Abstract
Abiotic stresses negatively influence the survival, biomass production, and yield of crops. Tolerance to diverse abiotic stresses in plants is regulated by multiple genes responding differently to various stress conditions. Genetic engineering approaches have helped develop transgenic crops with improved abiotic stress tolerance including yields. The dehydration-responsive element binding protein (DREB) is a stress-responsive transcription factor that modulates the expression of downstream stress-inducible genes, which confer simultaneous tolerance to multiple stresses. This review focuses on advances in the development of DREB transgenic crops and their characterization under various abiotic stress conditions. It further discusses the mechanistic aspects of abiotic stress tolerance, yield gain, the fate of transgenic plants under controlled and field conditions and future research directions toward commercialization of DREB transgenic crops.
Electronic supplementary material
The online version of this article (10.1007/s12298-019-00711-2) contains supplementary material, which is available to authorized users.
Keywords: Climate resilience, Dehydration-responsive element binding protein, Functional markers, Omics, Transgenic crop, Yield traits
Introduction
Abiotic stresses like drought, salinity, and temperature fluctuations, negatively affect the growth and development of plants, and are responsible for the reduced productivity of major crops and significant economic losses, especially in the changing climatic scenario (Bhatnagar-Mathur et al. 2008; Patel et al. 2017). Globally, drought and salinity affect around 20% and 22% of the arable land, respectively; resulting in more than 50% reduced crop productivity in the affected areas (Agarwal et al. 2017; Bhatnagar-Mathur et al. 2008; Sarkar et al. 2016). Abiotic stresses can trigger various morpho-physiological alterations, such as leaf-wilting, leaf-abscission, reduction in relative water content (RWC), a decrease in chlorophyll content and membrane stability, increased electrolyte leakage (EL), and generation of reactive oxygen species (ROS) (Patel et al. 2016). Therefore, stress tolerant varieties of major crops need to be developed to enhance their productivity under multiple abiotic stresses (Bosamia et al. 2015; Sarkar et al. 2014).
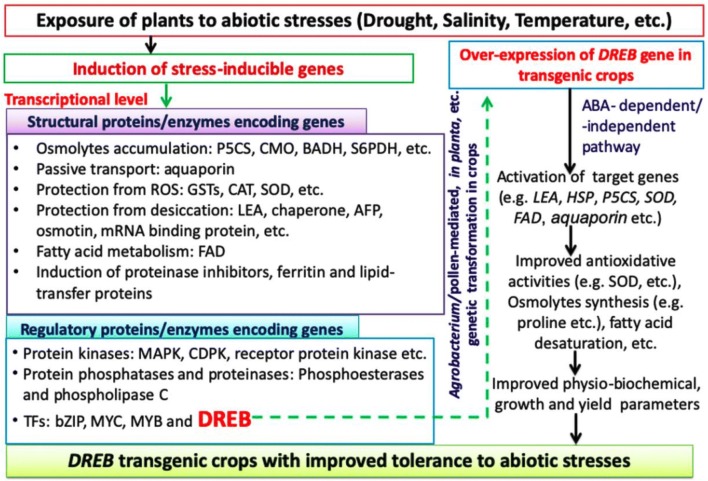
Both classical and marker-assisted breeding methods have been employed to improve the abiotic stress tolerance of crops but have resulted in limited success (Sarkar et al. 2014). Stress tolerant quantitative trait loci (QTL) have been used to develop a stress-tolerant, high yielding variety (Bhatnagar-Mathur et al. 2008). The complex response mechanism of plants to various abiotic stresses involves the expression of multiple stress-inducible genes, which result in morphological, cellular, biochemical and molecular changes. Stress-inducible genes can be grouped into structural and regulatory proteins encoding genes (Fig. 1). Structural genes code for proteins that have structural, enzymatic, or other functions. Regulatory genes code for proteins that control the expression of other genes.
Fig. 1.
An outline of the induction of various inducible genes involving abiotic stress response pathways and mechanisms of DREB-regulated abiotic stress tolerance in transgenic plants. Superoxide dismutase (SOD), glutathione-S-transferases (GSTs), pyrroline-5-carboxylate synthase (P5CS), choline monooxygenase (CMO), betaine aldehyde dehydrogenase (BADH), sorbitol-6-phosphate dehydrogenase (S6PDH), catalase (CAT), mitogen-activated protein kinase (MAPK), calcium-dependent protein kinase (CDPK), glutathione reductase (GR), ascorbate peroxidase (APX), dehydroascorbate reductase (DHAR), monodehydroascorbate reductase (MDHAR), cold-regulated (COR), dehydrin (DHN), late embryogenesis abundant (LEA), antifreeze protein (AFP), fatty acid desaturase (FAD), basic leucine zipper domain (bZIP), myelocytomatosis related proteins (MYC), myelocytomatosis related proteins (MYB) and heat shock protein (HSP)
Since abiotic-stress tolerance is polygenic in nature, and stress responses in plants have overlapping signaling mechanisms, transferring any single-action gene will likely not be sufficient to induce the desired level of tolerance (i.e. the level at which transgenic plant can show optimal growth and yield). Regulons, that are regulated as a unit by a single gene coding a transcription factor (TF), can be harnessed to simultaneously induce the expression of multiple stress-inducible genes. The dehydration-responsive element (DRE) binding protein (DREB) or CRT (C-Repeat) binding factor (CBF) is a trans-acting TF, which activates multiple downstream stress-inducible genes via the cis-acting DRE/CRT motifs on their respective promoter regions (Yamaguchi-Shinozaki and Shinozaki 1994). DREBs belong to the ERF (ethylene-responsive element binding factor) family of TFs and consist of two main subclasses—DREB1/CBF and DREB2—that respectively mediate cold and salinity/dehydration tolerance (Liu et al. 1998; Agarwal et al. 2006).
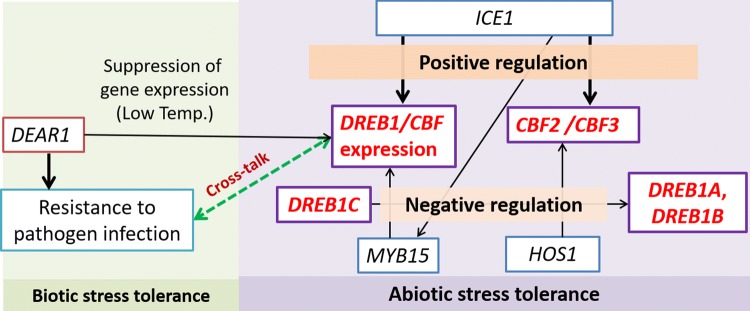
Expression of DREB1 or its target genes are regulated by up-stream TFs like ICE1 (Chinnusamy et al. 2003), HOS1 (Lee et al. 2001), DEAR1 (Tsutsui et al. 2009), and others (Akhtar et al. 2012) (Fig. 2). The DREB gene is the master regulator of an array of downstream stress-inducible genes, and therefore a promising candidate for its utilization in engineering stress-tolerant transgenic crops. Transcriptome, proteome, epigenome, and metabolome studies have shown that the DREB can regulate various stress-responsive pathways in transgenic crops (Table S1). Some epigenomic analyses have shown the involvement of various microRNAs in the fine-tuning of DREB-regulated expression of the downstream gene(s) (Hackenberg et al. 2012; Jiang et al. 2017). Furthermore, a recent meta-analysis has shown that the promoter sequence, type, and duration of stress and genus of donor and recipient plants can significantly influence the DREB-regulated pathways under drought stress (DS) (Dong et al. 2017).
