Abstract
Male infertility due to spermatogenesis defects affects millions of men worldwide. However, the genetic etiology of the vast majority remains unclear. Here we describe three men with primary infertility due to multiple morphological abnormalities of the sperm flagella (MMAF) from two unrelated Han Chinese families. We performed whole-exome sequencing (WES) and Sanger sequencing on the proband of family 1, and found that he carried novel compound heterozygous missense mutations in dynein axonemal heavy chain 6 (DNAH6) that resulted in the substitution of a conserved amino acid residue and co-segregated with the MMAF phenotype in this family. Papanicolaou staining and transmission electron microscopy analysis revealed morphological and ultrastructural abnormalities in the sperm flagella in carriers of these genetic variants. Immunostaining experiments showed that DNAH6 was localized in the sperm tail. This is the first report identifying novel recessive mutations in DNAH6 as a cause of MMAF. These findings expand the spectrum of known MMAF mutations and phenotypes and provide information that can be useful for genetic and reproductive counseling of MMAF patients.
Subject terms: Genetic testing, Infertility
Introduction
Infertility is a major health concern that affects more than 20 million men worldwide1. Factors contributing to male infertility include genetic disorders, urogenital infections, and immunological or hormonal abnormalities. Male infertility caused by sperm flagellar defects usually present with asthenozoospermia and teratozoospermia, which can also be observed in men with primary ciliary dyskinesia (PCD), a multisystem disorder caused by dysfunction of motile cilium and flagellum, leading to chronic rhinosinusitis, bronchiectasis, or heterotaxis2. However, some patients show similar sperm abnormalities but no other PCD manifestations, a condition known as multiple morphological abnormalities of the sperm flagella (MMAF)3–8.
MMAF is a characteristic form of severe asthenozoospermia defined by the presence of spermatozoa in the ejaculate with mosaic morphological abnormalities of the flagella—e.g., absent, short, bent, coiled, and irregular flagella6,9. Up to 20% of MMAF cases have a genetic origin10. However, to date only a few genes related to spermatogenesis or ciliogenesis—including DNAH1, AKAP4, CAFP43, and CAFP445,6,8,11,12—have been identified as causative factors of MMAF in humans. Thus, the genetic factors underlying most MMAF cases are unknown.
DNAH6 contains 77 exons and encodes a 4158-amino acid protein that belongs to the dynein protein family13,14, which includes multiple microtubule-associated motor protein complexes and plays an important role in ciliary movement and cell division15. DNAH6 mutations were recently identified in a heterogeneous group of heterotaxy patients with abnormalities in respiratory tract cilia, which contributed to the development of PCD16. Additionally, DNAH6 mutations were shown to be responsible for male infertility and premature ovarian insufficiency in humans17–19. Variants of DNAH6 are also associated with changes in lung function in cystic fibrosis patients20. Different DNAH6 mutations lead to distinct phenotypes; therefore, it is possible that they can contribute to the development of MMAF without PCD manifestations.
To evaluate this possibility, in this study, we examined 10 individuals with MMAF without PCD manifestations. We first identified novel compound heterozygous mutations in DNAH6 in three infertile men with MMAF.
Results
Clinical data
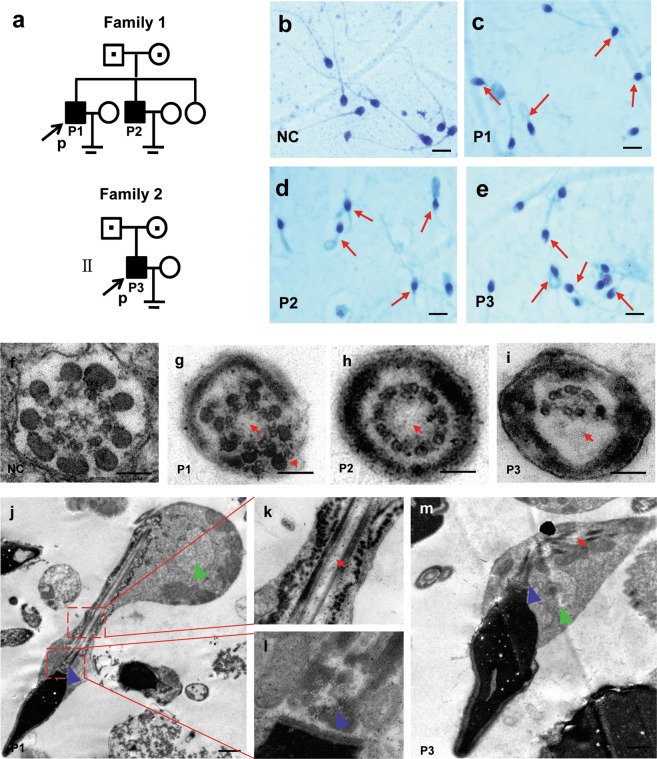
