent normal tissues expressing high levels (A), tissues near tumor tissues expressing intermediate levels (B) and ccRCC tissues expressing low levels of Mul1 (C) collected from 11 patients with ccRCC in our hospital. Left half is an image with low‐resolution (100×) and right half with high‐resolution (400×). (D) Plot showing a comparison of immunoreactivity scores of Mul1 between normal and cancer tissues from ccRCC patients as stated above. Mean and SD for normal tissues are 4.91 ± 1.04 and those for cancer tissues are 0.18 ± 0.60. ***P ≤ .001. Sample size is 11. (E,F) Immunoblot assays (E) and quantification (F) showing levels of Mul1 in tumor (T1, T2 and T3) and their respective adjacent normal tissues (N1, N2 and N3) from three ccRCC patients from our hospital. *P ≤ .05. (G) Full immunohistochemical image showing expression levels of Mul1 in a tissue microarray containing 100 ccRCC and 50 non‐cancerous tissues. (H‐K) Representative images showing adjacent normal tissues (H,I) and ccRCC tissues (J,K) with low‐resolution (100×) (H,J) or high resolution (400×) (I,K). (L) Plot showing a comparison of immunoreactivity scores of Mul1 between normal (n = 50) and cancer tissues (n = 100) as shown in G. Mean and SD for normal tissues are 2.78 ± 0.91 and those for cancer tissues are 0.91 ± 0.75. ***P ≤ .001
3.2. Reduced expression of Mul1 predicts poor prognosis of human patients with ccRCC
We examined the levels of Mul1 mRNA and protein in cultured cells including HK‐2 (human kidney 2), an immortalized proximal tubular cell line derived from a normal kidney, cell line 769‐P from primary clear cell adenocarcinoma, and ACHN from a patient with metastatic renal adenocarcinoma. The levels of Mul1 in cancer cells 769‐P and ACHN were lower than those in normal renal cell HK‐2 as indicated by levels of both protein and mRNA (Figure 2A‐C). The consistent low expression of both Mul1 mRNA and protein in ccRCC tissues prompted us to analyze the data deposited in TCGA database which contains information on the levels of Mul1 mRNA in cancer tissues from 534 ccRCC patients (Table 2). In agreement with results related to protein levels, patients diagnosed with higher clinical stage and higher degree of tumor invasion expressed lower levels of Mul1 mRNA and protein (Tables 1 and 2). Further analyses indicated that patients with lower levels of Mul1 mRNA had significantly poorer overall and disease‐free survival than those with higher levels of Mul1 mRNA (Figure 2D,E). In addition, univariate analysis and further multivariate analysis indicated that lower levels of Mul1 mRNA but higher degree of tumor invasion and pathological grade showed higher hazard ratio to disease‐free survival (Table 3). Therefore, reduced levels of Mul1 predict more malignant ccRCC and poor prognosis of ccRCC patients.
Figure 2.
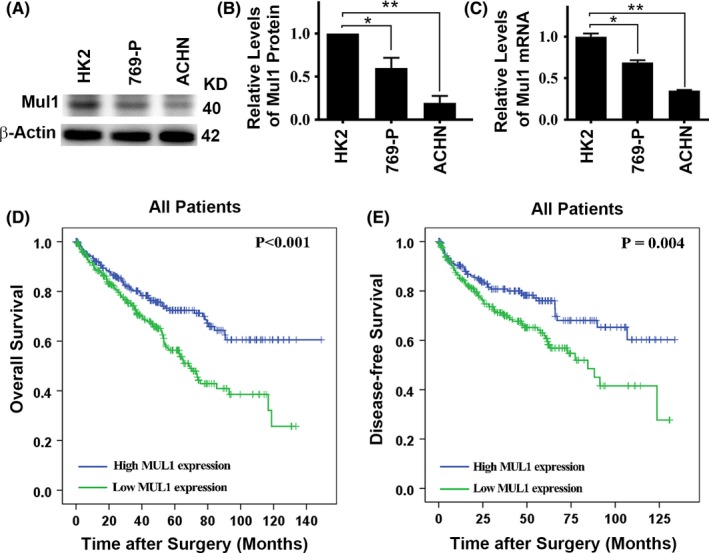
Relationship between levels of mitochondrial E3 ubiquitin ligase 1 (Mul1) mRNA in clear cell renal cell carcinoma (ccRCC) tissues and survival of ccRCC patients. (A,B) Representative immunoblot results (A) and quantification (B) showing levels of Mul1 protein in different types of renal cells. (C) Plot of relative levels of Mul1 mRNA in the same types of renal cells as shown in A,B. (D,E) Kaplan‐Meier survival curves showing overall survival time (D) and disease‐free survival (E) after surgery of ccRCC patients with high or low levels of Mul1 mRNA. Significance of difference between the two groups was estimated by chi‐squared test. *P < .05; **P < .01.
