Abstract
NOP2/Sun domain family, member 2 (NSUN2) is a nuclear RNA methyl‐transferase catalyzing 5‐methylcytosine formation. Evidence shows that NSUN2 is correlated with cell unlimited proliferation. However, its functional role in gallbladder carcinoma (GBC), which is the most common biliary tract malignancy and has a poor prognosis, remains to be determined. Here we found that NSUN2 was highly expressed in GBC tissues as well as cell lines. NSUN2 silencing repressed GBC cell proliferation and tumorigenesis both in vitro and in vivo. Conversely, upregulation of NSUN2 enhanced GBC cell growth and colony formation. We further discovered that RPL6 was a closely interacting partner with NSUN2. Silencing RPL6 resulted in insufficient NSUN2 translational level and accumulative NSUN2 transcriptional level. Exogenous expression of NSUN2 partially rescued the effect of RPL6 in gallbladder cancer progression. Taken together, our data provided novel mechanic insights into the function of NSUN2 in GBC, thus pointing to NSUN2 as a potential and effective therapeutic approach to GBC treatment.
Keywords: gallbladder carcinoma, NOP2/Sun domain family member 2, ribosomal protein L6, translational regulation, tumor progression
1. INTRODUCTION
Gallbladder carcinoma (GBC) remains the most common biliary tract malignancy.1 Although it has a relatively low incidence (affecting fewer than 2/100 000 individuals), GBC patients progress rapidly and have relatively high invasion and metastasis risks due to the lack of a serosal layer of gallbladder adjacent to the liver.1, 2 Surgical resection is the only effective procedure for gallbladder carcinoma.3 However, most patients receive late diagnoses and unsatisfactory treatments, which is attributed to non‐specific early symptoms. Despite more chemotherapy as well as target therapy strategies being carried out clinically, the overall 5‐year survival rate has not seen an obvious rise.4 Finding effective molecular targets of treatment has always been the priority issue for malignant diseases. Therefore, an extensive understanding of the molecular mechanisms underlying the initiation and progression of gallbladder carcinoma will be helpful for the development of effective adjuvant therapy.
NOP2/Sun domain family, member 2 (NSUN2, also known as MYC‐induced SUN domain‐containing protein [Misu]), a nuclear RNA methyltransferase, is the main enzyme catalyzing 5‐methylcytosine formation with covalent bonds in mRNA and noncoding RNA.5, 6, 7 M5C promotes mRNA export, with NSUN2 as the methyltransferase and ALYREF as the m5C reader.6 This modification is necessary to stabilize the anticodon‐codon pairing and translate the mRNA correctly, during which NSUN2 transfers the methyl group supplied by S‐adenosyl‐methionine (SAM) to the cytosine‐C5 of the target mRNA, with Cys321 as the catalytic amino acid and Cys271 as the releasing amino acid.5 Frye and Watt (2006)7 report that NSUN2 mediates Myc‐induced cell proliferation and growth, acting as a novel downstream target of the MYC pathway. In addition, NSUN2 is a component of the mitotic spindle and can stabilize it, leading to smooth cell division in tumor cells.8 Up to present, several studies have indicated that NSUN2 plays an important role in head and neck squamous carcinoma,9 and colon,7, 10 oral, esophagus, stomach, liver, pancreas, and breast cancer.10 However, the functional role of NSUN2 in GBC has not been elucidated.
In this report, we demonstrated for the first time that NSUN2 was highly correlated with the advanced clinical stage of gallbladder carcinoma. NSUN2 promoted the proliferation and tumorigenesis of GBC cells both in vitro and in vivo. Moreover, we found that NSUN2 closely interacted with RPL6, which partially regulated the NSUN2 protein level. NSUN2 promoted gallbladder carcinoma progress via its interacting partner RPL6, which is a new orchestration involved in GBC development and could be a potential therapeutic approach for effective GBC treatment.
2. MATERIALS AND METHODS
2.1. Patients and specimens
A total of 95 gallbladder carcinoma patients and 103 cholecystitis patients who underwent radical cholecystectomy as primary treatment at the Department of General Surgery, Xinhua Hospital affiliated to Shanghai Jiao Tong University School of Medicine (Shanghai, China) were enrolled in this study. Formalin‐fixed, paraffin‐embedded tissues were used to construct a tissue microarray (TMA) for immunohistochemistry (IHC) experiments. In addition, 46 pairs of GBC tissues and corresponding non‐cancerous tissues were collected during surgery and immediately placed in liquid nitrogen for further whole RNA extraction. The present study was approved by the Ethics Committee of Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, China. Written informed consent was obtained from all patients.
2.2. Immunohistochemical staining and evaluation
Immunohistochemistry staining was performed as previously described.11 Briefly, TMA were incubated with antibodies against NSUN2 (20854‐1‐AP, Proteintech), followed by HRP‐conjugated secondary antibody and visualization by DAB. IHC staining was conducted based on the intensity of the staining as well as the percentage of positive‐staining cells. NSUN2 staining intensity or percentage was divided into four degrees and scores (3, strong; 2, moderate; 1, weak; and 0, negative; and 3, >75%; 2, 50%‐75%; 1, 25%‐50%; 0, <25%). The final IHC score was decided by adding the intensity degree and the percentage score. Four persons independently evaluated the IHC staining of TMA.
2.3. Reagents and cell culture
The GBC cell line GBC‐SD was purchased from the cell bank of the Shanghai Institute for Biological Science, Chinese Academy of Sciences (Shanghai, China). NOZ and OCUG‐1 were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). EHGB‐1 and SGC‐996 were generous gifts from the Eastern Hepatobiliary Surgical Hospital and Institute, The Second Military University (Shanghai, China) and Tongji University Medical College (Shanghai, China), respectively. NOZ cells were maintained in William's medium (GNM41250, Genom). SGC‐996 cells were cultured in RPMI‐1640 medium (SH30809.01B, HyClone). The other three GBC cell lines were maintained in high‐glucose DMEM (11965‐092, Gibco). All the culture media were supplemented with 10% FBS (10270, Gibco) and 1% penicillin/streptomycin (15140‐122, Gibco). The human gallbladder epithelium cell line HGEpC was homemade from a cholecystitis patient and cultured in EpiCM complete medium (4101, ScienCell). All the cells were cultured at 37°C in a humidified incubator containing 5% CO2.
