Abstract
Acute lymphoblastic leukemia (ALL) accounts for 20% of all adult leukemias and is the most common leukemia during childhood (80%). We present data on cytogenetics of ALL from a tertiary centre in India correlating it with clinical factors. Karyotyping of bone marrow samples of 204 patients with newly diagnosed ALL was performed with standard G-banding technique. Clinical data of patients was obtained from case records. Survival was estimated using Kaplan–Meir curves and compared by the log-rank test. Univariate and multivariate analysis was done for survival with age, sex, immunophenotype, hyperleukocytosis, risk type, remission status and cytogenetics. The most common karyotypes observed were normal in 39.7% (N = 81), hyperdiploidy in 12.7% (N = 26), t(9;22) in 4.4% (N = 9), t(1;19) in 3.9% (N = 8). Adults with ALL had worse survival compared with pediatric patients (HR 3.62; 2.03–6.45 95% CI, p < 0.001). Patients not in morphologic remission after induction chemotherapy fared poorly (HR 4.86; 2.67–8.84 95% CI, p < 0.001). Patients with favourable cytogenetics had better overall survival (HR 0.36; 0.12–1.05 95% CI, p < 0.05). On multivariate analysis, achievement of morphologic remission emerged as single most significant predictor of survival (p < 0.001). MLL gene rearrangement and t(12;21) were seen less commonly as compared to Western data. However, incidence rates of various cytogenetic abnormalities were similar to that reported from other centres from India. Age, morphologic remission at end of induction chemotherapy and favourable cytogenetics correlated significantly with survival.
Keywords: Acute lymphoblastic leukemia, Cytogenetics, Karyotype, India, Overall survival
Introduction
Acute lymphoblastic leukemia (ALL) accounts for 20% of all adult leukemias, but is the most common leukemia in childhood (80%) [1]. The age adjusted standardised incidence rate of ALL in the Madras Metropolitan Tumor registry is 3.5 in males and 2.2 in females per 1,00,000 population [2]. The most important prognostic indicators in ALL are age, white blood cell count, immunophenotype, bone marrow status at end of induction chemotherapy and karyotype [3]. A number of recurring cytogenetic abnormalities are associated with distinct immunologic phenotypes of ALL and have characteristic outcomes [4, 5].
These recurring chromosomal abnormalities can be either numerical abnormalities (gain or loss) or structural abnormalities like translocations, inversions or deletions and can be observed in up to 80% of patients [6]. In addition to having prognostic significance, cytogenetic analysis also helps in the diagnosis and classification of subtypes of ALL and has been incorporated into the WHO classification of acute leukemias [7]. Chromosomal abnormalities like t(12;21)(p12;q22), t(1;19)(q23;p13), hyperdiploidy, t(9;22)(q34;q11) and t(4;11)(q21;23) are among the most commonly observed [8–11].
The translocation t(9;22) is observed in about 2–5% of children compared to about 20–25% in adults and is associated with worse prognosis [12]. Up to 25% of children and 3% of adults with ALL harbour the translocation t(12;21), which is associated with a favourable outcome [13]. A hyperdiploid karyotype (chromosome number = 51–66) is found in 30–40% of children as compared to 2–10% in adults and usually indicates good prognosis [14]. MLL gene rearrangement, the translocation t(4;11) is present in up to 60% of infants, but is rarely observed in adults [8–11]. Cytogenetic analysis plays a key role in diagnosis, risk adapted treatment and in identifying patients who would respond well to treatment. Karyotyping is recommended to be performed at presentation and at relapse [15].
We reviewed the results of cytogenetics in 204 patients with ALL registered at Cancer Institute (WIA), between January 2014 and December 2017. We describe the various chromosomal abnormalities and analyse their prognostic impact with all the clinical factors. To the best of our knowledge this is one of the largest studies of cytogenetics in patients with ALL and correlation with response to chemotherapy and survival from India.
Materials and Methods
Patient Samples
About 572 patients with newly diagnosed ALL were treated between January 2014 and December 2017 at Cancer Institute (WIA), Chennai, Tamil Nadu, India. The diagnosis of ALL was established on the basis of physical examination, complete blood counts, bone marrow morphology and immunophenotyping. Patients who had received at least one month of induction chemotherapy according to the BFM-95 protocol and whose bone marrow (BM) or peripheral blood were processed successfully at the cytogenetics laboratory were considered for further analysis. About 204 patients satisfied the above criteria and were included in the study. The treatment protocol was initially BFM-95 for both adults and pediatric patients without any change according to risk but subsequently was adapted based on response from August 2014. All patients underwent assessment at the end of initial induction chemotherapy to document remission. Demographic and clinical data of patients including follow up details were taken from case records. Follow up data was censored on 1st of June, 2018. Assessment for Minimal Residual Disease was not performed in this group of patients.