Table 3.
Prognostic value of Mul1 expression on disease‐free survival by Cox proportional hazards model
| Variable | Disease‐free survival | |
|---|---|---|
| HR (95% CI) | P‐value | |
| Univariate analysis | ||
| Age, y (≥60 vs <60) | 1.820 (1.330‐2.491) | .000** |
| Gender (Male vs Female) | 1.058 (0.778‐1.440) | .719 |
| Tumor invasion (T1‐T2 vs T3‐T4) | 3.164 (2.339‐4.281) | .000** |
| Lymph node stage (N0 vs N1) | 3.386 (1.797‐6.377) | .000* |
| Pathological grade (G1‐G2 vs G3‐G4) | 2.612 (1.860‐3.669) | .000* |
| Clinical stage (S1‐S2 vs S3‐S4) | 3.853 (2.810‐5.283) | .000** |
| Distant metastasis (M0 vs M1) | 4.348 (3.187‐5.930) | .000** |
| Mul1 expression (low vs high) | 0.552 (0.402‐0.757) | .000** |
| Multivariate analysis | ||
| Mul1 expression (low vs high) | 0.663 (0.479‐0.918) | .013* |
| Tumor invasion (T1‐T2 vs T3‐T4) | 2.308 (1.672‐3.186) | .000** |
| Age, y (≥60 vs <60) | 1.605 (1.168‐2.208) | .004** |
| Pathological grade (G1‐G2 vs G3‐G4) | 1.765 (1.227‐2.540) | .002** |
Abbreviation: CI, confidence interval; HR, hazard ratio; Mul1, mitochondrial E3 ubiquitin ligase 1.
*P < .05; **P < .01.
3.3. Mitochondrial E3 ubiquitin ligase 1 inhibits the growth and migration of ccRCC cells
In order to test the roles of Mul1 in tumorigenesis, we generated stable cell lines with reduced or overexpressed levels of Mul1 in the background of cancer cell 769‐P. Treating 769‐P cells with two types of Mul1‐specific siRNA molecule led to successful suppression of Mul1 protein (Figure 3A,B), whereas adding a plasmid carrying the Mul1 gene did not increase the levels of Mul1 protein (Figure 3C,D). Further measurements with RT‐PCR indicated that the treatment with siRNA led to a significant reduction whereas treatment with plasmid led to a significant increase in the levels of Mul1 mRNA (Figure 3E,F).
Figure 3.
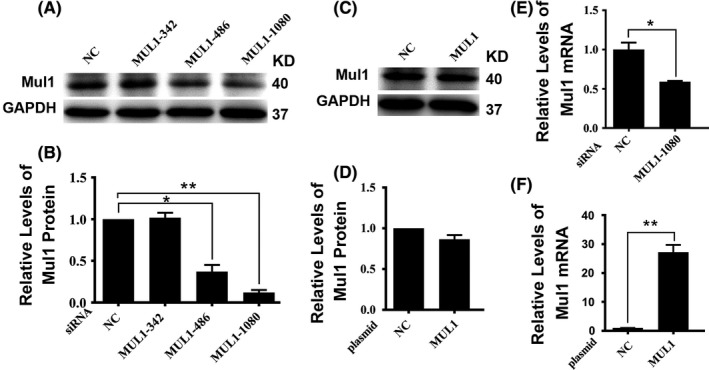
Expression of mitochondrial E3 ubiquitin ligase 1 (Mul1) in engineered 769‐P cells. (A‐D) Representative immunoblot results (A,C) and their respective quantification (B,D) showing levels of Mul1 protein in control cells (NC) and stable cell lines transfected with different Mul1‐specific siRNAs (A,B) or control cells (NC) and stable cell lines transfected with Mul1 expressing plasmid (C,D). (E,F) Plots showing a comparison of levels of Mul1 mRNA between control (NC) and stable cell line with siRNA 1080 (E) and between control (NC) and stable cell line with Mul1 plasmid (F). *P ≤ .05; and **P ≤ .01
To test the impact of Mul1 on growth and migration, we conducted CCK‐8 proliferation assay and wound‐healing experiments using stable cell lines. Suppressing the expression of Mul1 led to a significant increase in cell growth rates (Figure 4A). Because of the inefficiency to increase the levels of Mul1 protein by overexpression, cells transfected with Mul1 expression plasmid did not show any significant impact on growth as expected (Figure 4B). Similarly, suppressing expression of Mul1 promoted cell migration (Figure 4C,D), but the overexpression of Mul1 did not change the migration rates as expected (Figure 4C,E). In general, Mul1 inhibits the growth and migration of ccRCC cells.