Fig. 2.
Regulation of DREB1 gene(s) by various signaling pathways under abiotic/biotic stress. ICE1 positively regulates expression of DREB1/CBF, HOS1 negatively regulates expression of CBF2 and CBF3 genes by interfering with the functioning of ICE1 protein, MYB15 negatively regulates expression of DREB1/CBF, ICE1 negatively regulates expression of MYB15 and positively regulates expression of DREB1/CBF, DREB1C negatively regulates the expression of DREB1A and DREB1B genes, DEAR1 lead to suppression of DREB1/CBF expression under low-temperature stress and mediates the cross-talk between biotic and abiotic stress response pathways and confers resistance to pathogens in plants. HOS: high expression of somatically responsive; ICE1: inducer of CBF expression 1 protein; EAR: ethylene response factor-associated amphiphilic repression
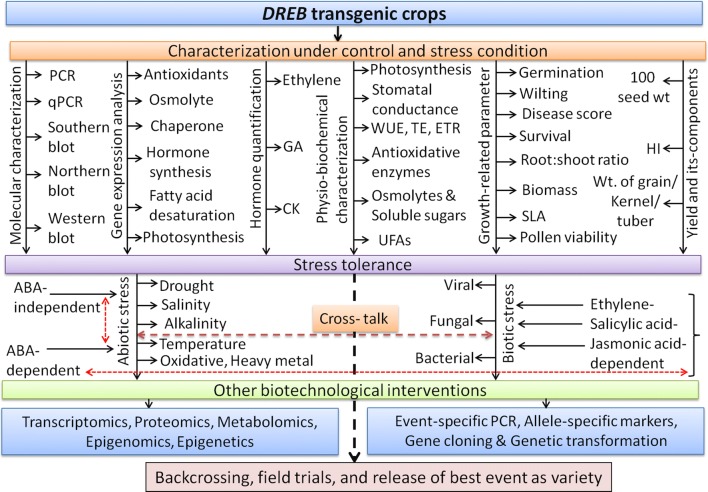
So far, the reports and reviews on the use of the DREB regulon have mainly focused on the functional validation of novel DREB genes and identifying the upstream and downstream regulatory mechanisms of DREB regulon in various transgenic plants under various abiotic stresses. This review was aimed to summarize the following aspects of DREB transgenic crops: (i) Detailed characterization of DREB transgenic plants; (ii) The fate of DREB transgenic crops under both confinement facility and field like conditions; (iii) Potential use of DREB transgenic plants in ongoing breeding programs (Fig. 3, Table S2).
Fig. 3.
Schematic representation of integrated approaches for the development and characterization of DREB transgenic crops for stress tolerance. GA: Gibberellic acid; HI: Harvest index; CK: Cytokinin; PCR: Polymerase chain reaction; WUE: Water use efficiency; TE: Transpiration rate; ETR: Electron transport rate; SLA: Specific leaf area; UFAs: Unsaturated fatty acids, qPCR: Quantitative PCR
The DREB regulon and the ABA-associated gene cascades
Abscisic acid (ABA) has its role in not only regulating the stomatal functions but also in coordinating various stress-responsive pathways. The DREB1/CBF and DREB2 proteins are known to regulate the ABA-independent-pathway, while the DRE/CRT cis-element also functions in an ABA-dependent-pathway, indicating a cross-talk between the two (Agarwal et al. 2006; Agarwal and Jha 2010; Agarwal et al. 2017). Expression analysis of downstream stress-inducible genes in DREB transgenic plants and promoter analysis of these target genes elucidated the molecular mechanisms involved in cross-talk between ABA-dependent and ABA-independent abiotic stress responsive pathways (Narusaka et al. 2003; Oh et al. 2005). However, exogenous ABA treatment could not induce the expression of a heterologous HvCBF4 gene in transgenic rice, indicating the latter’s possible role in the ABA-independent pathway (Oh et al. 2007). ARAG1, an ABA-responsive DREB gene in transgenic rice, mediates DS tolerance via the ABA-dependent signaling pathway (Zhao et al. 2010). DREB1 transgenic rice showed stomatal closure under DS via the ABA-dependent pathway (Datta et al. 2012), and exogenous ABA up-regulated StDREB1 in transgenic potato (Bouaziz et al. 2012), thus indicated causative role of ABA-dependent DREB regulon to abiotic stress responses.
Three AtABF3-target genes—Hsp70, PP2Ca, and receptor kinase—are induced by AtCBF3 in transgenic rice indicating a partial overlap between the ABA-dependent and independent pathways (Oh et al. 2005). However, many abiotic stress-inducible genes (e.g., rd29B) are specifically regulated by ABA, while others like COR15a are not, indicating parallel involvement of the ABA-dependent and independent pathways (Yang et al. 2011a; Zhang et al. 2013). The promoter of rd29A contains a DRE/CRT element that also functions as a coupling element (CE) of ABRE (abscisic acid response element), which mediates the ABA responsiveness of stress-inducible genes. Both these elements are interdependent in the stress-induced expression of rd29A, which can be activated by both ABA-dependent and independent pathways (Narusaka et al. 2003). Nevertheless, limited information is available regarding the underlying mechanisms of DREB-induced abiotic stress tolerance.
DS tolerance in DREB transgenic plants
The most common approach used to evaluate DS tolerance is potting the plants in dry soil (low water potential), which mimics in-field drought conditions. Pellegrineschi et al. (2004) compared the performance of the DREB1A lines and wild type (WT) variety of wheat by withholding water for 15 days and re-watering the plants until maturity. Similar strategies were also used for the AtDREB1A tomato and rice cultivars (Rai et al. 2013; Ravikumar et al. 2014). The DS tolerance of the AtDREB1A peanut lines was evaluated under progressively declining soil moisture regimes or varying vapor pressure deficits in pots/lysimeters/field, which can help quantify the severity of DS (Bhatnagar-Mathur et al. 2007, 2014; Sarkar et al. 2016).
Heterologous expression of DREB1A in transgenic rice helped in alleviating the deleterious effects of DS even at the reproductive stage as indicated by better morpho-physiological traits than WT varieties. For example, AtDREB1A rice (T3) showed greater tolerance in the booting and anthesis stages compared to the WT variety and displayed stomatal closure under DS indicating DREB-dependent regulation of stomatal behavior (Datta et al. 2012). Similarly, high chlorophyll SPAD value and more greenish leaves were observed in AtDREB1A rice, indicating better yield and ‘stay green’ traits across various transgenic lines as compared to the WT variety under DS (Ravikumar et al. 2014).
Pollen-viability and bud-sprouting are important agronomic traits that should be evaluated under stress conditions. Although the agronomic traits of TsCBF1 and WT maize were not significantly different under control conditions, most transgenic lines grew taller (10–15%), showed more tassel branches, shorter anthesis-silking interval (ASI), and better pollen viability than the WT during DS (Zhang et al. 2010). Furthermore, Sakuma et al. (2006) showed constitutive expression of DREB2A gene, if there is a deletion in the negative regulatory domain consisting of 29 amino acids. Also, the AtDREB2A CA sugarcane showed not only improved DS tolerance but also enhanced initial bud sprouting and higher sucrose content without biomass penalty (Reis et al. 2014).