Routine semen and sperm morphology analyses were carried out for ten patients presenting with severe asthenozoospermia resulting from a combination of multiple morphological defects of the sperm flagella including: absent, short, bent, coiled or irregular width; eight had 100% immobile spermatozoa and two had sperm motility <10% (Fig. 1a–e and Table 1). None of the subjects showed any other PCD-associated symptoms. There were almost no spermatozoa (1.5%–6.0%) with normal morphology in the patients’ ejaculate; short, absent, and coiled flagella were the most frequently observed phenotypes (Table 1). The ultrastructure of patients’ spermatozoa (P1, P2, and P3) by transmission electron microscopy (TEM) frequently revealed absence of the central pair complex (CPC) of microtubules (Fig. 1f–m), other defects were occasionally seen, including peripheral microtubule doublets or disorganization of outer dense fibers (Fig. 1f–m). Longitudinal sections showed that the disorganized fibrous sheath or mitochondrial sheath, and a lack of axonemal CPC in the sperm flagella of patients, and tails with cytoplamic mass containing unassembled components of the sperm flagellum were frequently observed. Yet, the head-tail connection region of the sperm was morphologically normal, including 9 regularly arranged triplets, the closely attached striated column and the vault at the base body (Fig. 1f–m). Taken together, these three patients were diagnosed with the MMAF syndrome according to previously established criteria6.
Figure 1.
Pedigree of the two families analyzed in this study, and morphological and ultrastructural analysis of patient spermatozoa. (a) Filled and open symbols indicate the affected individuals and their unaffected relatives, respectively. A dot in the middle of a symbol indicates a heterozygous carrier. Probands are indicated with black arrows. (b–e) Papanicolaou staining of spermatozoa smears from the seminal fluid of the normal control (NC) (b) and the three patients (P1, P2, and P3) (c–e). Morphological abnormalities of sperm flagella were observed (red arrows), including absent, short, bent, coiled, and irregular flagella. Scale bars = 50 μm. (f–m) Ultrastructure analysis of spermatozoa obtained from the NC and patients by TEM. (f–i) Axonemal cross-sections of sperm flagella in NC, P1, P2, and P3. Absence of central microtubules and peripheral microtubule doublets (red arrow) or disorganization of outer dense fibers (red arrowhead) were observed. (j–m) Longitudinal sections of sperm flagella in NC, P1, and P2 showed that the disorganized fibrous sheath or mitochondrial sheath, and a lack of axonemal CPC in the sperm flagella of patients, and tails with a cytoplasmic mass were shown (green arrowhead). The sperm neck area including proximal centriole, striated column, vault (blue arrowhead) are visible. Scale bars = 0.1 μm.
Table 1.
Semen characteristics and sperm morphology in the ten patients under light microscopy.
| Semen parameters | P1a | P2a | P3a | P4c | P5b | P6c | P7b | P8c | P9c | P10c |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | 41 | 34 | 35 | 46 | 26 | 35 | 28 | 27 | 43 | 36 |
| Volume (ml) | 4.7 | 2.5 | 2.9 | 2.3 | 3.2 | 2.5 | 3.2 | 2.2 | 2.5 | 3.1 |
| Concentration (106 ml−1) | 10.1 | 6.8 | 11.3 | 25.3 | 10.1 | 11.5 | 20.6 | 42.1 | 20.5 | 8.2 |
| Progressive motility (%) | 0 | 5.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Motility (%) | 0 | 8.8 | 0 | 0 | 0 | 0 | 3.5 | 0 | 0 | 0 |
| Normal flagella (%) | 2.4 | 6.0 | 2.7 | 2.6 | 3.7 | 1.8 | 2.3 | 4.7 | 1.5 | 3.0 |
| Absent flagella (%) | 21.0 | 19.0 | 22.3 | 18.2 | 23.1 | 20.2 | 16.7 | 16.3 | 13.5 | 24.0 |
| Short flagella (%) | 51.0 | 48.5 | 45.6 | 52.8 | 54.9 | 59.5 | 64.5 | 59.0 | 68.0 | 50.5 |
| Coiled flagella (%) | 12.5 | 17.5 | 14.4 | 12.5 | 10.3 | 9.5 | 6.5 | 8.5 | 7.5 | 12.5 |
| Bent flagella (%) | 5.1 | 3.0 | 5.5 | 5.5 | 2.0 | 3.5 | 4.5 | 5.0 | 4.5 | 3.0 |
| Irregular width (%) | 8.0 | 6.0 | 9.5 | 8.4 | 6.0 | 5.5 | 5.5 | 6.5 | 5.0 | 7.0 |
| Affected spermatozoa (%) | 97.6 | 94.0 | 97.3 | 97.4 | 96.3 | 98.2 | 97.7 | 95.3 | 98.5 | 97.0 |
Note: aRepresent that the genetic cause of these patients might be DNAH6 mutations; brepresent that the genetic cause of these patients might be DNAH1 mutations; crepresent that the genetic causes of these patients remain unknown.