Cytogenetic Analysis
Conventional cytogenetic analysis was performed using a standardized protocol. The bone marrow samples were cultured in Rosewell Park Memorial Institute (RPMI) 1640 basal medium, containing 10% fetal calf serum for 24 h at 37 °C. We did not use any stimulated peripheral blood lymphocytes for cytogenetic analysis. The samples were then treated with 0.1 µg/ml of colcemid to arrest cells in the metaphase stage of mitosis. After harvesting the cells with hypotonic potassium chloride solution and fixing with Carnoy’s fixative, the chromosomes were stained and analysed using the standard Giemsa trypsin banding technique [16]. At least 20 metaphases were analysed and the karyotypes were described according to International System for Human Cytogenetic Nomenclature (ISCN 2009). Structural abnormalities were considered when at least two metaphases had the same type of aberration. For numerical abnormalities to be reported, at least three metaphases had to show the same aberration. IKAROS chromosomal karyotyping software (Metasystem company) was used to capture and analyse the chromosome images using a Carl Zeiss Axio10 microscope. A normal GTG banding technique was used with a band level of 450–500. No high resolution banding techniques were employed for this study. Correlation with individual cytogenetic abnormalities was not feasible because of the low absolute number of patients with a specific abnormality. Hence, we grouped karyotypes like hyperdiploidy, t(12;21), t(1;19) which are historically associated with favourable outcomes into one “good” cytogenetics group. Hyperdiploidy was defined when the chromosome number was 51–66. Similarly t(9;22), hypodiploidy, complex karyotype and poor morphology were considered together as a “bad” cytogenetics group. Complex karyotype is defined when there are 5 or more chromosomal abnormalities. Wherever possible cytogenetic analysis was repeated at relapse. In patients whose marrow showed normal cytogenetics, no further evaluation by FISH or molecular methods were performed.
Statistical Analysis
Statistical analysis was performed using SPSS17. Statistical significance was calculated using Chi square test. Survival was estimated using Kaplan–Meir curves [17] and compared by the log-rank test [18, 19]. Overall survival (OS) was calculated from diagnosis until death due to any cause. Relapse free survival (RFS) was calculated from date of diagnosis until recurrence. Examination of bone marrow at the end of initial month of induction was taken to document status of remission. Remission was defined when the hemogram was normal and there were < 5% blasts in the bone marrow.
Results
Patient Characteristics
A total of 204 patients were eligible for analysis. Pediatric patients (age 1–17 years) constituted 68.6% (N = 140) and the rest 31.4% were adults (N = 64). The median age was 8 years (range 1–17 years) for the pediatric population and 26.5 (range 18–60 years) for adults. About two-thirds (N = 134, 65.7%) were male patients as compared to females (N = 70, 34.3%). Immunophenotyping revealed 63.7% of the leukemias to be of B cell origin and 36.3% to be of T-cell in origin. Hyperleucocytosis at presentation was defined as total count greater than 50,000 in B-ALL and greater than 1,00,000 in T-ALL. Hyperleucocytosis was present in 82 patients (40.2%). According to NCI risk classification, 32% (N = 65) were standard risk and 68% (N = 139) were high risk (Table 1).
Table 1.
