Abstract
Alternaria leaf blight is major fungal disease of summer groundnut, causes significant loss of haulm and pod yield. Aims of this study were to understand the role of metabolites and phenylpropanoid related enzymes in Alternaria leaf blight resistance and to find out metabolic marker for disease resistance. Alternaria leaf blight resistant (GPBD4 and CS186) and susceptible genotypes (GG2 and TPG41) of groundnut were grown in pots during rabi-summer 2015. Groundnut plants were infected with Alternaria alternata (Fr.) Keissler at 40 days after sowing. 5 days after infection, upper second leaves were collected from both control and infected plants for analysis. A total of 67 metabolites comprising sugars, sugar alcohols, amino acids, organic acids, fatty acids, sterols and phenolic were identified using gas chromatography–mass spectrometry (non-targeted metabolomics). Constitutive levels of alpha-d-galactoside, d-mannitol, d-erythropentitol, glycine, and hexadecanoic acid were observed higher in resistant genotypes compared to susceptible genotypes. Moreover, arabinofuranose, cinnamic acid, 2-butendioic acid, and linoleic acid were observed only in resistant genotypes at both control and infected stage. In susceptible genotypes myo-inositol, glucose and fructose content was increased after infection with pathogen while decreased in resistant genotypes. Resistant genotypes had higher constitutive level of cinnamic and salicylic acid compared to susceptible genotypes. Non-infected leaves of resistant genotypes also had higher activities of phenylalanine ammonia lyase and tyrosine ammonia lyase activities. Our results suggest that metabolites specifically present in resistant genotypes impart defense mechanism against Alternaria pathogen and can be used as bio-marker for screening of germplasm.
Electronic supplementary material
The online version of this article (10.1007/s12298-019-00708-x) contains supplementary material, which is available to authorized users.
Keywords: Groundnut, Alternaria alternate, Metabolite profile, GC–MS, Ion-chromatography
Introduction
Alternaria leaf blight is becoming a major fungal disease of summer groundnut. Outbreak of this minor disease was observed in farmer’s fields in summer groundnut during 2009 in Saurashtra and Kuchchh regions of Gujarat, India. The main effect of the disease is defoliation of plants, thereby causing reduction in pod yield from 13 to 22% and the reduction in haulm yield from 24 to 63% (Kumar et al. 2012). Groundnut crop residues are widely used for feeding dairy animals. Leaf diseases are likely to affect the quantity and nutritive value of the crop residues, resulting in reductions in milk production. Furthermore, residues fetch low prices in the fodder markets. Being considered a minor disease, very less studies are reported.
Physiological and biochemical disease resistance is governed by various metabolic processes in plants. Various chemical detection platforms are required for targeted and non-targeted metabolites involved in plant defense (Heuberger et al. 2014). To elucidate the resistance mechanisms of genetic sources, metabolomics studies are important for plant systems. Carbohydrates, organic acids, amines/amino acids, and lipids are primary metabolites which are involved in plant defense mechanism (Rojas et al. 2014). Alteration in energy metabolism, nitrogen metabolism particularly involving amino acids (Scharte et al. 2005; Tavernier et al. 2007), and cellular homeostasis such as pH and redox status i.e. malate, ascorbate, tocopherol takes place in plants after pathogen attack (Heuberger et al. 2014). A diverse group of secondary metabolites i.e. terpenoids, phenolics, indoles, alkaloids, and glucosinolates are also responsible for mediating plant defense. These secondary metabolites are categorized into phytoanticipins (pre-infection) and phytoalexins (induced) that have a direct toxic effect on pathogens (Dixon 2001).
Metabolic profiling has contributed to better understanding of plant defense mechanism and to discriminate genotypes varying in their quantitative resistance against various pathogens i.e. resistance of wheat varieties to fusarium head blight (Hamzehzarghani et al. 2008); resistance of potato leaves against Phytophthora infestans (Abu-Nada et al. 2010; Hamzehzarghani et al. 2016); resistance of pearl millet to Sclerospora graminicola (Mahatma et al. 2009); resistance of barley to Fusarium graminearum (Kumaraswamy et al. 2011, 2012); resistance of soybean germplasm to Fusarium tucumaniae (Scandiani et al. 2015). Peroxidases (PODs) are associated with suberization and lignifications of host plant cells and in the oxidation of phenolic compounds in cell wall during the defense reaction against pathogenic agents (Welinder 1992). Other important enzymes which link primary metabolism to phenylpropanoid metabolism include phenylalanine ammonia lyase (PAL) and tyrosine ammonia lyase (TAL). PAL enzyme catalyzes a reaction using l-phenylalanine as a substrate, synthesizes trans-cinnamic acid and ammonia. Cinnamic acid is a precursor of phenylpropanoids which plays important functions in plant defense against pathogens and predators. Salicylic acid, an essential signal involved in plant systemic resistance, is also synthesized by PAL mediated reactions (Kim and Hwang 2014).
Thus, detecting the interactions among these metabolites and enzymes may significantly contribute to the understanding of the mechanism of host-resistance/susceptibility. However, there is lack of studies that relied on targeted/non-targeted metabolomics workflows of groundnut-pathogens interaction. Therefore, present study aimed to understand the role of non-targeted and targeted metabolites along with some defense related enzymes in groundnut showing Alterneria leaf blight disease.
Materials and methods
The seeds of groundnut genotypes CS186 and GPBD4 (Alterneria resistant), GG2 and TPG41 (Susceptible) were procured from Plant Breeding Section, ICAR-Directorate of Groundnut Research, Junagadh, Gujarat, India. These genotypes were selected based on previous report of Kumar et al. (2012). All the genotypes were raised in earthen pots under PII glass house to maintain uniform temperature and humidity during summer 2015. A potting mixture (Vertisol, sand and farm yard manure in a 2:1:1 ratio) with diammonium phosphate at 1 g kg−1 of soil was used. Each genotype was grown in six pots; three pots were kept for control and three for inoculation of Alterneria pathogen (i.e. two sets). Four plants were kept in each pot (20 kg soil capacity). The pathogen, Alternaria alternata (Fr.) Keissler isolated from field grown infected leaves of TPG41. Pathogen was cultured on potato dextrose agar (PDA) as a pure culture. The inoculum was sprayed on leaves in evening at 40 DAS. Leaf samples were taken 5 days after infection (dai) because conidial germination and successful infection of A. alternate occurs at 48–72 h after inoculation of spores and development of disease symptoms starts 5 days after inoculation (Pandey et al. 2003). The upper second leaves were taken from each plant for all the analysis. Three replications for each analysis were taken.