Expression of DREB in transgenic plants modulated the up-regulation of water transport protein, thus help in maintaining cellular water status under various abiotic stress conditions. AtDREB1B expression in Salvia miltiorrhiza up-regulated the aquaporin gene, which modulates water permeability in plant cells (Wei et al. 2016). DREB1A transgenic crops maintained a conservative pattern of water use under DS as indicated by enhancement in the transpiration efficiency (TE) over its WT genotypes. Transgenic peanut (Bhatnagar-Mathur et al. 2007) and chickpea (Anbazhagan et al. 2015b) cultivars overexpressing DREB1A showed a substantial increase in TE under DS. Devi et al. (2011) reported a significant correlation between TE and its surrogate traits such as specific leaf area (SLA) and SPAD chlorophyll meter readings (SCMR) but none between TE and ∆13C (carbon isotope discrimination), indicating that the latter cannot be used to assess TE under DS.
Root architecture is a critical parameter for evaluating DS tolerance since it is associated with water mining capacity. Ban et al. (2011) observed higher LbDREB gene expression in the tobacco roots compared to the leaves under DS. Furthermore, the DS-induced expression of AtDREB1A gene in transgenic peanut was also found associated with improved root/shoot ratio, which enhanced their harvest index (HI) compared to the WT genotype due to greater water uptake from the deeper soil layers (Jagana et al. 2012; Vadez et al. 2007). Under drought-like conditions, AtDREB1A peanut (Sarkar et al. 2016) and chickpea (Anbazhagan et al. 2015b) showed enhanced root growth in deeper soil layers, which might have resulted from improved DS tolerance and increased water extraction capacity. The DREB transgene, however, does not always improve all parameters related to stress tolerance and yield. The AtDREB1A peanut lines did show variations in RWC and total biomass under DS, but no significant difference was observed for pod-yield (Sarkar et al. 2014).
Similarly, the AtDREB1A soybean line did not outperform the WT variety in terms of yield, but the number of seeds per pod and the total number of pods were higher in the former under DS (de Paiva Rolla et al. 2014). The presence of transgene in the genome may not always show its expression in transgenic plants (Shavrukov et al. 2016). Various transgenic lines expressing the same DREB gene in same genetic background may show variations in gene expression pattern, physio-biochemical responses, yield and yield-components under abiotic stress conditions. This is likely due to the positional effects of transgene integration in the genome, copy numbers, positive pleiotropic effects of the tissue culture process, variations in promoter activity, the regulatory function of heterologous DREB gene itself and genotype × environment interactions (Anbazhagan et al. 2015a; Datta et al. 2012; Sarkar 2014).
Antioxidant mechanisms are an important line of defense against DS. Rai et al. (2013) reported a significant increase in the activity of antioxidant enzymes like superoxide dismutase (SOD), catalase (CAT), glutathione reductase (GR), dehydroascorbate reductase (DHAR), ascorbate peroxidase (APX), and monodehydroascorbate reductase (MDHAR) in the AtDREB1A tomato under DS, indicating better tolerance compared to WT variety. In addition, the levels of antioxidants such as ascorbic acid, glutathione, and their reduced forms were also higher in the transgenic tomato and increased with ROS levels (Rai et al. 2013). Although DREB upregulates the antioxidant enzyme genes, it does not play any causative role in improving the TE in AtDREB1A peanut under DS (Bhatnagar-Mathur et al. 2009), indicating that the antioxidant mechanism does not regulate the TE.
In addition to physio-biochemical assays, visual observations are used to characterize abiotic stress tolerance. The AtDREB1A transgenic rice and peanut showed less severe, as well as delayed wilting/curling of leaves during drought. After stress withdrawal, both AtDREB1A transgenic rice (Ravikumar et al. 2014) and peanut (Sarkar et al. 2014) showed speedy recovery compared to their respective WT varieties, possibly due to the up-regulation of genes involved in energy and carbohydrate metabolism (Paul et al. 2015).
Although, DREB transgenic crops are generally tolerant to DS with improved physio-biochemical and growth-related parameters, yet some transgenic events do show dwarfism and pleiotropic effects on leaf ultrastructure and phenology as effective water conservation mechanisms (Li et al. 2012; Tang et al. 2017). Such altered phenotypic traits could be due to the DREB-induced down-regulation of some phytohormones biosynthesis pathways. The DREB gene regulates the response to multiple abiotic stresses, which influences the crop physiology during its entire life cycle. Since drought and salinity are the most prevalent abiotic factors affecting the major crops, considerable efforts have aimed at developing transgenic crops tolerant to DS, followed by salinity stress (SS) (Fig. S1).
Phenotyping of stress tolerant DREB transgenic lines
The abiotic stress tolerance of transgenic plants is evaluated in terms of physiological parameters such as total water use, water use efficiency (WUE), transpiration, TE, RWC, membrane stability (Table S2), osmotic adjustment, surrogate traits of photosynthesis like SLA and SCMR, specific leaf nitrogen (SLN), photochemical efficiency, photosynthetic rate, and stomatal conductance (Anbazhagan et al. 2015a; Bhatnagar-Mathur et al. 2007; Devi et al. 2011; Reis et al. 2014; Sarkar et al. 2014). Under abiotic stress, the heterologous expression of DREB in various transgenic crops showed better stomatal regulation, water status, osmotic homeostasis, protection of photosynthetic apparatus and photosynthetic efficiency (Table S2) over its WT genotype (Datta et al. 2012; Oh et al. 2007; Sarkar et al. 2016). Delayed wilting or curling of leaves in the transgenic crop is a morphological sign of DS tolerance, as it signifies slower water usage due to lower water potential in the tissues (Sarkar et al. 2016). Hence, this phenotypic index could be used to assess the optimal level of stress tolerance in transgenic crops.
The leaf chlorophyll content, photochemical efficiency (Fv/Fm), chlorophyll fluorescence and electron transport rate (ETR) are related to photosynthetic rate and net photosynthesis in transgenic crops (Jin et al. 2010; Ravikumar et al. 2014; Yang et al. 2010). The decrease in chlorophyll SPAD values or chlorophyll content under abiotic stress is a likely result of its photo-oxidation and degradation (Farooq et al. 2009; Jin et al. 2010; Yang et al. 2010), and therefore can be considered a symptom of oxidative stress. Thus, the improved photosynthetic rate, ETR and turnover rates of dark reactions in DREB/CBF transgenic plants might be due to less photo-oxidative damage and degradation of photosynthetic machinery than WT cultivars under drought and cold stresses (Ravikumar et al. 2014; Yang et al. 2010). Taken together, quantitative estimation of various physio-biochemical and molecular (gene expression analysis) indicators can help elucidate the mechanisms underlying stress-tolerance in DREB transgenic crops (Bhatnagar-Mathur et al. 2007, 2014; de Paiva Rolla et al. 2014; Sarkar et al. 2016).
Salinity stress (SS) tolerance in DREB transgenic crops
SS refers to the stress caused by high levels of sodium chloride (NaCl), the major component of most saline soils. It can be simulated experimentally by irrigating the plants with saline water (in pots) or by transferring the seedlings to salt-containing media or nutrient solution having electrical conductivity (EC) > 4. The effects of SS should be evaluated both at the seedling and mature plant stages (Bhatnagar-Mathur et al. 2008). Datta et al. (2012) and Sarkar et al. (2014) observed an improved SS tolerance, physio-biochemical parameters and yield of DREB transgenic rice and peanut cultivars when exposed to NaCl (12–18 EC or 100–200 mM)-supplemented nutrient solution for 28 days and 12 days, respectively.