Figure 4.
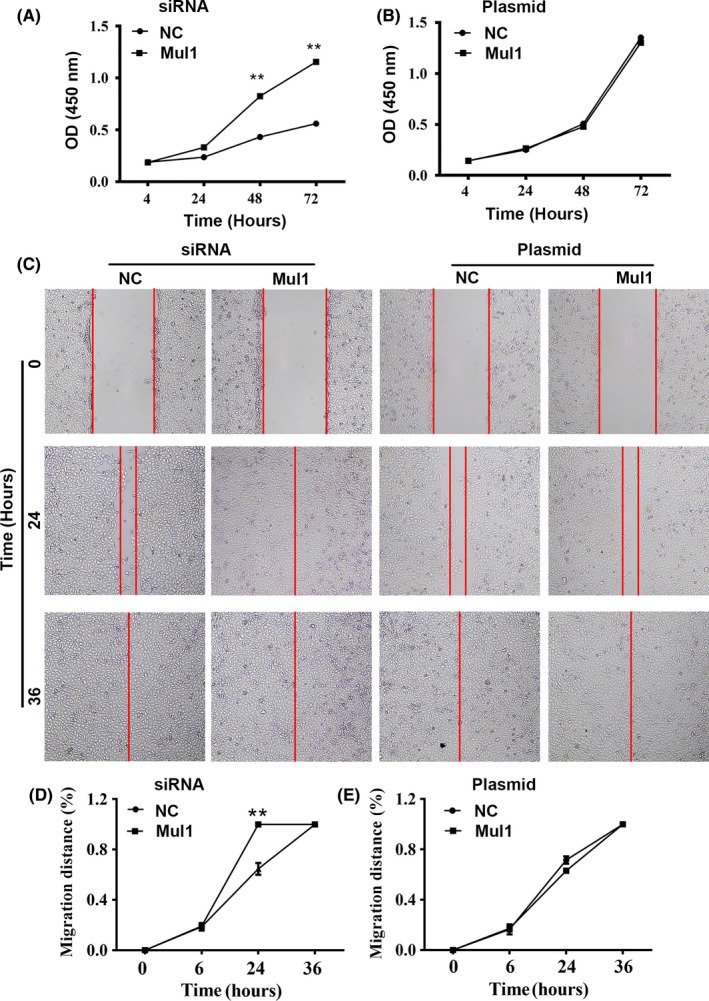
Impact of mitochondrial E3 ubiquitin ligase 1 (Mul1) on growth and migration of clear cell renal cell carcinoma (ccRCC) cells. (A,B) Plots showing a comparison of growth rates between control (NC) and cells with Mul1‐specific siRNA (A) or between control (NC) and cells with Mul1 plasmid (B). (C‐E) Representative images (C) and plots (D,E) showing a comparison of migration distances between cells as stated in A (D) or B (E). **P ≤ .01
3.4. Mitochondrial E3 ubiquitin ligase 1 promotes autophagy flux in ccRCC
Although it was observed that cells transfected with Mul1‐expressing plasmid showed higher levels of Mul1 mRNA (Figure 3F) but did not show increased levels of Mul1 protein (Figure 3C,D), levels of Mul1 protein did increase in cells expressing Mul1 in the presence of lysosomal inhibitor Bafilomycin A1 (Figure 5A,B). Levels of Mul1 protein in the presence of Bafilomycin A1 were reduced as expected when cells were treated with Mul1‐specific siRNA (Figure 5C,D). These results suggested that overexpressed Mul1 protein was degraded immediately and efficiently through the lysosomal system.