Non-targeted metabolite profiling using gas chromatography–mass spectrometry (GC–MS)
Alterneria infected and control leaves were taken at 5 dai for metabolite extraction. Metabolites were extracted and derivatized following the method described by Lisec et al. (2006) with some modifications. Upper second leaves were collected from control and infected plants and immediately immersed into liquid N for quenching of metabolism. The samples were stored at − 80 °C until further analysis. Groundnut leaves (200 mg) were homogenized in pestle and mortar with 2.8 ml of 100% methanol to inactivate enzymes. Samples were transferred in glass test tube, 60 µl of Ribitol (0.2 mg ml−1 stock in dH2O) was added as an internal quantitative standard and mixed by vortex for 10 s. Samples were kept for 10 min at 70 °C in a water bath. Samples were centrifuged for 10 min at 11,000g and supernatant was transferred to a glass vial. 1.5 ml of chloroform (− 20 °C) and 3.0 ml dH2O (4 °C) was added in each glass vial and vortexed for 10 s. These samples were centrifuged for 15 min at 5000g. Upper polar phase and lower non-polar phase were transferred into a separate glass tubes. Polar and non-polar phase were dried under nitrogen stream at 42 °C.
For derivatization, 100 µl methoxylamine hydrochloride solutions (20 mg ml−1 in pyridine) was added and kept in incubator shaker at 30 °C for 120 min. Carbonyl moieties of metabolites are protected by methoximation. Subsequently, 100 µl MSTFA (N-methyl-N-trimethylsilyl trifluoroacetamide) was added for derivatization of acidic protons and incubated at 37 °C for 90 min with gentle shaking (200 rpm). As a control, one derivatization reaction using an empty reaction tube was also prepared. One-microliter of derivatized samples were injected into a DB-17 MS capillary column (50%-phenyl)-methylpolysiloxane (30 m length, 0.25 mm I.D., 0.25 micron film thickness; Agilent Technologies Inc.) equipped in Shimadzu GC-2010 coupled with MS-QP2010 Plus. The temperature for the ion source was set to 230 °C, for the transfer line to 280 °C, and for the injector to 280 °C. The oven temperature was initially kept at 100 °C for 5 min, followed by increasing 5 °C min−1 to 290 °C for 1 min. Carrier gas (Helium) was used at a constant flow rate of 1 ml min−1. Instrument was calibrated daily using the default automatic calibration mode as recommended by the manufacturer. Chromatogram acquisition, peak deconvolution, and MS library searches were performed using GCMS Solution version 2.71, Shimadzu Corporation-Japan. Reagent peaks and peaks corresponding to column bleeding were excluded from further analyses. Metabolites were putatively identified by matching their mass spectra to spectra in NIST 147 library (National Institute of Standards and Technology, Gaithersburg, MD, USA). Baseline correction, alignment, peak picking, and integration were performed using the ACD/Spec Manager v.12.00 (Advanced Chemistry Development, Inc., ACD/Labs, Toronto, Canada). Data were exported as ‘‘.txt’’ files to MS Excel for the creation of data matrices and CSV comma delimited files were created for data analysis (Mahatma et al. 2018).
Sugar profiles of groundnut leaves using ion chromatography
Oligosaccharides from leaves (500 mg) of groundnut genotypes were extracted in 80% ethanol as described by Swami et al. (2015). Extracted sugar samples were filtered using membrane-filter, 25 µl of filtered sample was injected in the injection port of ion chromatograph (Dionex, ICS 3000). Sugars were separated using CarboPac PA10 analytical column which was supported by amino trap column and CarboPac PA10 guard column to remove other impurities. An isocratic mobile phase of 150 mM NaOH with a flow rate of 1 ml min−1 was used for elution. Different standard sugars i.e. glucose, fructose, myo-inositol, mannitol, trehalose, sucrose, lactose, raffinose, stachyose, and verbascose were purchased from Sigma-Aldrich for identification. An internal standard of lactose was used during the analysis. Different concentrations of these sugars were mixed in a standard mixture to resolve distinct peak of each sugar in chromatogram (Supplementary Fig S1). Chromeleon software supplied with the equipment was used for data integration (Mahatma et al. 2016).
Phenol profiling using Ion chromatograph (IC)
Extraction of phenolics from leaves
One gram of leaves were immersed into 80% HPLC grade methanol in screw cap glass tubes and stored in refrigerator at 4 °C for 48 h. Leaf phenolics were extracted and homogenized using pestle and mortar. Extracted samples were centrifuged at 10,000 rpm for 10 min. This extraction procedure was repeated four times on the same pellet. The supernatant from every centrifugation was collected in volumetric flask and made up to 25 ml with 80% methanol. The supernatants were evaporated to dryness under vacuum dryer at 50 °C and residue was dissolved in 1 ml mobile phase (Mahatma et al. 2011).
Extracted phenolics (25 µl) were filtered using nylon filter (25 mm × 0.45 µm) and injected by syringe in injection port of ion chromatograph. Phenolics were separated on Acclaim 120 C18 reverse phase column (5 µm, 4.6 × 250 mm). Gradient mobile phase of acetonitrile and acetic acid (2% v/v) was used with a flow rate of 0.5 ml−1 min (Supplementary Table S1). Column temperature was adjusted at 30 °C to get reproducible chromatogram. Separated phenolics were detected by UV detector at 280 nm. Standard phenolics i.e. catechol, chlorogenic acid, caffeic acid, cinnamic acid, coumaric acid, ferulic acid, gallic acid, resveratrol, salicylic acid and syringic acid were purchased from Sigma-Aldrich. These standards were used for identification of the phenolic compounds in groundnut leaves based on their retention time and confirmed by adding internal standards into the samples (Czerniewicz et al. 2008). Chromatogram of standard phenolics is given in Supplementary Fig S2. Concentration of the identified phenolics within plant material was expressed as mean values of three replications.