Identification of DNAH6 mutations by WES
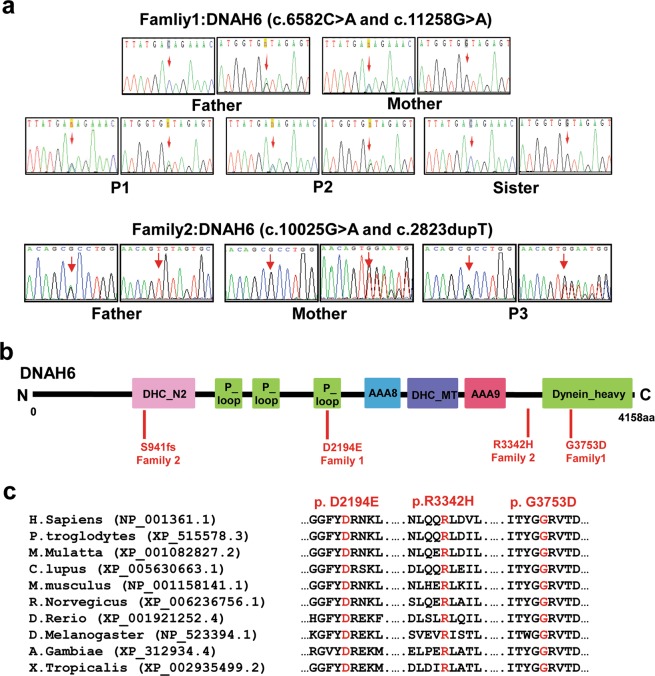
We screened for genes potentially causing MMAF in the proband (P1) by WES. We obtained 12.5 Gb of raw data with a mean depth of 159.28 folds for the target regions (Table S1). After mapping these data to the reference genome sequence (Hg 19), we identified 107,317 single nucleotide polymorphisms (SNPs) and 20,020 insertions/deletions (Indels) (Table S2). For rare inherited diseases, the frequency of possible pathogenic variants in the healthy population is very low. We filtered the WES results against a minor allele frequency >5% in publicly available SNP and Indel databases (Table S3); a total of 18,881 variants were retained, of which 362 were predicted to be deleterious. For infertile patients from a non-consanguineous family, homozygous or compound heterozygous variants are preferentially considered; based on these criteria, only five and eight variants, respectively, were retained. We focused on variants relevant to the phenotype in terms of expression and biological process, including model organisms with a male sterility phenotype similar to that observed in this family. Only one compound heterozygous variants of DNAH6 (NM_001370.1: c.6582 C > A, p. D2194E and c.11258 G > A, p. G3753D) satisfied these criteria (Table S4). After reviewing the literature, we concluded that these were most likely to be the disease-causing mutations.
We then performed Sanger sequencing and co-segregation analysis using the available DNA samples from family 1. The results showed that the affected brothers (P1 and P2) had the same compound heterozygous mutations (c.6582 C > A and c.11258 G > A) in DNAH6 (Fig. 2a and Table S5); their father carried one heterozygous variant (c.6582 C > A), while their mother carried the other (c.11258 G > A). Their unaffected sister (II-3) harbored the wild-type allele. The mode of inheritance was consistent with an autosomal recessive mode of inheritance.
Figure 2.
Segregation analysis and functional characterization of the DNAH6 mutation. (a) Sequence chromatograms for DNAH6 (c.6582 C > A and c.11258 G > A) in family 1, and DNAH6 (c.10025 G > A and c.2823dupT) in family 2. Both unaffected parents were heterozygous, whereas the patients (P1, P2, and P3) were compound heterozygous for these mutations and the unaffected sister harbored the wild-type variant. (b) Structure of DNAH6 protein; predicted functional domains are shown together with the position of novel mutations identified in MMAF patients. (c) Cross-species alignment showing strong conservation at positions 2194 and 3753 in DNAH6.
To validate our observations, we screened for DNAH6 mutations in another eight patients with MMAF (P3–P10) by WES. We detected another DNAH6 compound heterozygous mutation in P3 (family 2). Sanger sequencing confirmed that P3 harbored the compound heterozygous mutations c.10025 G > A and c.2823dupT in DNAH6, and segregation analysis demonstrated that these were inherited from the unaffected father (I-1) and mother (I-2), respectively (Fig. 2a). These DNAH6 mutations were not detected in the remaining seven patients. Notably, two of the seven patients (P5 and P7) were found to have novel mutations in the known MMAF gene DNAH1(NM_015512: c.G5105C: p.R1702P and c.11726_11727del: p.P3909fs for P5; c.8151-1 G > C and c.C12286T:p.R4096C for P7) (Table S5), indicating that MMAF is a genetically heterogeneous disorder.