Transgenic StDREB1 potato maintained large green leaves (stay-green trait) compared to the WT variety, which showed necrotic lesions and subsequently died after 20 days of SS (Bouaziz et al. 2013). The GmDREB1 transgenic wheat showed a higher level of osmolytes such as proline and glycine betaine for retaining macromolecule homeostasis, better seedling traits along with longer roots, higher fresh-weight, and more tillers than the WT variety, with increasing duration of SS (Jiang et al. 2014). The heterologous expression of the DREB gene might regulate the expression and function of ion transporters in a stress-specific manner for maintaining ion homeostasis at the cellular level. LbDREB transgenic tobacco showed significantly more K+/Na+ ratio than its WT counterparts under CuSO4 induced-oxidative stress, suggesting over-expression of DREB gene can significantly reduce Na+ content in transgenic tobacco under stress conditions (Ban et al. 2011). However, over-expression of PgDREB2A in transgenic tobacco could not activate ion exchange antiporters under SS (Agarwal et al. 2010).
Salinity-susceptibility index (Oraby and Ahmad 2012) and salt-injury index (Jiang et al. 2014) are critical parameters for evaluating SS. Constitutive expression of heterologous DREB genes, however, may result in salinity sensitivity as opposed to tolerance (Table S2), indicating the requirement of a stress-inducible promoter (Tang et al. 2017).
DREB imparted thermo-tolerance in transgenic crops
A number of studies have demonstrated thermo-tolerance in various DREB transgenic crops. DREB2A is a heat shock (H) stress-inducible gene containing an H element upstream of its promoter, which transcriptionally activates the downstream HsfA3, a heat shock TF which in turn activates the downstream heat shock proteins (HSPs) and confers thermo-tolerance (Sakuma et al. 2006; Schramm et al. 2008). The DREB transgenic tobacco showed short-term (6 h) tolerance to a sudden drop in temperature (4 °C) (Agarwal et al. 2010), as well as long-term tolerance (33 days) to gradually declining temperatures (12 °C to − 5 °C) under open field conditions (Wei et al. 2006). Similarly, the TaDREB2 and TaDREB3 transgenic barley showed less damage from frost stress and improved survival rate of 45–50% compared to the WT variety (Morran et al. 2011).
Other examples include the AtDREB1A tomato that showed improved stomatal conductance, transpiration rate, RWC, and osmotic adjustments compared to the WT variety when exposed to low temperatures (4, 6, and 8 °C) for one week (Shah et al. 2015). Also, MtDREB1C Medicago truncatula and China Rose cultivars showed better survival compared to their WT counterparts when exposed to − 6 °C for 10 h and − 6 °C for 60 h, respectively (Chen et al. 2010). The heterologous expression of CfCBF3 in tobacco also conferred tolerance to low-temperature stress by enhancing the level of total unsaturated fatty acids in the leaves, which helped protect its photosynthetic apparatus (Yang et al. 2011b).
In some cases, synthetic/modified DREB gene, rather than its native form have been used for achieving the desired level of ectopic gene expression and stress tolerance in transgenic crops (Reis et al. 2014; Wang et al. 2014). The RdreB1BI transgene was synthesized by overlap extension PCR on the DREB1B gene of rice using primers with optimized codons and decreased GC content, and induced low-temperature tolerance in transgenic strawberry (Wang et al. 2014). DREB also regulates the expression of heat shock proteins (HSPs), as well as photosynthetic/metabolic enzymes that confer high-temperature stress tolerance (Table S2, Fig. S1). Hong et al. (2009) reported high-temperature tolerance in AtDREB1A chrysanthemum when exposed to 45ºC for 36 h. However, the thermo-tolerance and overall improved physiological traits of various DREB transgenic crops have not been associated with an improved yield so far.
Multiple stress tolerance in DREB transgenic crops
Since crops are simultaneously exposed to multiple stresses under field conditions, it is of prime importance to develop multi-stress tolerant varieties of important crops to increase productivity. Various DREB transgenic lines do show tolerance to drought, salinity, alkalinity, and high/low temperature. For example, heterologous expression of AtDREB1A in peanut (Sarkar et al. 2014) and maize (Al-Abed et al. 2007) improved the RWC and membrane stability (low ion leakage) compared to the WT, thus indicating better cellular water status under drought, salt, and low-temperature stresses. Similarly, AtDREB1A peanut (Sarkar et al. 2014), AaDREB1 rice (Zong et al. 2016) and GhDREB wheat (Gao et al. 2009) also showed higher chlorophyll retention capacity, indicating a higher rate of photosynthesis, under drought, salt, and low-temperature conditions compared to their WT counterparts. The better performance of DREB transgenic plants could be attributed to the DREB-activated expression of downstream genes preventing the chlorophyll degradation and thereby maintaining the normal photosynthesis and energy metabolism under multiple stress conditions.
As discussed earlier (Fig. 1), osmolyte biosynthesis and antioxidant enzyme activity are vital responses underling physio-biochemical and growth-related improvements in transgenic plants under various abiotic stresses. For example, AtDREB1B potato (Movahedi et al. 2012), AtDREB1A Lolium perenne (Li et al. 2011), AaDREB1 rice (Zong et al. 2016) and IbCBF3 sweet-potato (Jin et al. 2017) showed higher proline levels, soluble sugar accumulation and antioxidant enzymatic activities due to the activation of macromolecule homeostasis and detoxification mechanisms under drought, salt and low-temperature stresses (Table S2). Also, AtDREB1A peanut showed better RWC under DS and SS than the WT variety; following stress withdrawal, the transgenic plants also recovered faster and performed better than the WT genotype (Sarkar et al. 2014). Furthermore, AtDREB1A peanut showed improved osmotic adjustment in terms of higher osmolality or lower osmotic potential under DS and SS (Sarkar et al. 2014), which again indicated the possibility of cross-talks among various stress-response mechanisms.
The constitutive co-expression of three stress-responsive TFs genes viz. AtDREB2A, AtHB7, and AtABF3 in peanut was reported conferring tolerance to various abiotic stresses (Pruthvi et al. 2014). However, since each transgene expression may increase the metabolic load on the plant, thus the incorporation of a single TF gene imparting the desired level of stress tolerance and yield under DS and SS might be a more feasible option (Sarkar 2014). In fact, studies on transgenic crops have demonstrated that individual physiological responses are inter-linked and eventually lead to stress tolerance, improved growth, and yield. For example, AtDREB1A peanut exposed to DS and SS showed elevated levels of proline, which resulted in better osmotic adjustment. Both phenomena increase water retention capacity and lower the level of ion leakage due to improved membrane integrity and better protection of photosynthetic mechanisms (Sarkar et al. 2014).
In addition, OsDREB1B tobacco shows improved resistance to tobacco streak virus and tolerance to drought, salt, low-temperature, as well as methyl viologen-induced oxidative stress (Gutha and Reddy 2008). Similarly, various transgenic crops expressing heterologous DREB gene also showed tolerance to oxidative stress in combination with biotic and abiotic stresses (Table S2) (Bouaziz et al. 2013; Charfeddine et al. 2015; Gutha and Reddy 2008; Pruthvi et al. 2014; Rai et al. 2013). Thus, the DREB transgenic crops show multi-stress tolerance likely due to the cross-talk between various biotic and abiotic stress-responsive mechanisms, and also due to the interaction between ABA-dependent and independent signal transduction pathways (Fig. S1).