Peroxidase (POX), phenylalanine ammonia lyase (PAL), and tyrosine ammonia lyase (TAL) enzyme assay
Extraction of enzymes
Enzymes were extracted in pre-chilled mortar and pestle with specific buffer. Peroxidase (POX; EC 1.11.1.7) enzyme from groundnut leaves was extracted in 50 mM sodium phosphate buffer (pH 7.4) following the method described by Mahatma et al. (2011). PAL (EC 4.3.1.5) and TAL (EC 4.3.1) enzymes were extracted in 50 mM borate–HCl buffer (pH 8.5) following the method described by Swami et al. (2015).
Enzyme assay
POX activity was assayed spectrophotometrically at 470 nm by measuring the increase in absorption due to the formation of tetraguaiacol. Assay reaction mixture contained 50 mM sodium phosphate buffer pH 7.0, 0.1 mM EDTA, 0.05 ml enzyme extract, 10 mM guaiacol, and 10 mM H2O2. Enzyme activity was expressed as mM guaiacol oxidised min−1 g−1 protein (Mahatma et al. 2011). PAL and TAL were assayed spectrophotometrically by measuring the amount of trans-cinnamic acid at 290 nm for PAL and the amount of p-coumarate at 333 nm for TAL using double beam UV–Vis spectrophotometer. The 3 ml reaction mixture consisted of either 6 µmol of l-phenylalanine (for PAL) or 5.5 µmol of l-tyrosine (for TAL), 500 µmol of Tris-HCI buffer (pH 8.0), and 100 µl of enzyme. Reaction mixtures were incubated for 90 min at 37 °C and reaction was stopped by the addition of 0.05 ml 5 N HCI (Beaudoin-Eagan and Thorpe 1985). Protein content of each enzyme extract was estimated by the method of Lowry et al. (1951). Standard curve of cinnamic acid and coumaric acid was prepared to express enzyme activity as µM cinnamic acid h−1 g−1 protein and µM coumaric acid h−1 g−1 protein for PAL and TAL activity, respectively.
Data processing and statistical analysis
MetaboAnalyst 3.0, an online statistical package was used for statistical and fold Change analysis (Xia et al. 2015). Peak areas of metabolites were taken into consideration for statistical analyses. Data were normalized with respect to the internal standards (adonitol) and pareto scaling. Chromatography peaks were considered significant where the signal to noise (S/N) ratio was > 50, the fold change (FC) was > 2.0 and p values were 0.05. A Partial Least Square-Discriminant Analysis (PLS-DA) was used to identify important metabolite and the outliers at each stage (Karpe et al. 2015). A heat map and dendrogram analysis employed to differentiate the relative levels and relationships of metabolites.
Triplicate data related to assay of enzymes, phenol and sugar profiles were analysed by two-way ANOVA and mean differences were compared by critical differences (LSD, genotype × treatment) at p < 0.05 for significance.
Results and discussion
Disease rating of groundnut genotypes
Disease symptoms appeared at 7 days after inoculation (Fig. 1a) and severity was determined by estimating the percent necrotic area on leaves. Disease rating (1–9) was given based on percent disease severity (Fig. 1b). Percent disease severity for CS186 and GBBD4 (resistant), GG2 and TPG41 (susceptible) was observed 10 and 4%, 35 and 50% respectively. Based on disease severity, disease rating 3.0, 2.0, 6.0 and 7.0 was given to CS186, GPBD4, GG2 and TPG41 respectively.
Fig. 1.
a Disease symptoms on the leaves of groundnut genotypes (Alternaria leaf spot resistant: CS186 and GPBD4; susceptible: GG2 and TPG41). b Disease rating (1–9) based on disease severity (%); where rating 1 is for < 1% disease severity, 2 for 1–5%, 3 for 6–10%, 4 for 11–20%, 5 for 21–30%, 6 for 31–40%, 7 for 41–60%, 8 for 61–80% and 9 for 81–100% disease severity
Non-targeted metabolic profiles using GC–MS
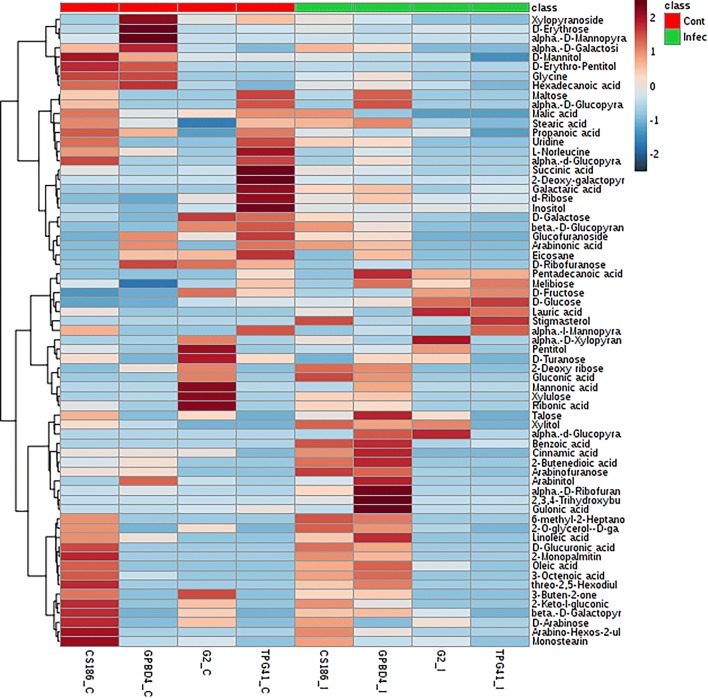
Non-targeted metabolic profiles using GC–MS revealed a total of 67 metabolites including amino acids, fatty acids, organic acids, sugars, sugar alcohols, sterols and phenolic compounds. Different metabolic response was observed in control and pathogen inoculated genotypes (Fig. 2). The partial least squares-discriminant analysis (PLS-DA) using variable importance in projection (VIP) measure was used to identify metabolites that are significant in discriminating the non-infected (control) and infected conditions. Control plants had higher levels of d-ribofuranose, d-mannose, malic acid, hexadecanoic acid, d-turanose, succinic acid, inositol and d-mannitol, whereas d-glucose, 2-butanedioic acid, 2,3,4- trihydroxybutyric acid, d-fructose, lauric acid, gulonic acid and xylitol level were found to be higher in leaves of infected plants (Supplementary Fig S3).