Baseline characteristics
| Characteristics | Total N = 204 | Percentage |
|---|---|---|
| Age | ||
| Pediatric | 140 | 68.6 |
| Adult | 64 | 31.4 |
| Phenotype | ||
| B-cell | 130 | 63.7 |
| T-cell | 74 | 36.3 |
| SEX | ||
| Male | 134 | 65.7 |
| Female | 70 | 34.3 |
| Hyperleucocytosis | ||
| Present | 82 | 40.2 |
| Absent | 122 | 59.8 |
| Risk type | ||
| Standard risk | 65 | 32 |
| High risk | 139 | 68 |
| Remission at the end of 1 month | ||
| Yes | 179 | 87.7 |
| No | 25 | 12.3 |
Cytogenetics
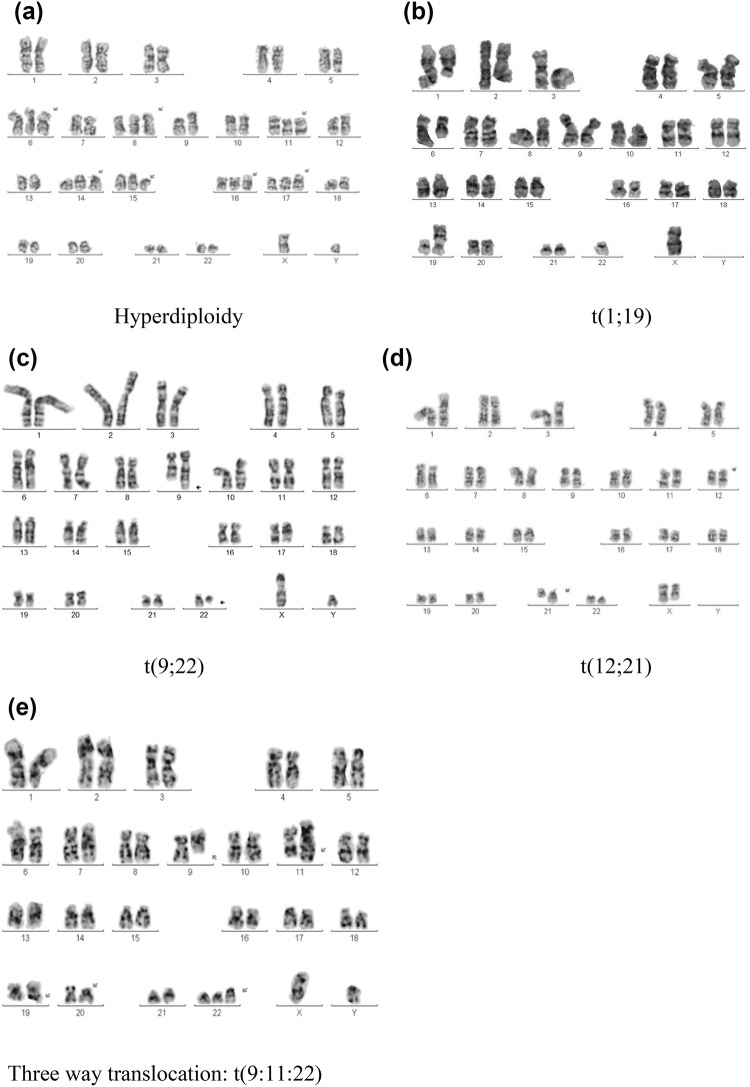
Normal diploid karyotype was the most common and was observed in 81(39.7%) patients. Bone marrow samples from 46 (22.5%) patients on culture showed poor morphology or did not have adequate metaphases. Hyperdiploidy (defined as chromosome number 51–66) was observed in 26 (12.7%) patients (Table 2). The other common cytogenetic abnormalities detected were t(9;22) in 9 (4.4%), t(1;19) in 8 (3.9%), t(12;21) in 2 patients (Table 2). Only 1 patient (age = 24 years) showed t(4;11). Rare 3 way translocations such as t(9;11;22), t(1;19;9;22) and t(1;8;14),(9;22) were also identified (Fig. 2).
Table 2.
Cytogenetics at presentation
| Karyotype | Total N = 204 | Percentage |
|---|---|---|
| Normal | 81 | 39.7 |
| Poor morphology of metaphase | 46 | 22.5 |
| Hyperdiploidy | 26 | 12.7 |
| Hypodiploidy | 2 | 1 |
| t(9;22) | 9 | 4.4 |
| t(1;19) | 8 | 3.9 |
| Del 6q | 6 | 2.9 |
| t(12;21) | 2 | 1 |
| Othersa | 17 | 8.3 |
| Add 19p | 3 | 1.5 |
| Complexb | 4 | 2 |
aOthers: del(12p), der(6), iso(9), add(8), del(1q), del(6q), t(1;16), del(17p), t(2;7)
bComplex : more than 5 abnormalities per metaphase
Fig. 2.
Karyotype of patients with hyperdiploidy a hyperdiploidy; b t(1;19); c t(9;22); d t(12;21); e t(9;11;22)
Chromosomal aberrations (numerical and structural) were observed in all the chromosomes except chromosome 3. Numerical abnormalities were most frequently observed involving chromosomes 4, 9, 10, 11, 16, 22 and X. Structural abnormalities mostly involved chromosomes 1, 6, 9, 12, 19 and 22. All the anomalies of chromosomes 1, 9 and 16 were present in the subtelomeric regions.
Other than common cytogenetic abnormalities, we also identified novel complex cytogenetic abnormalities in 5 patients. This includes 2 patients with chromosomal rearrangements t(9;22)(1;19) and t(1;8)(9;22), der(14). Dual translocation t(9;22)(1;19) was previously reported in three children with acute lymphoblastic leukaemia [20]. Two of these three patients had a poor outcome because of relapse. There are no previous reports of t(1;8)(9;22), der(14) translocation in patients with ALL.