Evaluation of yield-components and yield in DREB transgenic crops
The ultimate goal of developing stress-tolerant transgenic crops is to improve crop yield under harsh environmental conditions. However, most studies so far have only reported the short-term expression of heterologous DREB gene and survival of the transgenic plants under stress. In addition, the experimental acute stress used to study these transgenic plants does not reflect the in-field conditions. Therefore, the physiological, biochemical and growth-related data of transgenic plants may not always be reliable (Bhatnagar-Mathur et al. 2008). Thus, it is essential to determine the threshold level of any abiotic stress which a transgenic can tolerate and show optimal growth and yield as compared to its WT counterparts (Sarkar 2014).
Some reports have shown improved physio-biochemical characteristics and growth of DREB transgenic plants that resulted in improved yield, likely due to stress escape, avoidance, tolerance, and recovery mechanisms (Table S2). Yield parameters like daily transpiration (T), TE and HI have been recorded to determine the effect of DS on transgenic plants (Anbazhagan et al. 2015a, b; Bhatnagar-Mathur et al. 2010; Sarkar et al. 2014). Nevertheless, very few studies have evaluated transgenic crop yield as a function of stress tolerance under containment facility or open field conditions (Anbazhagan et al. 2015a; Bhatnagar-Mathur et al. 2014; de Paiva Rolla et al. 2014; Ravikumar et al. 2014; Sarkar et al. 2014) (Table S2). For example, AtDREB1A peanut (Bhatnagar-Mathur et al. 2014), rice (Ravikumar et al. 2014), and chickpea (Anbazhagan et al. 2015a) showed yield gain under DS and SS compared to the WT genotypes, without any yield penalty. Anbazhagan et al. (2015a) observed early flowering and seed setting in AtDREB1A transgenic chickpea line as compared to WT counterparts under DS, which was indicative of a drought escape mechanism.
Rice being an important cereal crop of the world, most studies on transgenic plants have focused on the development of high-yielding stress-tolerant rice cultivars (Fig. S2). DREB rice showed improved agronomic traits, such as an increase in the number of reproductive tillers and spikelets at the pre-flowering and vegetative stages, respectively, compared to the WT variety under DS and SS (Datta et al. 2012). As stated earlier, the DREB transgenic line could show multiple abiotic stress tolerance due to overlapping stress responsive mechanisms, with the exception of a few instances. For example, two TaDREB3 transgenic wheat lines (F3BC3) showed higher seed yield (18.9% and 21.5%) and better survival rate than WT varieties under DS, but no significant difference was observed in survival rate under low-temperature stress (Shavrukov et al. 2016).
On the contrary, AtDREB1A peanut showed improved HI, an important yield parameter under DS and SS (Bhatnagar-Mathur et al. 2014; Sarkar et al. 2014). In addition, improved RWC in AtDREB1A peanut was found positively correlated with improved HI (r = 0.64 and 0.91) compared to WT peanut under DS and SS (Sarkar et al. 2014). Therefore, improved physiological traits under abiotic stresses may improve the yield of crop plants.
Conclusions
Single gene action based transgenic approaches have not always shown much encouraging results due to the polygenic nature of abiotic stress-tolerance mechanisms. DREB gene was introduced into plants to develop stress-tolerant crops. This review discussed in-depth molecular, physio-biochemical mechanisms responsible for abiotic stress tolerance in transgenic crops and the fate of transgenic crops under confinement/contained facilities and field conditions (Table S2). This review also highlighted the use of DREB transgenic plants in back-cross breeding programs and future strategies towards commercialization of DREB transgenic crops (Fig. 3).
Although many DREB transgenic crops with varying degrees of abiotic and biotic stress tolerance have been developed worldwide, none of them are currently under large-scale cultivation (Table S2; Fig. S1, S2). In addition, limited reports are available showing improved yield under field conditions (Table S2). The recovery and yield of the transgenic plants after stress withdrawal are also critical parameters for evaluation of stress tolerant lines (Sarkar et al. 2014). Therefore, integrated approaches are needed to assess the tolerance and yield potential of the homozygous and stable DREB transgenic lines following the withdrawal of individual or multiple stresses under field-like conditions, in order to identify the best DREB transgenic lines for further characterization and large-scale cultivation (Fig. 3).
In addition, novel genetic engineering strategies like the use of stress-inducible, tissue-specific promoters to minimize excessive metabolic load on the transgenic crops, chloroplast transformation, and targeted and marker-free gene transfer can be introduced into the DREB regulon technology to nullify the environmental, ethical and health concerns associated with transgenic crops.
Future prospects
Advanced ‘omics’ approaches should be used to dissect the DREB regulon controlled downstream signaling pathways involved in stress responses in transgenic crops. The identification of novel genes that are up-regulated during stress and their respective promoters can help engineer new stress-tolerant crops with improved photosynthetic machinery, water conservation, WUE and root traits and cellular level tolerance, without any growth and yield penalty (Fig. 3). The downstream genes of DREB regulon that is up-regulated under abiotic stress could be mapped to the linkage groups of different transgenic crops. Furthermore, the identification of quantitative trait loci (QTL) based on bi-parental mapping populations or genome-wide association studies (GWAS) will help develop allele-specific markers (ASMs) of these candidate genes (Lata et al. 2011; Rao et al. 2015; Zhuang et al. 2015) and enable marker-assisted transgenic crop development for abiotic stress tolerance (Fig. 3).
The identification of novel promoters of stress-inducible genes could be utilized for driving expression of heterologous genes in transgenic plants. Further, the TaDREB3 gene was transferred into four elite bread wheat cultivars by repeated backcrossing and the resultant lines (F2BC3) showed 12–18% higher survival rates and yield than the WT varieties under DS. The backcross breeding programs also generated marker-free TaDREB3 transgenic wheat lines (F1BC2) due to recombination events that occurred in the genomic regions surrounding the transgene (Shavrukov et al. 2016). Such backcross-based gene introgression can also be extended to the other DREB based transgenic crops.
So far, very few abiotic stress-tolerant DREB transgenic plants have been tested to understand their resistance to biotic stresses (Table S2, Fig. S1). Hence, DREB transgenic crops need to be evaluated for their possible resistance to biotic stress and to unravel the in-depth molecular mechanisms responsible for cross-talks between biotic and abiotic stress responses. Further, each transgenic crop needs critical evaluation upon exposure to the appropriate level of stress, under confinement/contained facility or field trials (multi-year, multi-location), at the appropriate developmental stage, for evaluating its potential utility at commercial scale in near future.