Fig. 2.
Abundance of metabolites (log2) in Alternaria leaf spot resistant groundnut genotypes (CS186 and GPBD4) and susceptible genotypes (GG2 and TPG41) in control (non-infected) and infected condition shown by Heatmap analysis. C: control and I: infected (5 dai). On the log scale bar brick red colour indicates increased metabolite levels, and sky blue represents decreased levels
A heatmap analysis provides a powerful global visualization of alteration of metabolite levels due to infection. Heatmap analysis clearly distinguished metabolite levels in resistant and susceptible genotypes (Fig. 1). Constitutive levels of alpha-d-galactoside, d-mannitol, d-erythropentitol, glycine, and hexadecanoic acid were observed higher in resistant genotypes than that of susceptible genotypes. While higher levels of d-galactose, d-fructose and d-turanose were observed in susceptible genotypes. Interestingly, only resistant genotypes had arabinofuranose, 2-butendioic acid, and linoleic acid at both control and infected stage, but levels of these metabolites were further increased after infection. Benzoic acid and 2,3,4-trihydroxybutyric acid was detected only in resistant genotypes after infection. d-glucuronic acid, gulonic acid and 2-keto-l-gluconic acid were not observed in infected leaves of susceptible genotypes. These molecules are precursor of ascorbic acid, an antioxidant molecule (Isherwood et al. 1953; Saito et al. 1984). Similarly, a non-targeted GCMS study revealed 128 and 249 compounds including several sugars, organic acids, and amino acids in Arabidopsis after 9 and 24 h of infection with Alternaria brassicicola (Botanga et al. 2012). Significantly higher levels of trihydroxybutyrate, d-mannitol, linoleic acid, benzoic acid and propanoate were also reported in Phytophthora infestans inoculated leaves of resistant potato variety (Hamzehzarghani et al. 2016). Thus, implication of 2,3,4-trihydroxybutyric acid in resistant reaction can confer it a defense related metabolite. Cinnamic acid, a key compound of phenylpropanoid pathway plays an important role in defense against plant pathogens and abiotic stresses (Kim and Hwang 2014; Shinde et al. 2017). Benzoic and salicylic acids are active in signaling pathways and involved in plant defense as antifungal compounds (Mustafa and Verpoorte 2005). After infection, increased amount of linoleic acid in resistant genotypes triggers up-regulation of jasmonate signaling pathways and subsequent defense response against P. infestans (Hamzehzarghani et al. 2016). As a substrate of lipoxygenase (LOX) enzyme, linoleic acid also involves in generation of jasmonic acid, methyl jasmonic acid/or lipid peroxides, which act as signal molecules and co-ordinately amplify specific responses (Mhaske et al. 2013). In present study, arabinofuranose that is essential for biosynthesis of pectic polysaccharides (Hsieh et al. 2015) was observed to be higher in resistant genotypes. Being an integral part of plant cell walls, pectic substances determine the strength and flexibility of plant tissues (Chisholm et al. 2006). Moreover, they also impart defense mechanisms against plant pathogens and wounding (Voragen et al. 2009).
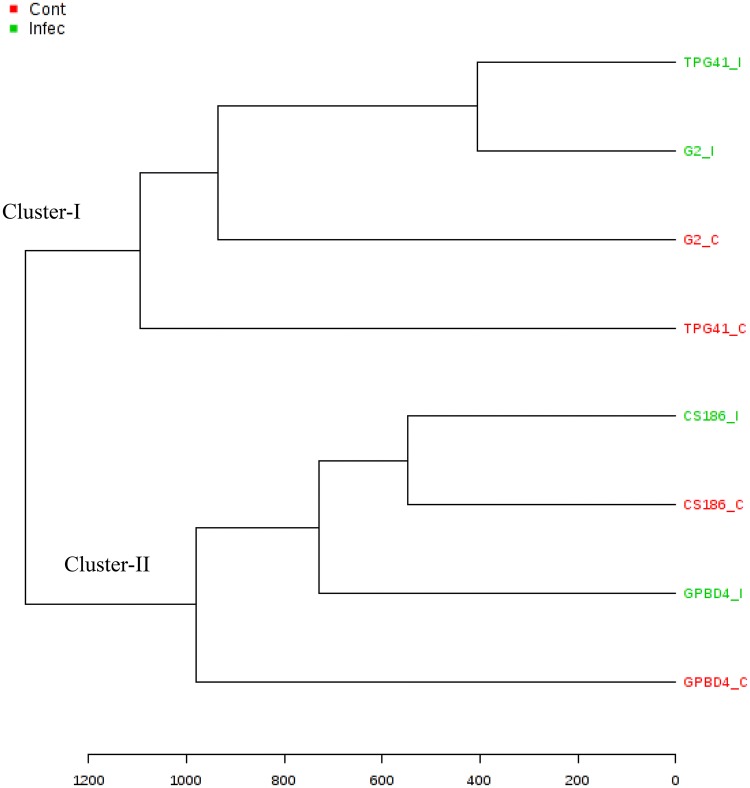
Metabolite data were further analysed for cluster formation. Based on metabolite pattern of genotypes, 2 clusters were formed in which susceptible genotypes shared the cluster-I and resistant genotypes occupied on cluster-II (Fig. 3). Non-infected leaves of susceptible genotypes TPG41 had quite different metabolites than that of infected leaves of TPG41 and GG2, thus shared different sub-cluster on cluster-I. Similarly, non-infected leaves of resistant genotypes GPBD4 had different metabolic pattern than infected leaves of GPBD4 and CS186 and non-infected leaves of CS186. Clustering pattern indicated that genotype CS186 had almost similar pattern of metabolites during infected and non-infected condition and thus shared same sub-cluster on cluster-II. Hence, these results imply that induced metabolic responses are less in CS 186.