Figure 5.

Impact of mitochondrial E3 ubiquitin ligase 1 (Mul1) on autophagy flux. (A‐D) Representative immunoblots (A,C) and plots (B,D) showing levels of Mul1 protein in cells with Mul1 plasmid (A,B) or siRNA (C,D) in the presence of Bafilomycin A1 (BAF). (E‐J) Representative immunoblots (E,H) and plots (F,G,I,J) showing levels of LC3‐II (F,I) or P62 (G,J) protein in cells with siRNA (E‐G) or Mul1 plasmid (H‐J) in the absence (Ctrl) or presence of Bafilomycin A1 (BAF). *P ≤ .05; and **P ≤ .01. (K‐M) Representative images showing adjacent normal tissues expressing normal levels (K), fibrotic tissues near clear cell renal cell carcinoma (ccRCC) tissues expressing low levels (L) and ccRCC tissues expressing high levels of adipose differentiation‐related protein (ADFP) (M) collected from 11 patients diagnosed with ccRCC in our hospital. (N) Plot showing a comparison of immunoreactivity scores of ADFP between normal and cancer tissues from ccRCC patients as stated above. **P ≤ .01. Sample size is 11
To further determine the cellular function of Mul1, we examined the levels of autophagy marker LC3‐II and P62 after bafilomycin treatment. Treating cells with Mul1‐specific siRNA to reduce the levels of Mul1 led to a reduction in autophagy flux as reflected by the reduced levels of LC3‐II in the presence of Bafilomycin A1 and an accumulation of aggresome marker P62 in the absence of Bafilomycin A1 (Figure 5E‐G). Overexpressing Mul1 caused an increase in autophagy flux as reflected by the increased levels of LC3‐II in the presence of Bafilomycin A1 and a dramatic reduction of aggresomes as reflected by the reduced levels of P62 in the presence of Bafilomycin A1 (Figure 5H‐J). Autophagy defects in the degradation of ADFP‐associated lipid droplets may enhance the initiation and development of ccRCC and reduce the survival of ccRCC patients.19 Mul1 deficiency in ccRCC tissues predicted a blockade of autophagy degradation of lipid droplets and an accumulation of their associated ADFP. Examination of levels of ADFP by histo‐immunochemical staining showed that levels of ADFP were significantly higher in ccRCC tissues than in adjacent normal or fibrotic tissues (Figure 5K‐N). Therefore, Mul1 promotes autophagy flux and its loss in tumor tissues may promote the development of ccRCC.
4. DISCUSSION
As a common urological malignancy, based on epidemiological data, ccRCC was shown to be caused by obesity, smoking, hypertension, chronic kidney disease and type 2 diabetes, suggesting the potential to prevent the development of ccRCC.2, 3, 4, 5 Established surgical treatments have shown good results for the treatment of localized ccRCC.6, 7, 8 However, the treatment for postoperative recurrence or concurrent metastasis of mccRCC still lacks any specific therapeutic drug that can improve the survival of ccRCC patients.
Currently, the relationship between autophagy and cancer is still unclear. In general, basal autophagy flux inhibits early tumor development by maintaining homeostasis and suppressing genomic damage events.33 Because the metabolic and biosynthesis requirements of tumor cells are significantly increased, it seems that cancer cells depend more on autophagy than do normal cells. Therefore, some suggest that autophagy promotes the development of advanced tumors.34 However, the latest research findings show that the development of cancer is often accompanied by the decline or loss of autophagy. When autophagy flux is blocked, cancer cells accumulate more genomic errors and become more malignant.35 Our previous results showed that promoting autophagy is beneficial to inhibit the development of hepatocellular carcinoma and ccRCC and to improve patient survival.35 It was found that autophagy flux was relatively reduced in ccRCC and negatively correlated with tumor clinical stage and pathological grade.17
Mitochondrial E3 ubiquitin ligase 1 was originally discovered as an E3 ligase of ubiquitination. It promotes autophagy degradation of mitochondria by interacting with a specific E2 ubiquitination‐binding enzyme and autophagy‐related protein GABARAP,25 or mediating the ubiquitination of autophagy upstream protein ULK1.27 Although Mul1 protein was immediately degraded potentially because of the Mul1‐enhanced autophagy flux, the autophagy flux is actually increased in cells overexpressing Mul1 as confirmed by increased levels of Mul1 mRNA. However, the reduced expression of Mul1 in ccRCC tissues compared to their adjacent normal renal tissues likely suggests true reductions in both mRNA and protein of Mul1 and autophagy flux. Loss of Mul1 expression and blockade of autophagy flux lead to the accumulation of P62‐associated protein aggresomes. P62 acts as a selective autophagy receptor protein to recruit ubiquitinated autophagy substrates in autophagosomes and transfers them to lysosomes for degradation.36, 37 Impairment of autophagy flux enhances ccRCC development by regulating degradation of hypoxia‐inducible factor 2α through P62.38 Suppression of Mul1 led to the enhancement of proliferation and migration of ccRCC cells, promotion of cancer malignancy and reduction of ccRCC patient survival. However, increased expression of Mul1 did not impact cell growth and migration but did enhance autophagy flux, suggesting that not the growth and migration of tumor cells, but the malignancy of ccRCC and its associated patient survival are directly related to autophagy flux. In summary, Mul1 promotes autophagy flux and its loss is associated with blockade of autophagy flux, accumulation of protein aggresomes, enhancement of ccRCC malignancy, and impairment on patient survival. The observations in the present study suggest that Mul1 might exert anti‐tumorigenic roles in certain stages of ccRCC development, although in vivo studies are required to reach a final conclusion.