Three patients had unusual three way translocations of t (9;11;22) (Fig. 2), t(8;10;14) + del(6q), and t(1:8:17) respectively. Translocation t(9;11;22) had been reported only in one patient with chronic myeloid leukaemia, who developed additional abnormalities upon treatment [21]. The other two triple translocations i.e. t(8;10;14) + del(6q) and t (1;8;17) have not been previously reported in any malignancy. To the best of our knowledge, we are the first to report these rare dual and triple translocations in patients with ALL.
Correlation with Outcome
Univariate Analysis
Bone marrow aspiration was repeated at the end of 1 month of induction chemotherapy and 179 (87.7%) patients achieved morphologic remission. Patients subsequently underwent consolidation and maintenance phases of treatment. Correlation with individual cytogenetic abnormalities was not feasible because of the low absolute number of patients with a specific abnormality. Hence, we grouped karyotypes like hyperdiploidy, t(12;21), t(1;19) which are historically associated with favourable outcomes into one “good” cytogenetics group. Similarly t(9;22), hypodiploidy, complex karyotype and poor morphology were considered together as a “bad” cytogenetics group. Poor morphology of metaphases and the inability to correctly predict the karyotype is due to various factors. In many reports this has been grouped together as a separate entity in terms of analysis of prognostic factors against outcome [22]. It is assumed that these contain complex karyotypes with aneuploidy and hence portend poor prognosis.
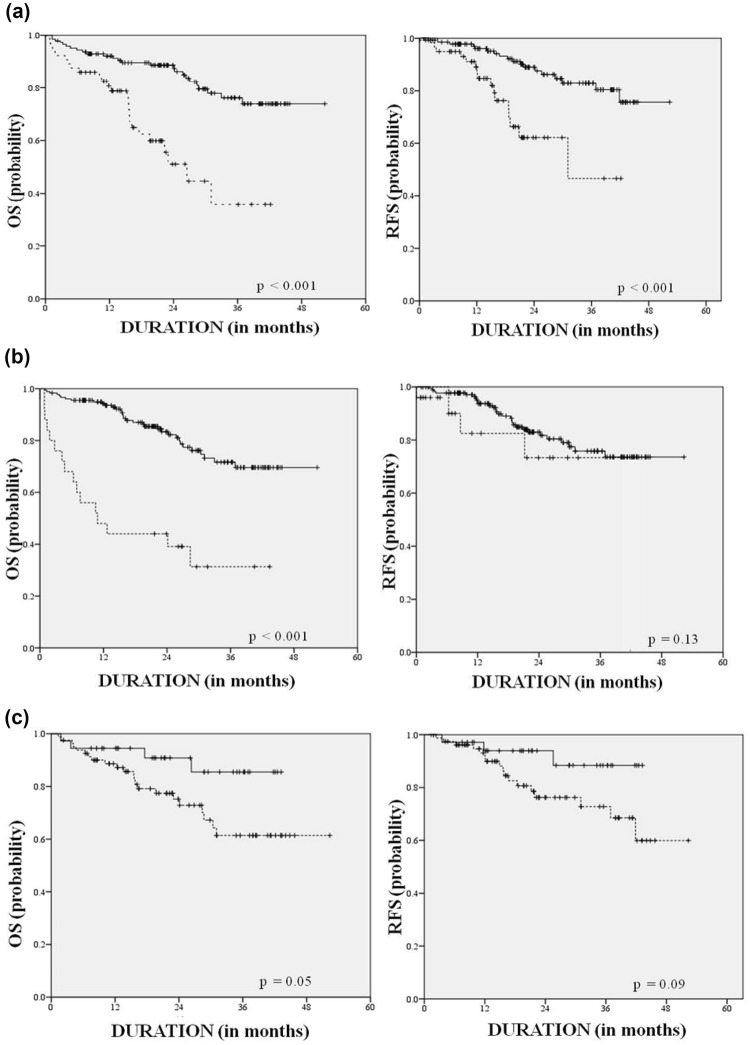
Adults with ALL had worse OS and RFS when compared to pediatric patients (HR for OS: 3.62; 2.03–6.45 95% CI, p < 0.001) even though adults comprised only 31% of the entire cohort. Patients who were not in morphological remission at the end of induction had worse OS (HR 4.86; 2.67–8.84 95% CI, p < 0.001). Patients with T cell phenotype (36%) seemed to fare worse, though not significantly, as compared to those with a B cell phenotype (HR for OS 1.36, 0.77–2.40 95% CI, p = 0.09) (Fig. 1). Males (65%) seemed to do better than females (HR for OS 0.78, 0.44–1.38 95% CI, p = 0.39). Patients who presented with hyperleucocytosis seemed to do better (not significant) than their counterparts (HR for OS 0.89; 0.53–1.72 CI 95%, p = 0.89). Presence of favourable cytogenetics was associated with better OS (HR 0.36, 0.12–1.05 95% CI, p < 0.05). Age alone was a significant factor with respect to relapse free survival (HR 3.60, 1.79–7.26 95% CI, p < 0.001) (Table 3).