2.4. Plasmid construction, siRNA, lentivirus and cell transfection
pcDNA3 vector and pIRES2‐EGFP vector containing human NSUN2 coding sequence were kindly gifted by Professor Yungui Yang from BIG, UCAS (Beijing, China). The human RPL6 CDS was subcloned into the EcoRI and XhoI sites of pCMV‐MYC vector (631604, Clontech). All the constructs were confirmed by DNA sequencing, and prepared with NucleoBond Xtra Midi Plasmid DNA Purification Kit (740410.50, Machery‐Nagel). siRNA against human NSUN2 and RPL6 were synthesized by GenePharma. Cells were transfected with siRNA or plasmids using RFect siRNA Transfection Reagent (11013, Bio‐Tran) or ViaFect Reagent (E4982, Promega), according to the manufacturer's instructions. In the preparation for the NSUN2 IP‐MS experiment, we transfected plasmids into NOZ cells by electroporation using an Electro Square Porator (ECM 830, BTX Harvard). Co‐transfection of siRNA duplexes and plasmids was accomplished by Lipofectamine 2000 Transfection Reagent (11668027, Invitrogen).
The sequence of siRNA against NSUN2 was packaged into a pFH1UGW lentivirus core vector containing an EGFP reporter gene, produced by Genechem. The viruses were used to infect cells in the presence of polybrene. Forty‐eight hours after the infection, NOZ and GBC‐SD cells were cultured in medium containing 1 μg/mL puromycin for the selection of stable clones. All the sequences are listed in Table S1.
2.5. RNA extraction and quantitative real‐time PCR
Total RNA was extracted from tissue samples or cells using TRIzol Reagent (15592026, Invitrogen). Then, the cDNA library was prepared with a PrimeScript RT Reagent Kit with gDNA Eraser (#RR047A, TaKaRa). Quantitative real‐time PCR experiments were performed using TB Green Premix Ex Taq (RR420A, TaKaRa) and the StepOne Plus system (Applied Biosystems) with GAPDH as an endogenous control. The relative mRNA levels were quantified using the 2(‐Delta Delta C(T)) method.12 Primers used are listed in Table S1.
2.6. Cellular proliferation and tumorigenesis assay
As described previously,11 NOZ and GBC‐SD cells were transfected with siRNA or plasmids of NSUN2 for 48 h, and then seeded in the 96‐well‐plates (800 cells/well for NOZ; 1000 cells/well for GBC‐SD). Cell proliferation assay was performed on days 0, 1, 2, 3, 4 and 5 using the Cell Counting Kit‐8 reagent (40203ES80, Yeasen). For colony formation assays, NOZ and GBC‐SD cells were seeded in 6‐well plates (400 cells/well for NOZ; 500 cells/well for GBC‐SD) and maintained for 10‐14 days. Cells were then fixed with 4% paraformaldehyde (G1101, Servicebio) and stained with crystal violet solution (60506ES60, Yeasen). Those colonies containing over 50 cells were counted.
2.7. Xenografted animals
All nude nu/nu mice, aged 4‐6 weeks, were obtained from the Shanghai Laboratory Animal Center of the Chinese Academy of Science (Shanghai, China). The mice were housed in specific pathogen‐free conditions and fed under standard conditions. 1 × 106 NOZ cells (shCTRL, shNSUN2) or 3 × 106 GBC‐SD cells (shCTRL, shNSUN2) were injected subcutaneously into the right axilla of the nude mice (n = 5 per group). We measured the size of the tumors every week after inoculation, approximately calculated as follows: tumor volume = 0.5 × (tumor width)2 × (tumor length). After approximately 3‐4 weeks, mice were killed; the tumors were dissected and weighed. All animal studies were approved by the Ethics Committee of Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, China.
2.8. Tandem affinity purification and liquid chromatography with tandem mass spectrometry analysis
NOZ cells transiently transfected with pIRES2‐SFB‐NSUN2 were used for tandem affinity purification. Cells were lysed in NETN buffer (100 mmol/L NaCl, 20 mmol/L Tris‐HCl pH 8.0, 0.5% NP‐40, 1 mmol/L PMSF, 1 mmol/L Na3VO4, 10 mmol/L β‐glycerophosphate, 1 mmol/L NaF and 1 × Cocktail). Then, cell lysates were incubated with Anti‐FLAG M2 Affinity Gel (A2220, Sigma) and eluted with 2 mg/mL Flag peptide (B23112, Biotool). The eluates were further incubated with S‐protein‐agarose (69704, Novagen) and boiled with 2 × SDS loading buffer. The precipitates were subjected to SDS‐PAGE and visualized by Coomassie blue staining or silver staining. The identities of eluted proteins were revealed by liquid chromatography with tandem mass spectrometry (LC‐MS/MS) analysis by Shanghai Applied Protein Technology (Shanghai, China). Two independent biological replicates were performed, of which the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository13 with the dataset identifier PXD013152. Protein candidates are also listed in Table S2. Gene Ontology (GO) analysis was performed using the DAVID tool with default parameters.14
2.9. Western blot assay and co‐immunoprecipitation
Cell lysates were prepared in RIPA buffer (P0013C, Beyotime) with Protease Inhibitors Cocktail added (B14001, Bimake). Then, cell lysates or immunoprecipitates were subjected to SDS‐PAGE and transferred onto PVDF membranes. Antibodies against human NSUN2 were purchased from Proteintech (20854‐1‐AP), RPL6 from Abcam (ab213444), γ‐actin from Santa Cruz (sc‐65638), and mouse anti‐FLAG (#F1804), rabbit anti‐FLAG (#F7425) and rabbit anti‐MYC (#C3956) from Sigma‐Aldrich. For exogenous co‐immunoprecipitation, cells were transfected with Flag‐tagged NSUN2 together with Myc‐tagged RPL6 or truncated RPL6 plasmids. The supernatant was incubated with Anti‐FLAG M2 Magnetic Beads (M8823, Sigma) or Anti‐Myc‐tag mAb‐Magnetic Beads (M047‐11, MBL) for at least 2 h at 4°C. Endogenous immunoprecipitation was operated by incubation with Protein A/G Magnetic Beads (B23202, Bimake), which had been preincubated with primary antibody. The precipitated proteins were detected by antibodies as indicated. Incubation with Rabbit IgG (A7016, Beyotime) or Mouse IgG (A7028, Beyotime) were used for non‐specific control.