Alternatively, the transgenic crops can also be evaluated for their tolerance, growth, and yield under sub-optimal and/or optimal stress exposure for their entire life span (Wang et al. 2016). However, upscaling from transgenic development to field trials requires continuous and consorted efforts, scientific networking, adequate fund flow, and availability of land, technical expertise and biosafety clearance (Mishra et al. 2017; Sarkar et al. 2017). Event-specific qualitative and quantitative PCR-based detection assays need to be developed for meeting the demands for field trials and curb the use of unauthorized transgenic crops (Liu et al. 2014). The currently available stress-tolerant DREB-based transgenic crops ought to be made accessible for small and resource-poor farmers. However, political will and simplification of biosafety regulations are also simultaneously required to facilitate the commercialization of transgenic crops.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Agarwal PK, Jha B. Transcription factors in plants and ABA-dependent and independent abiotic stress signaling. Biol Plant. 2010;54(2):201–212. doi: 10.1007/s10535-010-0038-7. [DOI] [Google Scholar]
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Agarwal P, Agarwal PK, Joshi AJ, Sopory SK, Reddy MK. Overexpression of PgDREB2A transcription factor enhances abiotic stress. Mol Biol Rep. 2010;37:1125–1135. doi: 10.1007/s11033-009-9885-8. [DOI] [PubMed] [Google Scholar]
- Agarwal PK, Gupta K, Lopato S, Agarwal P. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J Exp Bot. 2017;68(9):2135–2148. doi: 10.1093/jxb/erx118. [DOI] [PubMed] [Google Scholar]
- Akhtar M, Jaiswal A, Taj G, Jaiswal JP, Qureshi MI, Singh NK. DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J Genet. 2012;91(3):385–395. doi: 10.1007/s12041-012-0201-3. [DOI] [PubMed] [Google Scholar]
- Al-Abed D, Madasamy P, Talla R, Goldman S, Rudrabhatla S. Genetic engineering of maize with the Arabidopsis DREB1A/CBF3 gene using split-seed explants. Crop Sci. 2007;47:2390–2401. doi: 10.2135/cropsci2006.11.0712. [DOI] [Google Scholar]
- Anbazhagan K, Bhatnagar-Mathur P, Sharma KK, Baddam R, Kavi Kishor PB, Vadez V. Changes in timing of water uptake and phenology favours yield gain in terminal water stressed chickpea AtDREB1A transgenics. Funct Plant Biol. 2015;42:84–94. doi: 10.1071/FP14115. [DOI] [PubMed] [Google Scholar]
- Anbazhagan K, Bhatnagar-Mathur P, Vadez V, Dumbala SR, Kavi Kishor PB, Sharma KK. DREB1A overexpression in transgenic chickpea alters key traits influencing plant water budget across water regimes. Plant Cell Rep. 2015;34:199–210. doi: 10.1007/s00299-014-1699-z. [DOI] [PubMed] [Google Scholar]
- Ban Q, Liu G, Wang Y. A DREB gene from Limonium bicolour mediates molecular and physiological responses to copper stress in transgenic tobacco. J Plant Physiol. 2011;168(5):449–458. doi: 10.1016/j.jplph.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P, Devi MJ, Reddy DS, Lavanya M, Vadez V, Serraj R, Yamaguchi-Shinozaki K, Sharma KK. Stress-inducible expression of AtDREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 2007;26:2071–2082. doi: 10.1007/s00299-007-0406-8. [DOI] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P, Vadez V, Sharma KK. Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep. 2008;27:411–424. doi: 10.1007/s00299-007-0474-9. [DOI] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P, Devi MJ, Vadez V, Sharma KK. Differential antioxidative responses in transgenic peanut bear no relationship to their superior transpiration efficiency under drought stress. J Plant Physiol. 2009;166:1207–1217. doi: 10.1016/j.jplph.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P, Rao JS, Vadez V, Sharma KK. Transgenic strategies for improved drought tolerance in legumes of semi-arid tropics. J Crop Improv. 2010;24(1):92–111. doi: 10.1080/15427520903337095. [DOI] [Google Scholar]
- Bhatnagar-Mathur P, Rao JS, Vadez V, Dumbala SR, Rathore A, Yamaguchi-Shinozaki K, Sharma KK. Transgenic peanut overexpressing the DREB1A transcription factor has higher yields under drought stress. Mol Breed. 2014;33:327–340. doi: 10.1007/s11032-013-9952-7. [DOI] [Google Scholar]
- Bosamia TC, Mishra GP, Radhakrishnan T, Dobaria JR. Novel and stress relevant EST derived SSR markers developed and validated in peanut. PLoS ONE. 2015;10(6):e0129127. doi: 10.1371/journal.pone.0129127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaziz D, Pirrello J, Ben Amor H, Hammami A, Charfeddine M, Dhieb A, Bouzayen M, Gargouri-Bouzid R. Ectopic expression of dehydration responsive element binding proteins (StDREB2) confers higher tolerance to salt stress in potato. Plant Physiol Biochem. 2012;60:98–108. doi: 10.1016/j.plaphy.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Bouaziz D, Pirrello J, Charfeddine M, Hammami A, Jbir R, Dhieb A, Bouzayen M, Gargouri-Bouzid R. Overexpression of StDREB1 transcription factor increases tolerance to salt in transgenic potato plants. Mol Biotechnol. 2013;54(3):803–817. doi: 10.1007/s12033-012-9628-2. [DOI] [PubMed] [Google Scholar]
- Charfeddine M, Bouaziz D, Charfeddine S, Hammami A, Ellouz ON, Bouzid RG. Overexpression of dehydration-responsive element-binding 1 protein (DREB1) in transgenic Solanum tuberosum enhances tolerance to biotic stress. Plant Biotechnol Rep. 2015;9:79–88. doi: 10.1007/s11816-015-0345-8. [DOI] [Google Scholar]
- Chen JR, Lü JJ, Rong L, Xiong XY, Wang TX, Chen SY, Guo LB, Wang HF. DREB1C from Medicago truncatula enhances freezing tolerance in transgenic M. truncatula and china rose (Rosa chinensis Jacq.) Plant Growth Reg. 2010;60(3):199–211. doi: 10.1007/s10725-009-9434-4. [DOI] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1, a regulator of cold induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Baisakh N, Ganguly M, Krishnan S, Shinozaki KY, Datta SK. Overexpression of Arabidopsis and rice stress genes inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol J. 2012;10:579–586. doi: 10.1111/j.1467-7652.2012.00688.x. [DOI] [PubMed] [Google Scholar]
- de Paiva Rolla AA, de Fátima Corrêa Carvalho J, Fuganti-Pagliarini R, Engels C, do Rio A, Marin SRR, de Oliveira MCN, Beneventi MA, Marcelino-Guimarães FC, Farias JRB, Neumaier N, Nakashima K, Yamaguchi-Shinozaki K, Nepomuceno AL. Phenotyping soybean plants transformed with rd29A: AtDREB1A for drought tolerance in the greenhouse and field. Transgen Res. 2014;23(1):75–87. doi: 10.1007/s11248-013-9723-6. [DOI] [PubMed] [Google Scholar]
- Devi MJ, Bhatnagar-Mathur P, Sharma KK, Serraj R, Anwar SY, Vadez V. Relationships between transpiration efficiency and its surrogate traits in the rd29A:DREB1A transgenic lines of groundnut. J Agron Crop Sci. 2011;197(4):249–328. doi: 10.1111/j.1439-037X.2011.00464.x. [DOI] [Google Scholar]