Fig. 1.
Overall and relapse free survival curves. The left panel is overall survival and right is relapse free survival. The overall difference between children (solid line) and adults (dotted line) (a); the difference between those who achieved remission at end of 1 month (solid line) versus those who had residual disease (dotted line) (b); the difference in survival between patients who had favourable (solid line) versus normal cytogenetics (dotted line) (c)
Table 3.
(a) Univariate analysis. (b) Multivariate analysis
| (a) Univariate analysis | |||||||
|---|---|---|---|---|---|---|---|
| N = 204 (%) | Overall survival | Relapse free survival | |||||
| HR | CI |
p Value (log rank) |
HR | CI |
p Value (log rank) |
||
| Age | |||||||
| Pediatric | 140 (68.6) | 1.000 | < 0.001 | 1.000 | < 0.001 | ||
| Adult | 64 (31.4) | 3.626 | 2.03–6.45 | 3.608 | 1.79−7.26 | ||
| Cell phenotype | |||||||
| B-cell | 130 (63.7) | 1.000 | 0.27 | 1.000 | 0.09 | ||
| T-cell | 74 (36.3) | 1.366 | 0.77–2.40 | 1.773 | 0.90−3.48 | ||
| Sex | |||||||
| Female | 70 (34.3) | 1.000 | 0.39 | 1.000 | 0.21 | ||
| Male | 134 (65.7) | 0.78 | 0.44–1.38 | 0.648 | 0.32−1.29 | ||
| Hyperleucocytosis | |||||||
| Absent | 122 (59.8) | 1.000 | 0.89 | 1.000 | 0.60 | ||
| Present | 82 (40.2) | 0.896 | 0.53–1.72 | 0.826 | 0.4−1.70 | ||
| Risk type | |||||||
| Standard risk | 65 (32) | 1.000 | 0.14 | 1.000 | 0.15 | ||
| High risk | 139 (68) | 1.61 | 0.85–3.04 | 1.78 | 0.8−3.95 | ||
| Remission | |||||||
| Yes | 179 (87.7) | 1.000 | < 0.001 | 1.000 | 0.13 | ||
| No | 25 (12.3) | 4.86 | 2.67–8.84 | 2.09 | 0.8−5.43 | ||
| Cytogenetics | |||||||
| Normal cytogenetics | 81 (39.7) | 1.000 | 1.000 | ||||
| Favourable | 36 (17.6) | 0.363 | 0.12–1.05 | 0.05 | 0.345 | 0.10–1.18 | 0.09 |
| Unfavourable | 61 (29.9) | 1.057 | 0.56–1.97 | 0.86 | 0.96 | 0.45–2.01 | 0.92 |
| (b) Multivariate analysis for overall survival | |||
|---|---|---|---|
| HR | CI | p Value (log rank) | |
| Age | |||
| Adult | 4.91 | ||
| Pediatric | 1.00 | 2.64–9.12 | < 0.001 |
| Morphologic remission | |||
| No | 6.90 | ||
| Yes | 1.00 | 3.65–13.02 | < 0.001 |
| Favourable karyotype | |||
| Normal cytogenetics | 1.000 | ||
| Favourable | 0.37 | 0.12–1.09 | 0.184 |
Multivariate Analysis
The significant factors in multivariate analysis were that paediatric patients with acute lymphoblastic leukemia did much better than adults (HR for OS 4.91, 2.64–9.12 95% CI, p < 0.001). Morphological complete remission at the end of the first month was also an independent predictor of overall survival (HR for OS 6.90, 3.65–13.02 95% CI, p < 0.001). Patients harbouring favourable cytogenetics did not have a significant impact on OS (HR 0.37, 0.12–1.09 95% CI, p = 0.18) (Table 3).
Discussion
Most of the existing data on cytogenetic abnormalities and their significance in ALL is from the UK-MRC and CALGB ALL trials [8, 9]. There are at least three reports from India, that describe results of karyotyping from both adult and pediatric patients respectively [23–25]. However, two of these reports have not evaluated the karyotyping data with other prognostic factors in relation to outcome [23, 24]. The more recent report evaluated the impact of cytogenetics only on pediatric ALL [25]. To the best of our knowledge, this is the first Indian report to correlate cytogenetics with response to chemotherapy and survival in both adult and pediatric patients with ALL.