2.10. Statistical analysis
All statistical analysis was performed with GraphPad Prism 7 or SPSS 21.0 software. Statistic data were presented as mean ± SD. Student's t test and one‐way analysis of variance (ANOVA) were conducted for comparing difference between groups. A P‐value less than 0.05 was considered statistically significant.
3. RESULTS
3.1. Highly expressed NSUN2 is associated with human gallbladder carcinoma progression
To investigate the clinical significance of NSUN2 in human gallbladder carcinoma, we first performed quantitative RT‐PCR experiments in 46 pairs of human GBC tumors and adjacent normal tissues. We found that NSUN2 was highly expressed in tumors than in normal tissues (Figures 1A,B). We also detected NSUN2 expression level in 95 human gallbladder cancer and 103 cholecystitis tissues by IHC staining. We found that NSUN2 was strongly stained in tumors than in cholecystitis (Figure 1C). The percentage of strongly and moderately stained specimens was significantly greater in GBC than in cholecystitis, while the quantity of weakly stained specimens was dramatically less in GBC than in cholecystitis (Figure 1D, Table 1). Furthermore, the protein levels of NSUN2 were measured in 11 pairs of human GBC specimens and their matched adjacent non‐tumor tissues by western blot (Figure 1E). Similarly, the results showed that NSUN2 was highly expressed in most tumor tissues than in their non‐tumor counterparts. Therefore, NSUN2 may play an important role in GBC progression.
Figure 1.
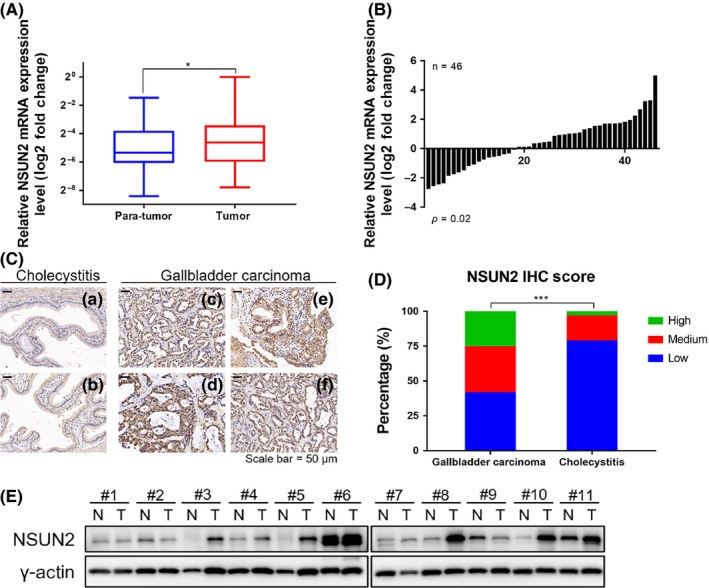
Clinical significance of NSUN2 in human gallbladder carcinoma. A, Box plots of the relative expression of NSUN2 in gall bladder cancer (GBC) tissues and their matched non‐tumor counterparts. NSUN2 expression was calculated based on the NSUN2/GADPH expression ratio (2−Δ CT). B, Comparison of the NSUN2 expression level between GBC tissues and their corresponding non‐tumor tissues. C, Representative immunohistochemistry (IHC) staining images of cholecystitis and gallbladder carcinoma tissues with antibodies against human NSUN2. a,b Relatively low expression level of NSUN2 in cholecystitis tissue; c‐f Relatively strong expression level of NSUN2 in GBC tissues (scale bar, 50 μm). D, Percentage of each staining score group of NSUN2 expression in patients with cholecystitis or gallbladder carcinoma. E, Protein expression of NSUN2 in representative primary GBC tissues (T) and their paired non‐tumor tissues (N)
Table 1.
Immunohistochemistry analysis of NSUN2 in gallbladder cancer
| Group | Number of cases | NSUN2 expression by immunohistochemistry | P‐value | |||
|---|---|---|---|---|---|---|
| Negative (0) | Weak (1‐2) | Moderate (3) | Strong (4‐6) | |||
| Cholecystitis | 103 | 56 | 25 | 12 | 10 | <0.001 |
| Gallbladder cancer | 95 | 15 | 25 | 10 | 45 | |
Bold value indicates P < 0.05.