Fig. 3.
Dendrogram showing clustering pattern (distance measure using euclidean, and clustering algorithm using ward) of Alternaria leaf spot resistant groundnut genotypes (CS186 and GPBD4) and susceptible genotypes (GG2 and TPG41). C: control and I: infected (5 dai)
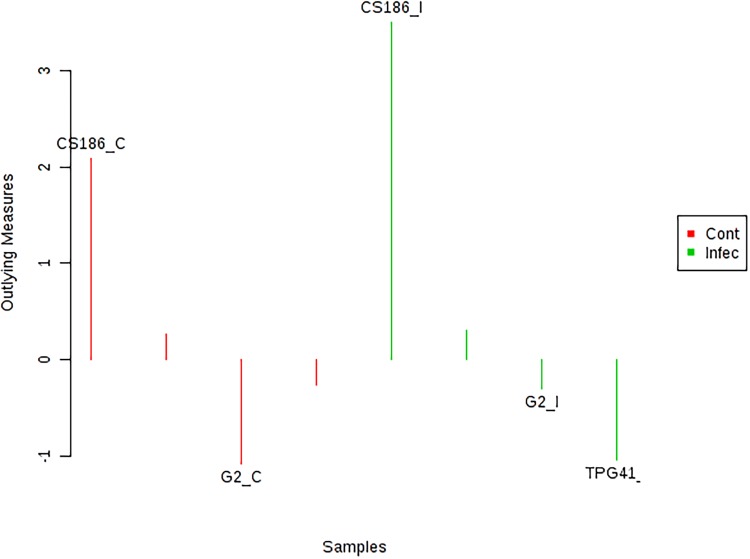
Metabolomics data were further analyzed using random forest to identify potential outliers (genotypes) in control and infected conditions. Resistant genotypes CS186 and GPBD4 were identified as positive outlier in control condition, however, CS186 had higher outlying measure than GPBD4. In infected condition, only genotype CS186 showed positive outlying measure (Fig. 4). Thus, these results suggest that genotype CS186 is distinct among other genotypes during control and infected condition.
Fig. 4.
Identification of potential outliers groundnut genotypes (Alternaria leaf spot resistant: CS186 and GPBD4; susceptible: GG2 and TPG41) at control (non-infected) and infected condition. C: control and I: infected (5 dai)
Targeted metabolites
Sugars
After infection, glucose, fructose and inositol contents were significantly reduced in resistant genotypes while induced in susceptible genotypes. Sucrose content increased in all genotypes after infection but pronounced increase was observed in susceptible genotypes (Table 1). Substantial changes in the carbohydrate content of cucumber plants have been observed during powdery mildew infection which may favour fungal development (Abood and Lösel 2003). Increased rate of soluble sugars in susceptible genotypes after infection suggests an increasing demand of sugars for spreading of the fungus and might regulate gene expression of the pathogen.
Table 1.
Changes in sugar profile of groundnut genotypes during Alternaria-groundnut interaction
| Genotype/treatment | Glucose (mg g−1) | Fructose (mg g−1) | Inositol (mg g−1) | Sucrose (µg g−1) | ||||
|---|---|---|---|---|---|---|---|---|
| Control | Infected | Control | Infected | Control | Infected | Control | Infected | |
| CS186 | 2.40b | 1.54b | 2.40bc | 1.51c | 2.90a | 2.70bc | 256.33c | 291.33c |
| GPBD4 | 1.42c | 1.33b | 1.56c | 1.35c | 3.09a | 2.29c | 231.67c | 343.67c |
| GG2 | 1.63c | 7.7a | 2.80bc | 10.63a | 2.20b | 3.14bc | 512.67b | 933.33b |
| TPG41 | 4.89a | 7.09a | 5.95a | 9.25b | 3.18a | 5.40a | 978.33a | 1393.00a |
All values are mean of three replications. Mean values with different letters (a, b, c and d) within the same column are statistically different according to Duncan’s multiple range test (p value < 0.05)
Similarly, higher glucose and sucrose content in apoplasmic fluids from Botrytis cinerea infected cotyledons of tomato was observed compared to 0 h control (Asai et al. 2015). These results indicated plant sugars are required for fungal growth at the early stage of infection, especially sucrose, to the plant-fungus interface. Pathogen induced invertase activity cleaves sucrose into glucose and fructose and is linked to enhanced sugar content in apoplast (Berger et al. 2007).
Phenols
Resistant genotypes of groundnut had higher content of cinnamic acid and salicylic acid in both non-infected and infected leaves. About 4 to fivefold higher content of cinnamic acid was observed in resistant genotypes at both control and infected stage. Interestingly, salicylic acid was not observed in non-infected leaves of both susceptible genotypes; however it was induced in GG2 after infection. Moreover, infected leaves of resistant genotypes also had 3 to fourfold higher levels of caffeic acid and coumaric acid. Ferulic acid was found to be higher in susceptible genotypes than that of resistant genotypes (Table 2).
Table 2.