Hyperdiploidy is said to be present when the number of chromosomes is greater than 50 and is associated with favourable prognosis (Fig. 2). Favourable prognosis is attributed to better sensitivity of these hyperdiploid cells to chemotherapeutic drugs especially antimetabolites. In the West, the incidence of hyperdiploidy in ALL varies from 7 to 10% in adults and 25–30% in children [14]. The incidence reported in Indian patients is higher—adults 24% and pediatric 44% [23]. The combined incidence of hyperdiploidy in the present report is 12.7%. High-hyperdiploidy defined as chromosome number 58–66 has been described in some studies to be associated with a more favourable outcome. This was observed in fifteen patients in our study. Age and WBC count at presentation were not correlated with favourable outcomes seen in patients with hyperdiploidy. Specific trisomies and early response to steroids were not analysed separately.
The fusion product TCF3-PBX1 results from the translocation t(1;19) (Fig. 2). This fusion product upregulates WNT gene transcription and was historically associated with poor prognosis. But with contemporary pediatric intense induction protocols, prognosis is better [26]. The incidence in the West is 3% (adults) and 5–6% (pediatric). Incidence in India is 4% and 7% respectively [23]. However, the combined incidence in this report is only 3.9%.
Philadelphia chromosome positive ALL is a distinct subtype of ALL with overall poor prognosis and high risk of relapse. This entity is characterised by t(9;22) resulting in the distinctly short chromosome 22 (Fig. 2). The breakpoint in ALL is different from that seen in CML in that it is more upstream in the BCR (Breakpoint Cluster Region) region forming a smaller 185–190 KD protein. These patients are usually treated with a tyrosine kinase inhibitor (TKI) incorporated into the induction chemotherapy protocol and early allogenic bone marrow transplantation [12]. The incidence in the West is 25–30% in adults and 2–5% in children with ALL [12]. The study from Tata Medical Centre reported similar incidence in Indian patients too. In our study, this abnormality was observed in 4.4% patients and all of them achieved a morphologic remission at the end of induction therapy which included imatinib. The current detection of bcr-abl fusion transcript by RT-PCR is more sensitive and is complementary to cytogenetics.
ETV6-RUNX1 fusion gene (previously TEL-AML1) arises from the translocation t(12;21). This is the most common genetic lesion in pediatric ALL (up to 25%). Late relapses are common in these patients, but these relapses are also extremely chemosensitive and overall prognosis is very good [23]. This translocation is cryptic and is usually not easily detected by conventional karyotyping due to similarity of 12p and 21q chromosomes (Fig. 2). The incidence in our study was very low (1%) compared to the West. This may in part be explained by the cryptic nature of this translocation which necessitates the use of FISH or PCR based techniques for identification.
MLL gene (11q23) rearrangements confer a worse prognosis in ALL. These leukemias co-express myeloid markers and patients fare badly even after HSCT [27]. This abnormality has been observed in 5–10% of Western and in up to 3% of Indian patients [23]. There was a single patient in our study who had the translocation t(4;11). This is similar to the observation at Tata Memorial Centre, Mumbai [23]. This patient had a good response to chemotherapy.
Statistical analysis was not performed separately for the pediatric and adult patients in view of the relatively small number of patients with a specific cytogenetic abnormality. Further, cytogenetic results were grouped into 3 categories—(1) normal; (2) “good”—comprising favourable karyotypes like hyperdiploidy, t(12;21) and t(1;19); 3) “bad”—comprising unfavourable cytogenetics like t(9;22), complex karyotype, hypodiploidy and poor morphology. With the above caveats, univariate analysis revealed significant association of overall survival (OS) with age, morphologic remission after induction chemotherapy and the presence of favourable cytogenetics. Overall survival was not significantly associated with gender, B/T cell phenotype, initial WBC count or risk assignment and presence of unfavourable cytogenetics. In contrast, another study from India showed that unfavourable karyotype correlated adversely with OS in multivariate analysis [25]. This may be explained by the different age groups in their report which was focused only on children. This study included more diverse age groups including young adults and elderly patients. Earlier studies on ALL from Cancer Institute (WIA) showed significant association between OS and risk group in adults [28] and a better Event Free Survival (EFS) in children with early response to chemotherapy and female sex [22].