3.2. NSUN2 promotes gallbladder carcinoma progression both in vitro and in vivo
To investigate the influence of NSUN2 on GBC progression, we first checked NSUN2 expression in five GBC cell lines named NOZ, GBC‐SD, SGC‐996, EHGB‐1 and OCUG‐1, as well as a gallbladder epithelium cell line named HGEpC. We found that NSUN2 was highly expressed both in mRNA (Figure 2A) and protein (Figure 2B). In the following study, we chose NOZ and GBC‐SD cell lines that had moderate highly expressed NSUN2 to perform both NSUN2 silencing and overexpression. Then, we detected the gallbladder cancer cell growth rate and colony formation ability upon NSUN2 depletion and overexpression in NOZ (Figure 2C,F) and in GBC‐SD (Figure S1A,D). Cells grew significantly slowly (Figure 2D) and formed fewer colonies (Figure 2E) after NSUN2 knockdown in NOZ cells, as was the case in GBC‐SD (Figure S1B,C). In contrast, cells grew dramatically faster (Figure 2G) and formed more colonies (Figure 2H) after we overexpressed NSUN2 in NOZ and GBC‐SD cell lines (Figure S1E,F). We also performed cell cycle analysis in NSUN2 depleted NOZ cells (Figure S1G), and the results indicated that NSUN2 depletion caused more NOZ cells to be blocked in phase G0/G1, preventing them entering into phase G2/M, which could be a possible mechanism to influence cell proliferation.
Figure 2.
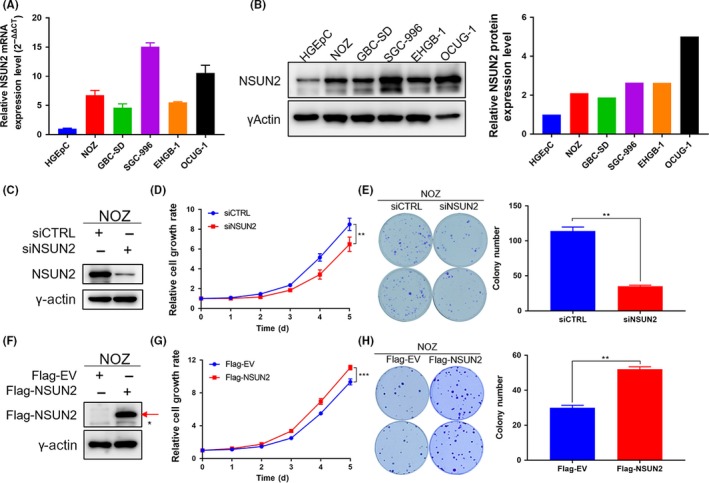
Highly expressed NSUN2 promotes NOZ cell proliferation and tumorigenesis in vitro. A, mRNA level of NSUN2 in gall bladder cancer (GBC) cell lines NOZ, GBC‐SD, SGC‐996, EHGB‐1, OCUG‐1 and the human gallbladder epithelium cell line HGEpC. B, Left panel shows the protein level of NSUN2 in GBC cell lines and the human gallbladder epithelium cell line. γ‐actin was used as the loading control. The right panel shows the quantitative results. C and F, Western blot analysis of the efficiency of NSUN2 knockdown or overexpression in NOZ cells. γ‐actin was used as the loading control. * indicates non‐specific bands and the red arrow represents Flag‐NSUN2. D and G, The relative cell growth rates were determined by CCK‐8 proliferation assays at various time points in NOZ cells. E and H, Representative images of colony formation with NSUN2 knockdown or overexpression induced by RNAi or plasmid transfection in NOZ cells. The numbers of colonies are presented in the right panel. *P < 0.05, **P < 0.01 and ***P < 0.001
Next, we constructed NSUN2 stably deficient NOZ and GBC‐SD cell lines using lentivirus containing shRNA against human NSUN2 (Figures 3A and S2A). We inoculated those cells subcutaneously into nude mice and monitored them for a few weeks. NSUN2‐deficient cells grew into tumors much more slowly than in the control group (Figures 3C and S2C). Moreover, NOZ and GBC‐SD cells with NSUN2 deficiency formed smaller tumors (Figures 3B,D and S2B,D). In summary, our results suggested that NSUN2 promoted gallbladder carcinoma development both in vitro and in vivo.
Figure 3.
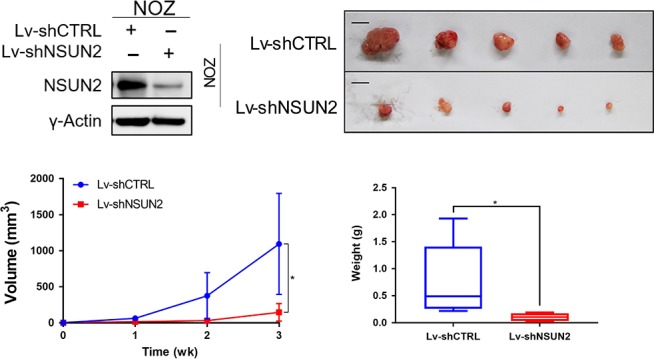
NSUN2 silencing retards NOZ cells tumorigenesis in vivo. A, Western blot analysis of the efficiency of NSUN2 silencing induced by lentivirus in NOZ cells. γ‐actin was used as the loading control. B, Representative examples of tumors formed in nude mice injected with the indicated NOZ cells. Scale bar = 1 cm. C, Tumor growth curves are summarized in the line chart. D, The average tumor weight in the subcutaneous xenograft model is illustrated. *P < 0.05, **P < 0.01 and ***P < 0.001
3.3. RPL6 interacts with NSUN2 and partially regulates its translation
To illuminate the mechanism of NSUN2 regulating GBC progression, we attempted to identify NSUN2's potential partners. We transfected the pIRES2‐SFB‐NSUN2 plasmids into NOZ cells and performed tandem affinity purification. We finally obtained many potentially interacting protein bands (Figure 4A) and identified those proteins by LC‐MS/MS analysis (Table S2). We used the gene list which we have got from MS to do GO pathway analysis. Gene pathways were improved in regard to in mesenchyme migration, cellular component biogenesis, nuclear division, cellular response to transforming growth factor beta stimulus, mitotic cell cycle, establishment of protein localization to endoplasmic reticulum, cell cycle G2/M phase transition, the nuclear‐transcribed mRNA catabolic process, etc (Figure 4B). Most of these results were consistent with the known functional role of NSUN2,6, 7, 8, 15 which further suggested an active role of NSUN2 in human GBC. One of the potential partners, ribosomal protein L6, (RPL6) attracted our attention. First, from the list we obtained from LC‐MS/MS analysis, we found a lot of interacting protein candidates that were correlated to the known functions of NSUN2; that is, mRNA translation (modifying cytosine‐5 in transfer RNA). From the Coomassie brilliant blue staining and silver staining results (Figure 4A), we can see an obvious band around the size of 35 kD. As a 34‐kD protein, RPL6 may have a strong interaction with NSUN2. RPL6 has been attracting increasing attention in relation to the treatment of malignant diseases. It is known to regulate the HDM2‐p53 pathway in order to inhibit cell growth.16 RPL6 is associated with drug resistance in gastric cancer,17 which is currently a hot topic in clinical treatment. Thus, we want to know whether they play a synergetic role or the two proteins function by regulating each other. More importantly, the role of RPL6 in GBC has not been elucidated. Therefore, we chose RPL6 as our target for further research.