Changes in phenolic profile of groundnut genotypes during Alternaria-groundnut interaction
| Genotype/treatment | Cinnamic acid (µg−1 g) | Caffeic acid (µg−1 g) | Coumaric acid (µg−1 g) | Ferulic acid (µg−1 g) | Salicylic acid (µg−1 g) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Infected | Control | Infected | Control | Infected | Control | Infected | Control | Infected | |
| CS186 | 10.00a | 12.70a | 2.30b | 6.17b | 2.20c | 8.30b | 8.90b | 5.30c | 16.77a | 20.50a |
| GPBD4 | 11.30a | 11.20b | 3.10a | 8.20a | 3.40b | 13.00a | 3.53c | 3.70c | 15.87a | 21.13a |
| GG2 | 2.20b | 3.50c | 1.13c | 2.20c | 1.40c | 1.20d | 9.77b | 9.63b | NDb | 12.1b |
| TPG41 | 2.50b | 3.50c | 2.50ab | NDd | 4.93a | 4.37c | 23.40a | 39.00a | NDb | NDc |
All values are mean of three replications. Mean values with different letters (a, b, c and d) within the same column are statistically different according to Duncan’s multiple range test (p value < 0.05)
Higher content of phenolic acids (cinnamic, coumaric, caffeic and salicylic acid) in Alternaria resistant groundnut genotypes further suggested that disease resistant plants may consist inherently higher amount of phenolics (Sztejnberg et al. 1983; Jadhav et al. 2013. Phenolics bind to fungal cell walls, make them more rigid and impermeable thus, hindering further growth or uptake of water and nutrients (Lattanzio et al. 2006). Furthermore, phenolics are the precursors of lignin, which acts as a general barrier for pathogen advancement (Bollin et al. 2010). Ponts et al. (2011) tested effect of coumaric, caffeic, ferulic, 4-hydroxybenzoic and syringic acids on toxin production by Fusarium graminearum. They observed that 4-hydroxybenzoic decreased toxin production while coumaric and ferulic acid increased it. Salicylic acid has been recognized a major immune-related signal in the plant. Salicylic acid potentiates the oxidative burst by leading to the production of reactive oxygen species which is ultimately required for systemic resistance (Borsani et al. 2001; Delaney et al. 1994). Absence of salicylic acid in non-infected leaves of susceptible genotypes imply role of salicylic acid in Alternaria resistance.
Enzymes
Both constitutive and induced levels of phenylalanine ammonia-lyase activities were observed to be significantly higher in resistant genotypes than that of susceptible genotypes. PAL activity was increased in resistant genotypes while decreased in susceptible genotypes after infection. Resistant genotypes had two to threefold higher PAL activities at constitutive level and 3 to fourfold higher at induced level compared to susceptible genotypes (Table 3). Non-infected leaves of resistant genotypes also had appreciably higher activities of TAL, that further induced about twofold in infected leaves (Table 3). A distinct trend of POX was not observed in resistant and susceptible genotypes at control but susceptible genotypes had significantly lower POX activity than that of resistant genotypes. Though, POX enzyme was induced in all genotypes after infection but more prominent induction was observed in resistant genotypes.
Table 3.
Changes in defense related enzymes activities of groundnut genotypes during Alternaria-groundnut interaction
| Genotype/treatment | PAL activity (µM cinnamic acid h−1 g−1 protein) | TAL activity (µM coumaric acid h−1 g−1 protein) | POX activity (mM guaiacol min−1 g−1 protein) | |||
|---|---|---|---|---|---|---|
| Control | Infected | Control | Infected | Control | Infected | |
| CS186 | 8.71a | 9.11a | 23.38b | 56.61b | 11.35d | 25.14a |
| GPBD4 | 6.33b | 8.37a | 45.72a | 85.00a | 15.90a | 24.02b |
| GG2 | 3.27c | 2.90b | 19.02c | 10.5d | 13.13c | 17.60c |
| TPG41 | 1.89d | 1.62b | 18.07c | 29.38c | 14.68b | 17.56c |
All values are mean of three replications. Mean values with different letters (a, b, c and d) within the same column are statistically different according to Duncan’s multiple range test (p value < 0.05)
In the first step of phenylpropanoid pathway, phenylalanine ammonia-lyase (PAL) catalyzes a reaction on l-phenylalanine which undergoes deamination to yield trans-cinnamate and ammonia. PAL activity is key point for the biosynthesis of lignin, suberin, phytoalexins, stilbenes, coumarins and other flavonoids (Zhang et al. 2017). Induction of PAL activity is also essential, if pre-formed antifungal phenolics are inadequate to retard the development of the infectious process (Shiraishi et al. 1995; Yedidia et al. 2003). Thus, higher activities of PAL and TAL in resistant genotypes trigger the phenylpropanoid pathway for higher accumulation of phenolics. Our results inferred that increased levels of phenolics provide an adequate substrate to oxidative reactions catalyzed by polyphenol oxidase and/or peroxidase. These enzymes utilize oxygen and produce toxic quinones which make the host unfavourable for the further infectious process of pathogens (Lattanzio et al. 2006).
Conclusions
Metabolomics study comprising non-targeted and targeted metabolite profiles clearly distinguished Alternaria leaf spot resistant and susceptible groundnut genotypes and thus can be used as a tool to discriminate Alterneria resistant and susceptible genotypes. Arabinofuranose, 2-butendioic acid, and linoleic acid were exclusively observed in resistant genotypes that can be used as bio-marker. Benzoic acid and 2,3,4-trihydroxybutyric acid was detected only in resistant genotypes after infection that may restrict pathogen development in resistant genotypes. Constitutive higher levels of cinnamic and salicylic acid may also act as pre-formed resistant molecules. These phenolics were further induced after infection with concomitant increase of PAL and TAL enzymes. Our result suggests that resistant genotypes had higher levels of cell wall synthesis and rigidity related compound (Arabinofuranose) along with phenylropanoids (cinnamic, coumaric caffeic and salicylicacid) that may restrict pathogen entry. Higher activities of POX in resistant genotypes may oxidize these phenolics into toxic substances for the extra-cellular enzymes produced by the pathogen. Overall, this study delineate the basic mechanism of Alternaria leaf spot resistance in groundnut, however further studies are needed for understanding of detail mechanism using expression profiles of specific pathways related genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors are grateful to the Director, ICAR-Directorate of Groundnut Research for all the support required to conduct this experiment. We thank Dr. S. K. Bera, Principal Scientist (Genetics and Cytogenetics), and Dr. Narendra Kumar, Scientist (Plant Breeding) ICAR-DGR for providing seeds of groundnut genotypes.