- Dong C, Ma Y, Wisniewski M, Cheng ZM. Meta-analysis of the effect of overexpression of CBF/DREB family genes on drought stress response. Environ Exp Bot. 2017;142:1–4. doi: 10.1016/j.envexpbot.2017.07.014. [DOI] [Google Scholar]
- Farooq M, Wahid A, Kobayashi N, Fujita D, Basra SMA. Plant drought stress: effects, mechanisms and management. Agron Sustain Dev. 2009;29(1):185–212. doi: 10.1051/agro:2008021. [DOI] [Google Scholar]
- Gao SQ, Chen M, Xia LQ, Xiu HJ, Xu ZS, Li LC, Zhao CP, Cheng XG, Ma YZ. A cotton (Gossypium hirsutum) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep. 2009;28:301–311. doi: 10.1007/s00299-008-0623-9. [DOI] [PubMed] [Google Scholar]
- Gutha LR, Reddy AR. Rice DREB1B promoter shows distinct stress- specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol Biol. 2008;68:533–555. doi: 10.1007/s11103-008-9391-8. [DOI] [PubMed] [Google Scholar]
- Hackenberg M, Shi B-J, Gustafson P, Langridge P. A transgenic transcription factor (TaDREB3) in barley affects the expression of microRNAs and other small non-coding RNAs. PLoS ONE. 2012;7(8):e42030. doi: 10.1371/journal.pone.0042030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Ma C, Yang Y, Wang T, Yamaguchi-Shinozaki K, Gao J. Over-expression of AtDREB1A in chrysanthemum enhances tolerance to heat stress. Plant Mol Biol. 2009;70:231–240. doi: 10.1007/s11103-009-9468-z. [DOI] [PubMed] [Google Scholar]
- Jagana SR, Vadez V, Bhatnagar-Mathur P, Narasu ML, Sharma KK. Better root:shoot ratio conferred enhanced harvest index in transgenic groundnut overexpressing the rd29A:DREB1A gene under intermittent drought stress in an outdoor lysimetric dry-down trial. J SAT Agric Res. 2012;10:1–7. [Google Scholar]
- Jiang Q, Hu Z, Zhang H, Ma Y. Overexpression of GmDREB1improves salt tolerance in transgenic wheat and leaf protein response to high salinity. Crop J. 2014;2:120–131. doi: 10.1016/j.cj.2014.02.003. [DOI] [Google Scholar]
- Jiang Q, Sun X, Niu F, Hu Z, Chen R, Zhang H. GmDREB1 overexpression affects the expression of microRNAs in GM wheat seeds. PLoS ONE. 2017;12(5):e0175924. doi: 10.1371/journal.pone.0175924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Chang Q, Li W, Yin D, Li Z, Wang D, Liu B, Liu L. Stress-inducible expression of GmDREB1 conferred salt tolerance in transgenic alfalfa. Plant Cell Tissue Organ Cult. 2010;100:219–227. doi: 10.1007/s11240-009-9628-5. [DOI] [Google Scholar]
- Jin R, Kim BH, Ji CY, Kim HS, Li HM, Kwak SS. Overexpressing IbCBF3 increases low temperature and drought stress tolerance in transgenic sweetpotato. Plant Physiol Biochem. 2017;118:45–54. doi: 10.1016/j.plaphy.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Lata C, Bhutty S, Bahadur RP, Majee M, Prasad M. Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)] J Exp Bot. 2011;62(10):3387–3401. doi: 10.1093/jxb/err016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cheng X, Liu J, Zeng H, Han L, Tang W. Heterologous expression of the Arabidopsis DREB1A/CBF3 gene enhances drought and freezing tolerance in transgenic Lolium perenne plants. Plant Biotech Rep. 2011;5(1):61–69. doi: 10.1007/s11816-010-0157-9. [DOI] [Google Scholar]
- Li J, Sima W, Ouyang B, Wang T, Ziaf K, Luo Z, Liu L, Li H, Chen M, Huang Y, Feng Y. Tomato SlDREB gene restricts leaf expansion and internode elongation by downregulating key genes for gibberellin biosynthesis. J Exp Bot. 2012;63(18):6407–6420. doi: 10.1093/jxb/ers295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively in Arabidopsis. Plant Cell. 1998;10(8):1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang M-H, Yu Y-B, Sun Z, Zhen Z, Gao X-J. Event-specific qualitative and quantitative detection in transgenic soybean OsDREB3 based on the 5′ flanking sequence. Food Biotechnol. 2014;28(1):63–78. doi: 10.1080/08905436.2013.871289. [DOI] [Google Scholar]
- Mishra GP, Singh B, Seth T, Singh AK, Halder J, Krishnan N, Tiwari SK, Singh PM. Biotechnological advancements and Begomovirus management in okra (Abelmoschus esculentus L.): status and perspectives. Front Plant Sci. 2017;8:360. doi: 10.3389/fpls.2017.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB⁄CBF factors. Plant Biotechnol J. 2011;9:230–249. doi: 10.1111/j.1467-7652.2010.00547.x. [DOI] [PubMed] [Google Scholar]
- Movahedi S, Sayed TBE, Alizade H, Ghobadi C, Yamchi A, Khaksar G. Constitutive expression of Arabidopsis DREB1B in transgenic potato enhances drought and freezing tolerance. Biol Plantarum. 2012;56(1):37–42. doi: 10.1007/s10535-012-0013-6. [DOI] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005;138:341–351. doi: 10.1104/pp.104.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Kwon C-W, Choi D-W, Song SI, Kim JK. Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol J. 2007;5:646–656. doi: 10.1111/j.1467-7652.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- Oraby H, Ahmad R. Physiological and biochemical changes of CBF3 transgenic oat in response to salinity stress. Plant Sci. 2012;185–186:331–339. doi: 10.1016/j.plantsci.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Patel KG, Mandaliya VB, Mishra GP, Dobaria JR, Radhakrishnan T. Transgenic peanut overexpressing mtlD gene confers enhanced salinity-stress tolerance via mannitol accumulation and differential antioxidative responses. Acta Physiol Plant. 2016;38:181. doi: 10.1007/s11738-016-2200-0. [DOI] [Google Scholar]
- Patel K, Thankappan R, Mishra GP, Mandalia V, Kumar A, Dobaria JR. Transgenic peanut (Arachis hypogaea L.) overexpressing mtlD gene showed improved photosynthetic, physio-biochemical and yield-parameters under soil-moisture deficit stress in lysimeter system. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Gayen D, Datta SK, Datta K. Dissecting root proteome of transgenic rice cultivars unravels metabolic alterations and accumulation of novel stress responsive proteins under drought stress. Plant Sci. 2015;234:133–143. doi: 10.1016/j.plantsci.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, Hoisington D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome. 2004;47:493–500. doi: 10.1139/g03-140. [DOI] [PubMed] [Google Scholar]