Figure 4.
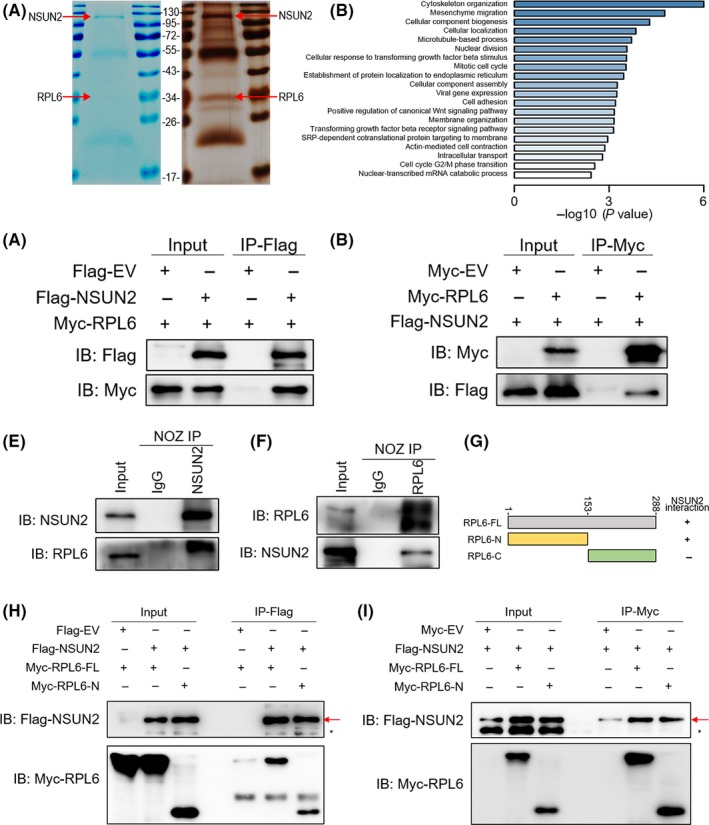
NSUN2 interacts with RPL6. A, Coomassie brilliant blue staining and silver staining of NSUN2 and its associated proteins. The red arrow represents SFB‐NSUN2 or endogenous RPL6. B, NSUN2‐associated proteins identified by IP‐MS were clustered by GO molecular function. C and D, Interaction between exogenous NSUN2 and RPL6. E and F, Interaction of endogenous NSUN2 and RPL6. NSUN2 and RPL6 showed a bona fide interaction between each other in NOZ cells. G, Schematic diagram of Myc‐RPL6 full‐length (RPL6‐FL), N‐terminal (RPL6‐N) and C‐terminal (RPL6‐C) constructs. The numbers represent amino acid residues. H and I, NOZ cells were co‐transfected with Flag‐NSUN2 and Myc‐RPL6 full‐length (Myc‐RPL6‐FL), N‐terminal (Myc‐RPL6‐N) or C‐terminal (Myc‐RPL6‐C) constructs as indicated. Interaction between NSUN2 and RPL6 depends on the N‐terminal of RPL6
Co‐immunoprecipitation experiments using lysates from NOZ and GBC‐SD cells co‐transfected with Flag‐NSUN2 and Myc‐RPL6 demonstrated that exogenously expressed RPL6 can be pulled down with Flag‐NSUN2 (Figure 4C), and vice versa (Figure 4D). To further validate this finding, endogenous NSUN2 was immunoprecipitated and RPL6 was tested by immunoblotting. Consistently, endogenous RPL6 could be pulled down with NSUN2 (Figures 4E and S3A). Endogenous NSUN2 could also be pulled down with RPL6 (Figures 4F and S3B). To further explore the exact region in which two proteins interacted with each other, we constructed plasmid vectors containing truncated N terminal (1‐153aa) and C terminal (154‐288aa) of RPL6 (Figure 4G). We co‐transfected Flag‐NSUN2 together with either full‐length, truncated N terminal or truncated C terminal Myc‐RPL6 into NOZ cells. Then, we found that both full‐length and N terminal Myc‐RPL6 can be detected after Flag immunoprecipitation (Figure 4H); Flag‐NSUN2 can be detected after Myc immunoprecipitation (Figure 4I). These results, altogether, supported a direct interaction between NSUN2 and RPL6.