DISCLOSURE
Authors declare no conflicts of interest for this article.
ACKNOWLEDGMENTS
This work was supported by funding from National Natural Science Foundation of China (81974392), Natural Science Foundation of Guangdong Province (2018A030313087) and Science and Technology Program of Guangzhou City of China (201607010157) to Guibin Xu. Medical Science and Technology Program of Guangzhou of China (20171A010325) to Haibo Zhao. Medical Science and Technology Program of Guangzhou of China (20171A011092), Medical Research Foundation of Guangdong Province of China (A2018093) to Zhengming Su and National Natural Science Foundation of China (81772931) to Leyuan Liu.
Yuan Y, Li X, Xu Y, et al. Mitochondrial E3 ubiquitin ligase 1 promotes autophagy flux to suppress the development of clear cell renal cell carcinomas. Cancer Sci. 2019;110:3533–3542. 10.1111/cas.14192
Yaoji Yuan, Xiezhao Li, Yuyu Xu contributed equally to this work.
Contributor Information
Leyuan Liu, Email: liuleyuan@hotmail.com.
Guibin Xu, Email: gyxgb@163.com.
REFERENCES
- 1. Dy GW, Gore JL, Forouzanfar MH, Naghavi M, Fitzmaurice C. Global burden of urologic cancers, 1990‐2013. Eur Urol. 2017;71:437‐446. [DOI] [PubMed] [Google Scholar]
- 2. Bergstrom A, Hsieh CC, Lindblad P, Lu CM, Cook NR, Wolk A. Obesity and renal cell cancer–a quantitative review. Br J Cancer. 2001;85:984‐990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cumberbatch MG, Rota M, Catto JW, La Vecchia C. The role of tobacco smoke in bladder and kidney carcinogenesis: a comparison of exposures and meta‐analysis of incidence and mortality risks. Eur Urol. 2016;70:458‐466. [DOI] [PubMed] [Google Scholar]
- 4. Hidayat K, Du X, Zou SY, Shi BM. Blood pressure and kidney cancer risk: meta‐analysis of prospective studies. J Hypertens. 2017;35:1333‐1344. [DOI] [PubMed] [Google Scholar]
- 5. Macleod LC, Hotaling JM, Wright JL, et al. Risk factors for renal cell carcinoma in the VITAL study. J Urol. 2013;190:1657‐1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. El Dib R, Touma NJ, Kapoor A. Cryoablation vs radiofrequency ablation for the treatment of renal cell carcinoma: a meta‐analysis of case series studies. BJU Int. 2012;110:510‐516. [DOI] [PubMed] [Google Scholar]
- 7. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67:913‐924. [DOI] [PubMed] [Google Scholar]
- 9. Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913‐1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861‐2873. [DOI] [PubMed] [Google Scholar]
- 11. Huang R, Xu Y, Wan W, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57:456‐466. [DOI] [PubMed] [Google Scholar]
- 12. Levy J, Cacheux W, Bara MA, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome‐influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015;17:1062‐1073. [DOI] [PubMed] [Google Scholar]
- 13. Santanam U, Banach‐Petrosky W, Abate‐Shen C, Shen MM, White E, DiPaola RS. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev. 2016;30:399‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strohecker AM, Guo JY, Karsli‐Uzunbas G, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E‐driven lung tumors. Cancer Discov. 2013;3:1272‐1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang A, Rajeshkumar NV, Wang X, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905‐913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yeo SK, Wen J, Chen S, Guan JL. Autophagy differentially regulates distinct breast cancer stem‐like cells in murine models via EGFR/Stat3 and Tgfbeta/Smad signaling. Cancer Res. 2016;76:3397‐3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng Q, Wang Z, Wang L, et al. Lower mRNA and protein expression levels of LC3 and Beclin1, markers of autophagy, were correlated with progression of renal clear cell carcinoma. Jpn J Clin Oncol. 2013;43:1261‐1268. [DOI] [PubMed] [Google Scholar]