Author contributions
MKM conceived, designed and performed the research and wrote the manuscript. LKT analyzed targeted metabolites and enzymes; KSJ and TPP performed pot experiment and inoculated disease. KJR and KHS did the GC–MS analysis and statistical analyses of GC–MS data; SKB did enzymes assay and statistical analyses of targeted metabolites and enzyme data. BAG contributed devices and interpretation of metabolites data. The authors declare that they have no conflict of interest.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abood JK, Lösel DM. Changes in carbohydrate composition of cucumber leaves during the development of powdery mildew infection. Plant Pathol. 2003;52:256–265. [Google Scholar]
- Abu-Nada Y, Kushalappa AC, Prasher S, Al-Mughrabi K, Murphy A. Metabolic profiling to phenotype potato varieties varying in horizontal resistance to leaf infection by Phytophthora infestans. Am J Plant Sci Biotechnol. 2010;4:55–64. [Google Scholar]
- Asai Y, Kobayashi Y, Kobayashi I. Increased expression of the tomato SISWEET15 gene during grey mold infection and the possible involvement of the sugar efflux to apoplasm in the disease susceptibility. J Plant Pathol Microbiol. 2015;7:329. doi: 10.4172/2157-7471.1000329. [DOI] [Google Scholar]
- Beaudoin-Eagan LD, Thorpe TA. Tyrosine and phenylalanine ammonia-lyase activities during shoot initiation in tobacco callus cultures. Plant Physiol. 1985;78:438–441. doi: 10.1104/pp.78.3.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Sinha AK, Roitsch T. Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. J Exp Bot. 2007;58:4019–4026. doi: 10.1093/jxb/erm298. [DOI] [PubMed] [Google Scholar]
- Bollin V, Kumaraswamy GK, Kushalappa AC, Choo TM, Dion Y, Rioux S, Faubert D, Hamzehzarghani H. Mass spectrometry based metabolomics application to identify quantitative resistance related metabolites in barley against Fusarium head blight. Mol Plant Pathol. 2010;11:769–782. doi: 10.1111/j.1364-3703.2010.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Valpuesta V, Botella MA. Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol. 2001;126:1024–1030. doi: 10.1104/pp.126.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botanga CJ, Bethke G, Chen Z, Gallie DR, Fiehn O, Glazebrook J. Metabolite profiling of arabidopsis inoculated with alternaria brassicicola reveals that ascorbate reduces disease severity. Mol Plant Microbe Interact. 2012;25:1628–1638. doi: 10.1094/MPMI-07-12-0179-R. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Czerniewicz P, Leszczyski B, Chrzanowski G. The chromatographic analysis (HPLC) of phenolic compounds from different host-plants of bird cherry-oat aphid. Herba Pol. 2008;54:19–23. [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Dixon RA. Natural products and plant disease resistance. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- Hamzehzarghani H, Paranidhara V, Abu-Nada Y, Kushalappa AC, Dion Y, Rioux S, Comeau A, Yaylayan V, Marshall W. Metabolic profiling coupled with statistical analyses for potential high throughput screening of quantitative resistance to fusarium head blight in wheat cultivars. Can J Plant Pathol. 2008;30:24–36. [Google Scholar]
- Hamzehzarghani H, Vikram A, Abu-Nada Y, Kushalappa AC. Tuber metabolic profiling of resistant and susceptible potato varieties challenged with Phytophthora infestans. Eur J Plant Pathol. 2016;145:277–287. [Google Scholar]
- Heuberger AL, Robison FM, Lyons SM, Broeckling CD, Prenni JE. Evaluating plant immunity using mass spectrometry-based metabolomics workflows. Fron Plant Sci. 2014;5:291. doi: 10.3389/fpls.2014.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YS, Zhang Q, Yap K, Shirley NJ, Lahnstein J, Nelson CJ, Burton RA, Millar AH, Bulone V, Fincher GB. Genetics, transcriptional profiles and catalytic properties of the udp-arabinose mutase family from Barley. Biochemistry. 2015;55:322–334. doi: 10.1021/acs.biochem.5b01055. [DOI] [PubMed] [Google Scholar]
- Isherwood FA, Chen YT, Mapson LW. Synthesis of l-ascorbic acid in plants and animals. Nature. 1953;171:348–349. doi: 10.1038/171348a0. [DOI] [PubMed] [Google Scholar]
- Jadhav PR, Mahatma MK, Mahatma L, Jha S, Parekh VB, Khandelwal V. Expression analysis of key genes of phenylpropanoid pathway and phenol profiling during Ricinus communis–Fusarium oxysporum f. sp. ricini interaction. Ind Crop Prod. 2013;50:456–461. [Google Scholar]
- Karpe AV, Beale DJ, Morrison PD, Harding IH, Palombo EA. Untargetedmetabolic profiling of Vitis vinifera during fungal degradation. FEMS Microbiol Lett. 2015;362:fnv060. doi: 10.1093/femsle/fnv060. [DOI] [PubMed] [Google Scholar]
- Kim DS, Hwang BK. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J Exp Bot. 2014;65:2295–2306. doi: 10.1093/jxb/eru109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Lukose C, Bagwan NB, Koradia VG, Padavi RD. Occurrence of Alternaria leaf blight of groundnut in Gujarat and reaction of some genotypes against the disease. Indian Phytopathol. 2012;65:25–30. [Google Scholar]
- Kumaraswamy KG, Ajjamada C, Kushalappa AC, Choo TM, Dion Y, Rioux S. Mass spectrometry based metabolomics to identify potential biomarkers for resistance in barley against fusarium head blight (Fusarium graminearum) J Chem Ecol. 2011;37:846–856. doi: 10.1007/s10886-011-9989-1. [DOI] [PubMed] [Google Scholar]
- Kumaraswamy KG, Kushalappa AC, Choo TM, Dion Y, Rioux S. Differential metabolic response of barley genotypes, varying in resistance, to trichothecene-producing and -nonproducing (tri5) isolates of Fusarium graminearum. Plant Pathol. 2012;61(509):521. [Google Scholar]