To further explore the relationship of NSUN2 and RPL6, we manipulated the expression of NSUN2 or RPL6 then detected the expression change of the other one, transcriptionally and translationally. mRNA level was significantly increased for NSUN2 upon RPL6 depletion (Figures 5A and S4A). However, NSUN2 had no change with RPL6 overexpression (Figures 5B and S4B). RPL6 displayed no difference in mRNA level upon either NSUN2 depletion or overexpression (Figures 5C,D and S4C,D). NSUN2 was dramatically decreased in protein upon RPL6 depletion (Figure 5E), but it did not increase dramatically upon RPL6 overexpression (Figure 5F). Finally, RPL6 protein level still had no change after either NSUN2 depletion or overexpression (Figure 5G and H). These results indicated that RPL6 was partially involved in the translation process of NSUN2, acting as an upstream regulator of NSUN2.
Figure 5.
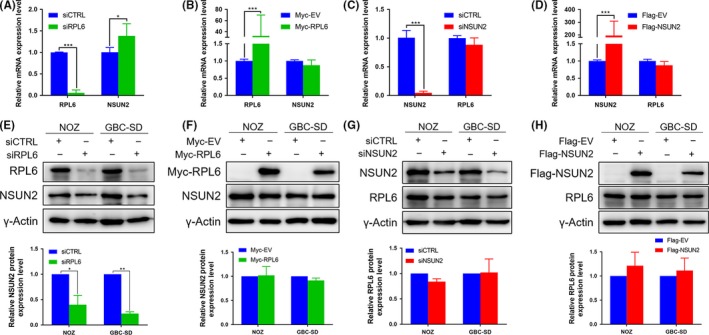
RPL6 is partially involved in the translation process of NSUN2. A‐D, Relative RPL6 and NSUN2 mRNA expression level of NOZ cells transfected with siCTRL, siRPL6 or siNSUN2; and with empty vector (Myc‐EV or Flag‐EV), Myc‐RPL6 or Flag‐NSUN2. E‐H, RPL6 and NSUN2 protein level of NOZ and GBC‐SD cells treated with siCTRL, siRPL6 or siNSUN2; and with empty vector (Myc‐EV or Flag‐EV), Myc‐RPL6 or Flag‐NSUN2. Lower panel shows the quantification of the results
3.4. NSUN2 promotes gallbladder carcinoma progression via RPL6
To investigate the effect of RPL6 on NSUN2 which promoted GBC progression, we performed cell proliferation and colony formation assays in NOZ and GBC‐SD cells with RPL6 depleted and NSUN2 rescued (Figures 6A and S5A). GBC cells grew significantly slowly upon RPL6 depletion and could be partially rescued by NSUN2 (Figures 6B and S5B). In addition, RPL6‐depleted cells formed fewer colonies, while cells could form relatively more colonies with NSUN2 rescued (Figures 6C,D and S5C,D). Those cells with NSUN2 overexpression grew slower and formed fewer colonies after RPL6 silencing. Given that the mRNA level of NSUN2 was relatively higher in gallbladder cancer than normal tissue, RPL6, to some degree, facilitated the translation process of NSUN2 through directly binding to NSUN2. In summary, the orchestration of NSUN2 and RPL6 led to an excessive tumorigenesis ability of GBC cells compared to normal cells (Figure 6E).
Figure 6.
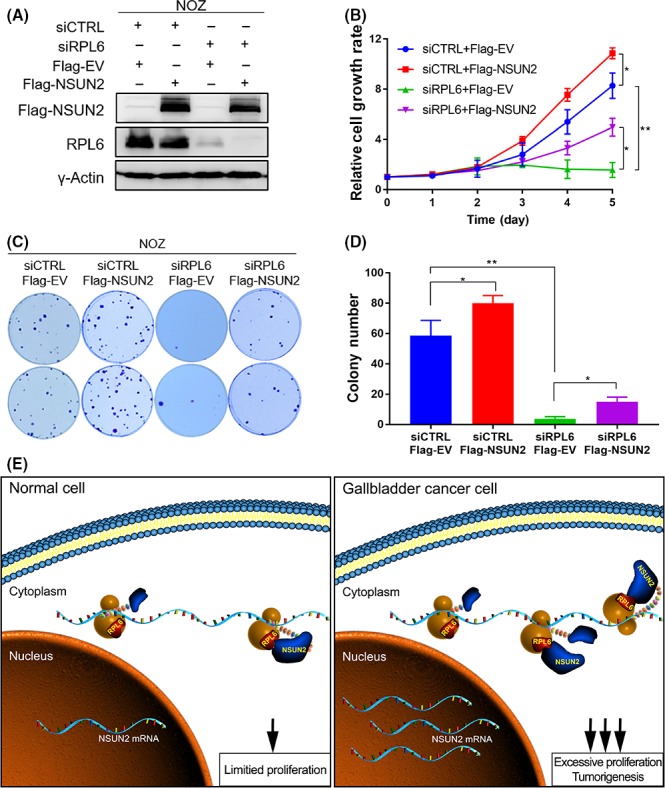
RPL6 is associated with NSUN2 to influence gallbladder cancer (GBC) cell tumorigenesis ability. A, Western blot analysis of the efficiency of RPL6 knockdown with NSUN2 rescue in NOZ cells. B, The relative NOZ cell growth rate was determined by CCK‐8 proliferation assays at various time points. C, Representative images of colony formation of NOZ cells treated with RPL6 knockdown and NSUN2 rescue induced by RNAi and Plasmid. D, The numbers of colonies were calculated from Figure C (*P < 0.05, **P < 0.01 and ***P < 0.001). E, Working model of NSUN2 and RPL6 interacting with each other to promote GBC cell tumorigenesis
4. DISCUSSION
There are currently no effective treatment strategies for GBC patients. The situation is particularly dire for those diagnosed at an advanced stage; these patients always have a poor prognosis.1, 18 For instance, median overall survival for patients with locally advanced or metastatic biliary tract cancer was only extended for 3 months after receiving cisplatin plus gemcitabine rather than gemcitabine alone.19 Thus, it is essential for us to find reliable tumor biomarkers for early detection and molecular targets for treatment. In our previous studies,18, 20 we identified recurrent mutations in the ErbB pathway and demonstrated that ERBB2/ERBB3 mutations promoted PD‐L1‐mediated immune escape in GBC. Therefore, we conducted a clinical trial treating advanced GBC patients with standard chemotherapy, GEMOX regimen, plus a selected targeted therapy base on gene tests. The results indicated that personalized medication improved therapeutic efficiency significantly.21 However, exploration of novel therapeutic targets with better efficacy is still urgently needed.