Another abnormality observed in 2.9% of our patients was del 6q. This was associated with variable response to chemotherapy [24, 29]. Rare 3 way variants such as t(9;11;22), t(1;19;9;22) and t(1;8;14),(9;22) were also identified (Fig. 2). Similar translocations have been reported in cases of CML previously but reports of these in ALL are exceedingly rare [30, 31]. Clinical implications of these are yet not fully understood.
We acknowledge that the sample size of the study is small compared to most Western studies and hence pediatric and adult populations have been analysed as a single group. Similarly, cytogenetic abnormalities were also grouped together into favourable and unfavourable categories rather than being analysed as separate abnormalities. This may preclude generalisation of the results. Another limitation is that advanced techniques such as FISH, MLPA and microarray CGH were not performed if the karyotype was normal. Nevertheless, this is the largest study from India which has evaluated karyotype of leukemic cells with outcome measures.
Conclusion
MLL gene rearrangement and t(12;21) were seen less commonly as compared to Western data. However incidence rates of various cytogenetic abnormalities were similar to that reported from other centres from India. Age, morphologic remission at end of induction chemotherapy and favourable cytogenetics correlated significantly with survival.
Compliance with Ethical Standards
Ethical Approval
This is a retrospective study. According to institutional policies ethical committee approval is not required for such studies.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pavan Reddy and Ramesh Shankar have contributed equally to this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Swaminathan R, Rama R, Shanta V. Childhood cancers in Chennai, India, 1990–2001: incidence and survival. Int J Cancer. 2008;122(11):2607–2611. doi: 10.1002/ijc.23428. [DOI] [PubMed] [Google Scholar]
- 3.Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J. 2017;7(6):e577. doi: 10.1038/bcj.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mrózek K, Harper DP, Aplan PD. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Hematol Oncol Clin North Am. 2009;23(5):991–1010. doi: 10.1016/j.hoc.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381(9881):1943–1955. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faderl S, Kantarjian HM, Talpaz M, Estrov Z. Clinical significance of cytogenetic abnormalities in adult acute lymphoblastic leukemia. Blood. 1998;91(11):3995–4019. doi: 10.1182/blood.V91.11.3995. [DOI] [PubMed] [Google Scholar]
- 7.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Beau MML, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 8.Moorman AV, Harrison CJ, Buck GAN, Richards SM, Secker-Walker LM, Martineau M, et al. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood. 2007;109(8):3189–3197. doi: 10.1182/blood-2006-10-051912. [DOI] [PubMed] [Google Scholar]
- 9.Wetzler M, Dodge RK, Mrózek K, Carroll AJ, Tantravahi R, Block AW, et al. Prospective karyotype analysis in adult acute lymphoblastic leukemia: the cancer and leukemia group B experience. Blood. 1999;93(11):3983–3993. [PubMed] [Google Scholar]
- 10.Moorman AV, Ensor HM, Richards SM, Chilton L, Schwab C, Kinsey SE, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11(5):429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- 11.Rowe JM, Buck G, Burnett AK, Chopra R, Wiernik PH, Richards SM, et al. Induction therapy for adults with acute lymphoblastic leukemia: results of more than 1500 patients from the international ALL trial: MRC UKALL XII/ECOG E2993. Blood. 2005;106(12):3760–3767. doi: 10.1182/blood-2005-04-1623. [DOI] [PubMed] [Google Scholar]
- 12.Jones LK, Saha V. Philadelphia positive acute lymphoblastic leukaemia of childhood. Br J Haematol. 2005;130(4):489–500. doi: 10.1111/j.1365-2141.2005.05611.x. [DOI] [PubMed] [Google Scholar]
- 13.Shurtleff SA, Buijs A, Behm FG, Rubnitz JE, Raimondi SC, Hancock ML, et al. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9(12):1985–1989. [PubMed] [Google Scholar]
- 14.Dastugue N, Suciu S, Plat G, Speleman F, Cavé H, Girard S, et al. Hyperdiploidy with 58-66 chromosomes in childhood B-acute lymphoblastic leukemia is highly curable: 58951 CLG-EORTC results. Blood. 2013;121(13):2415–2423. doi: 10.1182/blood-2012-06-437681. [DOI] [PubMed] [Google Scholar]