- Lattanzio V, Lattanzio VMT, Cardinali A. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. In: Imperato F, editor. Phytochemistry: advances in research. Trivandrum: Research Signposts; 2006. pp. 23–67. [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:1–10. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mahatma MK, Bhatnagar R, Dhandhukia P, Thakkar VR. Variation in metabolites constituent in leaves of downy mildew resistant and susceptible genotypes of pearl millet. Physiol Mol Biol Plants. 2009;15(3):249–255. doi: 10.1007/s12298-009-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahatma MK, Bhatnagar R, Mittal GK, Mahatma L. Phenol metabolism in downy mildew resistant and susceptible genotypes of pearl millet. Arch Phytopathol Plant Prot. 2011;44:623–636. [Google Scholar]
- Mahatma MK, Thawait LK, Bishi SK, Khatediya N, Rathnakumar AL, Lalwani HB, Misra JB. Nutritional composition and antioxidant activity of Spanish and Virginia groundnuts (Arachis hypogaea) J Food Sci Technol. 2016;53:2279–2286. doi: 10.1007/s13197-016-2187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahatma MK, Thawait LK, Jadon KS, Thirumalaisamy PP, Bishi SK, Jadav JK, Khatediya N, Golakiya BA. Metabolic profiles of groundnut (Arachis hypogaea L.) genotypes differing in Sclerotium rolfsii reaction. Eur J Plant Pathol. 2018;151:463–474. [Google Scholar]
- Mhaske SD, Mahatma MK, Jha S, Singh P, Ahmad T. Polyamine metabolism and lipoxygenase activity during Fusarium oxysporum f. sp. ricini-Castor interaction. Physiol Mol Biol Plant. 2013;19:323–331. doi: 10.1007/s12298-013-0172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa NR, Verpoorte R. Chorismate derived C6Cl compounds in plants. Planta. 2005;222:1–5. doi: 10.1007/s00425-005-1554-0. [DOI] [PubMed] [Google Scholar]
- Pandey KK, Pandey PK, Kalloo G, Banerjee MK. Resistance to early blight of tomato with respect to various parameters of disease epidemics. J Gen Plant Pathol. 2003;69:364–371. [Google Scholar]
- Ponts N, Pinson-Gadais L, Boutigny AL, Barreau C, Richard-Forget F. Cinnamic-derived acids significantly affect Fusarium graminearum growth and in vitro synthesis of type B Trichothecenes. Phytopathol. 2011;101:929–934. doi: 10.1094/PHYTO-09-10-0230. [DOI] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Tzin V, Mysore K. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front Plant Sci. 2014;5:17. doi: 10.3389/fpls.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Morita SI, Kasai Z. Synthesis of l(+) tartaric acid from 5-keto-d-gluconic acid in pelargonium. Plant Cell Physiol. 1984;25:1223–1232. doi: 10.1104/pp.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandiani MM, Luque AG, Razori MV, Casalini LC, Aoki T, Odonnell K, Cervigni GDL, Spampinato CP. Metabolic profiles of soybean roots during early stages of Fusarium tucumaniae infection. J Exp Bot. 2015;66:391–402. doi: 10.1093/jxb/eru432. [DOI] [PubMed] [Google Scholar]
- Scharte J, Schon H, Weis E. Photosynthesis and carbohydrate metabolism in tobacco leaves during an incompatible interaction with Phytophthora nicotianae. Plant, Cell Environ. 2005;28:1421–1435. [Google Scholar]
- Shinde BA, Dholakia BB, Hussain K, Panda S, Meir S, Rogachev I, Aharoni A, Giri AP, Kamble AC. Dynamic metabolic reprogramming of steroidal glycol-alkaloid and phenylpropanoid biosynthesis may impart early blight resistance in wild tomato (Solanum arcanum Peralta) Plant Mol Biol. 2017;95:411–423. doi: 10.1007/s11103-017-0660-2. [DOI] [PubMed] [Google Scholar]
- Shiraishi T, Yamada T, Nicholson RL, Kunoh H. Phenylalanine ammonia-lyase in barley: activity enhancement in response to Erysiphe graminis f.sp. hordei (race 1) a pathogen, and Erysiphe pisi, a nonpathogen. Physiol Mol Phytopathol. 1995;101:929–934. [Google Scholar]
- Swami RM, Mahatma MK, Parekh MJ, Kalariya KA, Mahatma L. Alteration of metabolites and polyphenol oxidase activity in wilt resistant and susceptible pigeonpea genotypes during Fusarium udum infection. Indian J Agric Biochem. 2015;28:18–23. [Google Scholar]
- Sztejnberg A, Azaizia H, Chet I. The possible role of phenolic compounds in resistance of horticultural crops to Dematophora necatrix hartig. J Phytopathol. 1983;107:318–332. [Google Scholar]
- Tavernier V, Cadiou S, Pageau K, Lauge R, Reisdorf-Cren M, Langin T, Masclaux-Daubresse C. The plant nitrogen mobilization promoted by Colletotrichum lindemuthianumin Phaseolus leaves depends on fungus pathogenicity. J Exp Bot. 2007;58:3351–3360. doi: 10.1093/jxb/erm182. [DOI] [PubMed] [Google Scholar]
- Voragen AGJ, Coenen GJ, Verhoef RP, Schols HA. Pectin, a versatile polysaccharide present in plant cell walls. Struct Chem. 2009;20:263–275. [Google Scholar]
- Welinder KG. Plant peroxidases: structure, function relationships. In: Penel C, Gaspar T, Greppin H, editors. Plant peroxidases 1980–1990, topics and detailed literature on molecular, biochemical and physiological aspects. Geneva: Université de Genève; 1992. pp. 1–24. [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia I, Shoresh M, Kerem Z, Benhamou N, Kapulnik Y, Chet I. Concomitant induction of systemic resistance to Pseudomonas syringae pv. lachrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of phytoalexins. Appl Environ Microb. 2003;69:7343–7353. doi: 10.1128/AEM.69.12.7343-7353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang X, Zhang F, Dong L, Wu J, Cheng Q, Qi D, Yan X, Jiang L, Fan S, Li N, Li D, Xu P, Zhang S. Phenylalanine ammonia-lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci Rep. 2017;7:7242. doi: 10.1038/s41598-017-07832-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.