- 18. Takamura A, Komatsu M, Hara T, et al. Autophagy‐deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795‐800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu G, Jiang Y, Xiao Y, et al. Fast clearance of lipid droplets through MAP1S‐activated autophagy suppresses clear cell renal cell carcinomas and promotes patient survival. Oncotarget. 2016;7:6255‐6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deng F, Ma YX, Liang L, Zhang P, Feng J. The pro‐apoptosis effect of sinomenine in renal carcinoma via inducing autophagy through inactivating PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2018;97:1269‐1274. [DOI] [PubMed] [Google Scholar]
- 21. Verma SP, Agarwal A, Das P. Sodium butyrate induces cell death by autophagy and reactivates a tumor suppressor gene DIRAS1 in renal cell carcinoma cell line UOK146. In Vitro Cell Dev Biol Anim. 2018;54:295‐303. [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y, Fan Y, Huang S, et al. Thymoquinone inhibits the metastasis of renal cell cancer cells by inducing autophagy via AMPK/mTOR signaling pathway. Cancer Sci. 2018;109:3865‐3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528‐542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li W, Bengtson MH, Ulbrich A, et al. Genome‐wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE. 2008;3:e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peng J, Ren KD, Yang J, Luo XJ. Mitochondrial E3 ubiquitin ligase 1: a key enzyme in regulation of mitochondrial dynamics and functions. Mitochondrion. 2016;28:49‐53. [DOI] [PubMed] [Google Scholar]
- 26. Zhang B, Huang J, Li HL, et al. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 2008;18:900‐910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J, Qi W, Chen G, et al. Mitochondrial outer‐membrane E3 ligase MUL1 ubiquitinates ULK1 and regulates selenite‐induced mitophagy. Autophagy. 2015;11:1216‐1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Archer SL. Mitochondrial dynamics–mitochondrial fission and fusion in human diseases. N Engl J Med. 2013;369:2236‐2251. [DOI] [PubMed] [Google Scholar]
- 29. Tang FL, Liu W, Hu JX, et al. VPS35 deficiency or mutation causes dopaminergic neuronal loss by impairing mitochondrial fusion and function. Cell Rep. 2015;12:1631‐1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yun J, Puri R, Yang H, et al. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. eLife. 2014;3:e01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doiron K, Goyon V, Coyaud E, Rajapakse S, Raught B, McBride HM. The dynamic interacting landscape of MAPL reveals essential functions for SUMOylation in innate immunity. Sci Rep. 2017;7:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim SY, Kim HJ, Kang SU, et al. Non‐thermal plasma induces AKT degradation through turn‐on the MUL1 E3 ligase in head and neck cancer. Oncotarget. 2015;6:33382‐33396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim SY, Kim HJ, Kim HJ, et al. HSPA5 negatively regulates lysosomal activity through ubiquitination of MUL1 in head and neck cancer. Autophagy. 2018;14:385‐403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. White E. Deconvoluting the context‐dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nassour J, Radford R, Correia A, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659‐663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847‐22857. [DOI] [PubMed] [Google Scholar]
- 37. Lamark T, Svenning S, Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61:609‐624. [DOI] [PubMed] [Google Scholar]
- 38. Liu XD, Yao J, Tripathi DN, et al. Autophagy mediates HIF2alpha degradation and suppresses renal tumorigenesis. Oncogene. 2015;34:2450‐2460. [DOI] [PMC free article] [PubMed] [Google Scholar]