We also found that some genes, microRNAs (miRNAs) and long non‐coding RNAs (lncRNAs) were related to GBC progression. One of these genes, SPOCK1, a potential cancer prognostic marker, increased the proliferation and metastasis of GBC cells by activating the PI3K/AKT pathway.22 In addition, KPNA2 promoted the development of GBC differentially regulated by E2F1 and E2F7.23 For non‐coding RNA, we found that LncRNA‐PAGBC acted as a microRNA sponge in promoting gallbladder tumorigenesis.24 These molecules, including NSUN2, could be a potential target for GBC treatment.
Several studies have shown that NSUN2 is implicated in cell proliferation, stem cell differentiation, testis differentiation and human cancers.7, 8, 9, 10, 15 Among these functions, the role of NSUN2 in cell proliferation was described first by Frye and Watt,7 who found that the expression of NSUN2 was elevated upon MYC activation and varied throughout the cell cycle. NSUN2 decorated the spindle in mitosis and was critical for correct spindle assembly. Another study25 described NSUN2 as a substrate of Aurora‐B kinase, which phosphorylated NSUN2 inhibiting its RNA methyl‐transferase activity and the assembly of nucleolar RNA processing machinery during mitosis. By methylating specific subsets of RNA (at m5C), such as the CDK1 3′UTR26 and the P27 5′ UTR,27 NSUN2 is a critical regulator of the cell proliferative state. In terms of tumor malignancy, the NSUN2 gene copy number was increased in oral and colorectal cancers,10 and NSUN2 were implicated in 5‐FU sensitivity in HeLa cells.28
Herein, we for the first time identified the m5C methyltransferase NSUN2 as a tumor accelerator in GBC, and this gene was highly expressed in GBC as compared with normal or cholecystitis tissues. NSUN2 enhanced the GBC cell proliferation rate and colony formation ability. It is known that NSUN2 can affect small RNA expression through methyltransferase modification.29 We also detected the expression of some proliferation‐related proteins such as p53, c‐myc and CDK1 (data not shown). It turned out that NSUN2 played an important role in the dynamics of mRNA of those genes and further regulated their proteins’ level in order to promote GBC cell proliferation. Further study is needed to show how NSUN2 functions. We also identified NSUN2 interacting proteins in a gallbladder carcinoma cell line using LC‐MS/MS analysis and performed GO pathway analysis. Candidates were mostly involved in the mitotic cell cycle, nuclear division, establishment of protein localization to endoplasmic reticulum and nuclear‐transcribed mRNA catabolic process pathways. The candidates list also indicates the potential interacting partners that may facilitate NSUN2 promoting GBC tumorigenesis.
Among the list, RPL6 were found to play potential roles in tumor progression. RPL6 was identified as an upregulated gene in multidrug‐resistant gastric cancer cells.30 Yang et al31 (2019) found that RPL6 could be recruited to DNA damage sites and regulated the DNA damage response, which suggests its nuclear localization. Then, focusing on RPL6, we confirmed that NSUN2 can directly interact with RPL6. Next, we examined the NSUN2 expression level upon manipulating the RPL6 expression. Surprisingly, NSUN2 showed contradictory expression levels in transcription and translation upon RPL6 depletion, which we assumed was due to the different regulatory function of RPL6. RPL6 was probably an essential component during NSUN2 translation because depletion of RPL6 leads to less NSUN2 protein, which induced the cells to accumulate more NSUN2 mRNA in a negative feedback way. In other words, NSUN2 mRNA could be translated into proteins opportunely when there were adequate RPL6 proteins in tumor cells. In that case, compared to gallbladder carcinoma cells, normal cells do not transcribe excessive NSUN2 mRNA, and RPL6 maintains NSUN2 proteins at a limited level, which prevents cells from unlimited proliferation. Although the results indicated that RPL6 was partially involved in the translation process of NSUN2, determining whether RPL6 directly acts in the translation process of NSUN2 is urgent. Further analysis which could directly measure the NSUN2 mRNA translation process is needed.
Then we wonder whether RPL6 can regulate the tumorigenesis function of NSUN2. Consistently, after we depleted RPL6 and rescued with NSUN2, the cell tumorigenesis ability recovered partially. Rescue with NSUN2 mRNA could not totally reverse that influence, which again indicated RPL6's critical role in NSUN2 biological function. Because RPL6 plays a vital role in gastric cancer multidrug resistance,30 more experiments are needed to confirm whether NSUN2 contributes to chemotherapy resistance in GBC.
In summary, we characterized a novel tumorigenesis function of the m5C methyltransferase NSUN2 in human gallbladder carcinoma. The highly expressed NSUN2, through closely coordinating with RPL6, promoted GBC cells proliferation and tumorigenesis both in vitro and in vivo. Collectively, our study provides a valuable resource for deciphering the biological significance of NSUN2. This newly discovered orchestration also helped explain the regulation of cell proliferation by NSUN2 in GBC, which makes it an important marker during GBC progression. Therefore, NSUN2 could be a potential therapy approach for GBC and provide a better prognosis for GBC patients.