- Pruthvi V, Narasimhan R, Nataraja KN. Simultaneous expression of abiotic stress responsive transcription factors, AtDREB2A, AtHB7and AtABF3 improves salinity and drought tolerance in peanut (Arachis hypogaea L.) PLoS ONE. 2014;9(12):e111152. doi: 10.1371/journal.pone.011115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai GK, Rai NP, Rathaur S, Kumar S, Major S. Expression of rd29A::AtDREB1A/CBF3 in tomato alleviates drought-induced oxidative stress by regulating key enzymatic and non-enzymatic antioxidants. Plant Physiol Biochem. 2013;69:90–100. doi: 10.1016/j.plaphy.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Rao ES, Kadirvel P, Symonds RC, Geethanjali S, Thontadarya RN, Ebert AW. Variations in DREB1A and VP1.1 genes show association with salt tolerance traits in wild tomato (Solanum pimpinellifolium) PLoS ONE. 2015;10(7):e0132535. doi: 10.1371/journal.pone.0132535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar G, Manimaran P, Voleti SR, Subrahmanyam D, Sundaram RM, Bansal KC, Viraktamath BC, Balachandran SM. Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice. Transgen Res. 2014;23(3):421–439. doi: 10.1007/s11248-013-9776-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis RR, da Cunha BADB, Martinsa PK, Martinsa MTB, Alekcevetch JC, Chalfun-Júnior A, Andrade AC, Ribeiro AP, Qind F, Mizoie J, Yamaguchi-Shinozaki K, Nakashima K, Carvalho JDFC, de Sousa CAF, Nepomucenof AL, Kobayashi AK, Molinari HBC. Induced over-expression of AtDREB2A CA improves drought tolerance in sugarcane. Plant Sci. 2014;221:59–68. doi: 10.1016/j.plantsci.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA. 2006;103(49):18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T (2014) Development of transgenic resistance to abiotic stress in groundnut using AtDREB1A gene through Agrobacterium mediated genetic transformation. Ph.D. Thesis, Saurashtra University, Rajkot, Gujarat, India
- Sarkar T, Radhakrishnan T, Kumar A, Mishra GP, Dobaria JR. Heterologus expression of AtDREB1A gene in transgenic peanut conferred tolerance to drought and salinity stresses. PLoS ONE. 2014;9(12):e110507. doi: 10.1371/journal.pone.0110507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T, Radhakrishnan T, Kumar A, Mishra GP, Dobaria JR. Stress inducible expression of AtDREB1A transcription factor in transgenic peanut (Arachis hypogaea L.) conferred tolerance to soil-moisture deficit stress. Front Plant Sci. 2016;7:935. doi: 10.3389/fpls.2016.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T, Mogili T, Sivaprasad V. Improvement of abiotic stress adaptive traits in mulberry (Morus spp.): an update on biotechnological interventions. 3 Biotech. 2017;7:214. doi: 10.1007/s13205-017-0829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Elizabeth Vierling E, Koskull-Doäring PV. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008;53:264–274. doi: 10.1111/j.1365-313X.2007.03334.x. [DOI] [PubMed] [Google Scholar]
- Shah SH, Ali S, Jan SA, Jalal-Ud-Din Ali GM. Piercing and incubation method of in planta transformation producing stable transgenic plants by overexpressing DREB1A gene in tomato (Solanum lycopersicum Mill.) Plant Cell Tissue Organ Cult. 2015;120:1139–1157. doi: 10.1007/s11240-014-0670-6. [DOI] [Google Scholar]
- Shavrukov Y, Baho M, Lopato S, Langridge P. The TaDREB3 transgene transferred by conventional crossings to different genetic backgrounds of bread wheat improves drought tolerance. Plant Biotechnol J. 2016;14:313–322. doi: 10.1111/pbi.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Liu K, Zhang J, Li X, Xu K, Zhang Y, Qi J, Yu D, Wang J, Lil C. JcDREB2, a physic nut AP2/ERF gene, alters plant growth and salinity stress responses in transgenic rice. Front Plant Sci. 2017;8:306. doi: 10.3389/fpls.2017.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Kato W, Asada Y, Sako K, Sato T, Sonoda Y, Kidokoro S, Yamaguchi-Shinozaki K, Tamaoki M, Arakawa K, Ichikawa T, Nakazawa M, Seki M, Shinozaki K, Matsui M, Ikeda A, Yamaguchi J. DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J Plant Res. 2009;122:633–643. doi: 10.1007/s10265-009-0252-6. [DOI] [PubMed] [Google Scholar]
- Vadez V, Rao S, Sharma KK, Bhatnagar-Mathur P, Jyotsna DM. DREB1A allows for more water uptake in groundnut by a large modification in the root/shoot ratio under water deficit. J SAT Agric Res. 2007;5(1):1–5. [Google Scholar]
- Wang F, Gao ZH, Qiao YS, Mi L, Li JF, Zhang Z, Lin Z-L, Gu X-B (2014) RDREB1BI gene expression driven by the stress-induced promoter RD29A enhances tolerance to cold stress in benihope strawberry. In: ISHS acta horticulturae 1049: VII international strawberry symposium. 10.17660/actahortic.2014.1049.159
- Wang H, Wang H, Shao H, Tang X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front Plant Sci. 2016;7:67. doi: 10.3389/fpls.2016.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Sun Z, Han L, Yu L. Transgenic tobacco plants over-expressing Arabidopsis transcriptional factor CBF1 show morphological and biochemical characteristics associated with cold tolerance. Asian J Plant Sci. 2006;5:932–939. doi: 10.3923/ajps.2006.932.939. [DOI] [Google Scholar]
- Wei T, Deng K, Gao Y, Liu Y, Yang M, Zhang L, Zheng X, Wang C, Song W, Chen C, Zhang Y. Arabidopsis DREB1B in transgenic Salvia miltiorrhiza increased tolerance to drought stress without stunting growth. Plant Physiol Biochem. 2016;104:17–28. doi: 10.1016/j.plaphy.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Wang R, Meng JJ, Bi YP, Xu PL, Guo F, Wan SB, He QW, Li XG. Overexpression of Arabidopsis CBF1 gene in transgenic tobacco alleviates photoinhibition of PSII and PSI during chilling stress under low irradiance. J Plant Physiol. 2010;167(7):534–539. doi: 10.1016/j.jplph.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Yang S, Tang XF, Ma NN, Wang LY, Meng QW. Heterology expression of the sweet pepper CBF3 gene confers elevated tolerance to chilling stress in transgenic tobacco. J Plant Physiol. 2011;168(15):1804–1812. doi: 10.1016/j.jplph.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Yang W, Liu XD, Chi XJ, Wu CA, Li YZ, Song LL, Liu XM, Wang YF, Wang FW, Zhang C, Liu Y. Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta. 2011;233(2):219–229. doi: 10.1007/s00425-010-1279-6. [DOI] [PubMed] [Google Scholar]
- Zhang S, Li N, Gao F, Yang A, Zhang J. Over-expression of TsCBF gene confers improved drought tolerance in transgenic maize. Mol Breed. 2010;26:455–465. doi: 10.1007/s11032-009-9385-5. [DOI] [Google Scholar]
- Zhang X-X, Tang Y-J, Ma O-B, Yang C-Y, Mu Y-H, Suo H-C, et al. OsDREB2A, a rice transcription factor, significantly affects salt tolerance in transgenic soybean. PLoS ONE. 2013;8(12):e83011. doi: 10.1371/journal.pone.0083011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Hu Y, Chong K, Wang T. ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice. Ann Bot. 2010;105:401–409. doi: 10.1093/aob/mcp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Wang F, Xu Z-S, Xiong A-S. Microarray analysis of different expression profiles between wild-type and transgenic rice seedlings overexpression OsDREB1BI gene. Biologia. 2015;70(6):760–770. doi: 10.1515/biolog-2015-0092. [DOI] [Google Scholar]
- Zong J-M, Li X-W, Zhou Y-H, Wang F-W, Wang N, Dong Y-Y, Yuan Y-X, Chen H, Liu X-M, Yao N, Li H-Y. The AaDREB1 transcription factor from the cold-tolerant plant Adonis amurensis enhances abiotic stress tolerance in transgenic plant. Int J Mol Sci. 2016;17:611. doi: 10.3390/ijms17040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.