- 15.Johansson B, Mertens F, Mitelman F. Clinical and biological importance of cytogenetic abnormalities in childhood and adult acute lymphoblastic leukemia. Ann Med. 2004;36(7):492–503. doi: 10.1080/07853890410018808. [DOI] [PubMed] [Google Scholar]
- 16.Wan TSK. Cancer cytogenetics: methodology revisited. Ann Lab Med. 2014;34(6):413–425. doi: 10.3343/alm.2014.34.6.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282):457. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 18.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50(3):163–170. [PubMed] [Google Scholar]
- 19.Peto R, Peto J. Asymptotically efficient rank invariant test procedures. J R Stat Soc Ser A (General) 1972;135(2):185–207. doi: 10.2307/2344317. [DOI] [Google Scholar]
- 20.Griffin TC, Tomlinson GE, Raimondi SC, Sandoval C, Timmons CF, Rosenfield C, et al. Childhood acute lymphoblastic leukemia with both t(1;19) and t(9;22) Leukemia. 1992;6(6):535–540. [PubMed] [Google Scholar]
- 21.Aliano S, Cirmena G, Fugazza G, Bruzzone R, Palermo C, Sessarego M. Standard and variant Philadelphia translocation in a CML patient with different sensitivity to imatinib therapy. Leuk Res Rep. 2013;2(2):75–78. doi: 10.1016/j.lrr.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radhakrishnan V, Gupta S, Ganesan P, Rajendranath R, Ganesan TS, Rajalekshmy KR, et al. Acute lymphoblastic leukemia: a single center experience with Berlin, Frankfurt, and Munster-95 protocol. Indian J Med Paediatr Oncol. 2015;36(4):261–264. doi: 10.4103/0971-5851.171552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amare PSK, Jain H, Kabre S, Deshpande Y, Pawar P, Banavali S, et al. Cytogenetic profile in 7209 Indian patients with de novo acute leukemia: a single centre study from India. J Cancer Ther. 2016;07(07):530. doi: 10.4236/jct.2016.77056. [DOI] [Google Scholar]
- 24.Mazloumi SHM, Madhumathi DS, Appaji L, Prasannakumari null Combined study of cytogenetics and fluorescence in situ hybridization (FISH) analysis in childhood acute lymphoblastic leukemia (ALL) in a tertiary cancer centre in South India. Asian Pac J Cancer Prev. 2012;13(8):3825–3827. doi: 10.7314/APJCP.2012.13.8.3825. [DOI] [PubMed] [Google Scholar]
- 25.Chennamaneni R, Gundeti S, Konatam ML, Bala S, Kumar A, Srinivas L. Impact of cytogenetics on outcomes in pediatric acute lymphoblastic leukemia. South Asian J Cancer. 2018;7(4):263. doi: 10.4103/sajc.sajc_13_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uckun FM, Sensel MG, Sather HN, Gaynon PS, Arthur DC, Lange BJ, et al. Clinical significance of translocation t(1;19) in childhood acute lymphoblastic leukemia in the context of contemporary therapies: a report from the Children’s Cancer Group. J Clin Oncol. 1998;16(2):527–535. doi: 10.1200/JCO.1998.16.2.527. [DOI] [PubMed] [Google Scholar]
- 27.Pui C-H, Gaynon PS, Boyett JM, Chessells JM, Baruchel A, Kamps W, et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet. 2002;359(9321):1909–1915. doi: 10.1016/S0140-6736(02)08782-2. [DOI] [PubMed] [Google Scholar]
- 28.Ganesan P, Sagar TG, Kannan K, Radhakrishnan V, Dhanushkodi M, Swaminathan R, et al. Acute lymphoblastic leukemia in Young adults treated with intensive “pediatric” type protocol. Indian J Hematol Blood Transfus. 2018;34(3):422–429. doi: 10.1007/s12288-017-0892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mancini M, Vegna ML, Castoldi GL, Mecucci C, Spirito F, Elia L, et al. Partial deletions of long arm of chromosome 6: biologic and clinical implications in adult acute lymphoblastic leukemia. Leukemia. 2002;16(10):2055–2061. doi: 10.1038/sj.leu.2402640. [DOI] [PubMed] [Google Scholar]
- 30.Othman MAK, Rincic M, Melo JB, Carreira IM, Alhourani E, Hunstig F, et al. A novel cryptic three-way translocation t(2;9;18)(p23.2;p21.3;q21.33) with deletion of tumor suppressor genes in 9p21.3 and 13q14 in a T-cell acute lymphoblastic leukemia. Leuk Res Treat. 2014;2014:357123. doi: 10.1155/2014/357123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S-G, Park TS, Won SC, Song J, Lee K-A, Choi JR, et al. Three-way translocation involving MLL, MLLT1, and a novel third partner, NRXN1, in a patient with acute lymphoblastic leukemia and t(2;19;11) (p12;p13.3;q23) Cancer Genet Cytogenet. 2010;197(1):32–38. doi: 10.1016/j.cancergencyto.2009.10.009. [DOI] [PubMed] [Google Scholar]