DISCLOSURE
The authors have no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
We would like to thank Professor Yungui Yang from BIG, UCAS, China for kindly gifting us with the pcDNA3 and pIRES2‐EGFP containing NSUN2 plasmids. This work was supported by the National Natural Science Foundation of China (No. 31701124, 31620103910, 81874181 and 91440203), the China Postdoctoral Science Foundation (No. 2017M61025), the Shanghai Science and Technology Committee Rising‐Star Program (No. 19QA1405900), Shanghai Key Laboratory of Biliary Tract Disease Research Foundation (No. 17DZ2260200), the General Surgery Construction Program of Shanghai Municipal Health Commission (No. 2017ZZ02011), the Emerging Frontier Program of Hospital Development Center (No. SHDC12018107) and the Postdoctoral Initial Foundation of Xinhua Hospital, Affiliated to Shanghai Jiao Tong University School of Medicine (No. 2018BSH02).
Gao Y, Wang Z, Zhu Y, et al. NOP2/Sun RNA methyltransferase 2 promotes tumor progression via its interacting partner RPL6 in gallbladder carcinoma. Cancer Sci. 2019;110:3510‐3519. 10.1111/cas.14190
Yuan Gao, Zheng Wang and Yidi Zhu contributed equally to this work.
Contributor Information
Yingbin Liu, Email: liuyingbin@xinhuamed.com.cn.
Yajuan Hao, Email: haoyajuan@xinhuamed.com.cn.
REFERENCES
- 1. Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: recent update. World J Gastroenterol. 2017;23:3978‐3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol. 2014;6:99‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Misra S, Chaturvedi A, Misra NC, Sharma ID. Carcinoma of the gallbladder. Lancet Oncol. 2003;4:167‐176. [DOI] [PubMed] [Google Scholar]
- 4. Zeng H, Chen W, Zheng R, et al. Changing cancer survival in China during 2003‐15: a pooled analysis of 17 population‐based cancer registries. Lancet Glob Health. 2018;6:e555‐e567. [DOI] [PubMed] [Google Scholar]
- 5. Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang X, Yang Y, Sun BF, et al. 5‐methylcytosine promotes mRNA export ‐ NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc‐induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971‐981. [DOI] [PubMed] [Google Scholar]
- 8. Hussain S, Benavente SB, Nascimento E, et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186:27‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Invest. 2018;36:246‐253. [DOI] [PubMed] [Google Scholar]
- 10. Okamoto M, Hirata S, Sato S, et al. Frequent increased gene copy number and high protein expression of tRNA (cytosine‐5‐)‐methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31:660‐671. [DOI] [PubMed] [Google Scholar]
- 11. Zhang F, Xiang S, Cao Y, et al. EIF3D promotes gallbladder cancer development by stabilizing GRK2 kinase and activating PI3K‐AKT signaling pathway. Cell Death Dis. 2017;8:e2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods. 2001;25:402‐408. [DOI] [PubMed] [Google Scholar]
- 13. Ma J, Chen T, Wu S, et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47:D1211‐D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44‐57. [DOI] [PubMed] [Google Scholar]
- 15. Wang W. mRNA methylation by NSUN2 in cell proliferation. Wiley Interdiscip Rev RNA. 2016;7:838‐842. [DOI] [PubMed] [Google Scholar]
- 16. Bai D, Zhang J, Xiao W, Zheng X. Regulation of the HDM2‐p53 pathway by ribosomal protein L6 in response to ribosomal stress. Nucleic Acids Res. 2014;42:1799‐1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu Q, Gou Y, Wang Q, et al. Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PLoS ONE. 2011;6:e26401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li M, Liu F, Zhang F, et al. Genomic ERBB2/ERBB3 mutations promote PD‐L1‐mediated immune escape in gallbladder cancer: a whole‐exome sequencing analysis. Gut. 2018;68:1024‐1033. [DOI] [PubMed] [Google Scholar]
- 19. Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273‐1281. [DOI] [PubMed] [Google Scholar]
- 20. Li M, Zhang Z, Li X, et al. Whole‐exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet. 2014;46:872‐876. [DOI] [PubMed] [Google Scholar]
- 21. Bao R, Lyu W, Li M, Gong W, Liu Y. Clinical EFFICACY of GEMOX regimen combined with targeted therapy for advanced gallbladder cancer. Chin J Dig Surg. 2019;18:140‐145. [Google Scholar]
- 22. Shu Y‐J, Weng H, Ye Y‐Y, et al. SPOCK1 as a potential cancer prognostic marker promotes the proliferation and metastasis of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol Cancer. 2015;14:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiang S, Wang Z, Ye Y, et al. E2F1 and E2F7 differentially regulate KPNA2 to promote the development of gallbladder cancer. Oncogene. 2019;38:1269‐1281. [DOI] [PubMed] [Google Scholar]
- 24. Wu XS, Wang F, Li HF, et al. LncRNA‐PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18:1837‐1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakita‐Suto S, Kanda A, Suzuki F, Sato S, Takata T, Tatsuka M. Aurora‐B regulates RNA methyltransferase NSUN2. Mol Biol Cell. 2007;18:1107‐1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xing J, Yi J, Cai X, et al. NSun2 promotes cell growth via elevating cyclin‐dependent Kinase 1 translation. Mol Cell Biol. 2015;35:4043‐4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang H, Fan X, Xing J, et al. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging (Albany NY). 2015;7:1143‐1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Okamoto M, Fujiwara M, Hori M, et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5‐fluorouracil in HeLa cells. PLoS Genet. 2014;10:e1004639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hussain S, Sajini AA, Blanco S, et al. NSun2‐mediated cytosine‐5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du J, Shi Y, Pan Y, et al. Regulation of multidrug resistance by ribosomal protein l6 in gastric cancer cells. Cancer Biol Ther. 2005;4:242‐247. [DOI] [PubMed] [Google Scholar]
- 31. Yang C, Zang W, Ji Y, Li T, Yang Y, Zheng X. Ribosomal protein L6 (RPL6) is recruited to DNA damage sites in a poly(ADP‐ribose) polymerase‐dependent manner and regulates the DNA damage response. J Biol Chem. 2019;294:2827‐2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