Impact and expression profile of DNAH6 mutations
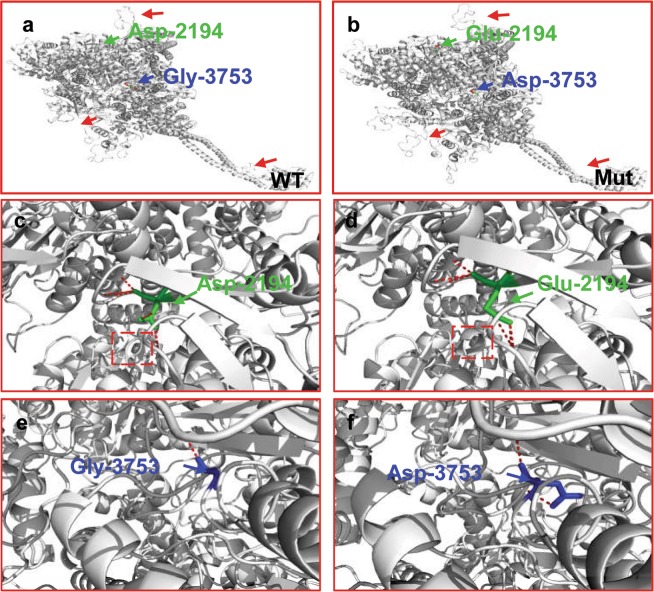
DNAH6 variants identified in the unrelated patients were located on predicted functional domains (Fig. 2b) and resulted in the substitution of conserved residues (Fig. 2c). All of the mutations were predicted to be deleterious by in silico analysis (Table S5). The three-dimensional structure of DNAH6 (1006–4158 amino acid residues) was modeled to analyze the impact of DNAH6 Asp2194Glu and p.Gly3753Asp in P1. The mutations changed part of the protein structure, including the β-sheets (Fig. 3a,b) and an α-helix (Fig. 3c,f), generating a misfolded protein with potentially reduced ATPase activity and microtubule-binding capacity.
Figure 3.
Modeling of wild-type and mutant DNAH6. Molecular structures of wild-type and mutant DNAH6 proteins were modeled with SWISS-MODEL software based on the template of dynein heavy chain, cytoplasmic (3vkh.pdb). (a,b) Structures of wild-type and mutant DNAH6 proteins; β-sheets (red arrows) are altered compared to the wild-type protein. (c,d) Enlarged view of the Asp-2194 site in wild-type and mutant DNAH6 protein. (e,f) Enlarged view of the Gly-3753 site in wild-type and mutant DNAH6 protein. Both Asp2194Glu and Gly3753Asp mutations of DNAH6 altered the structure of the protein. The positions of mutations are highlighted in blue.
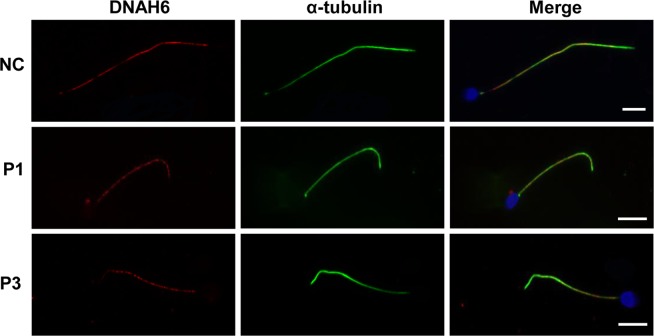
A reverse transcription PCR analysis showed that DNAH6 transcript was highly expressed in ciliated tissues in both adult humans and mice including testis, ovary, brain, and lung, and was weakly expressed in human stomach and kidney tissues. No expression was detected in other tissues such as eye, heart, and liver (Fig. S1). We also determined the expression and location of DNAH6 protein in the spermatozoa of patients (P1 and P3) by immunofluorescence analysis (Fig. 4). DNAH6 were localized in the sperm tail of normal spermatozoa (Fig. 4), and the expression level of DNAH6 in patients’ spermatozoa was not changed compared to the normal controls (Fig. 4).
Figure 4.
Immunolabeling of spermatozoa samples from DNAH6-mutant patients and normal controls (NC). Immunostaining of acetylated α-tubulin (green) and DNAH6 (red) in spermatozoa of NC and patients (P1 and P3). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). DNAH6 is specifically expressed in the sperm tail. Sperm tails in patients were short compared to those in the NC. Scale bars = 5 μm.
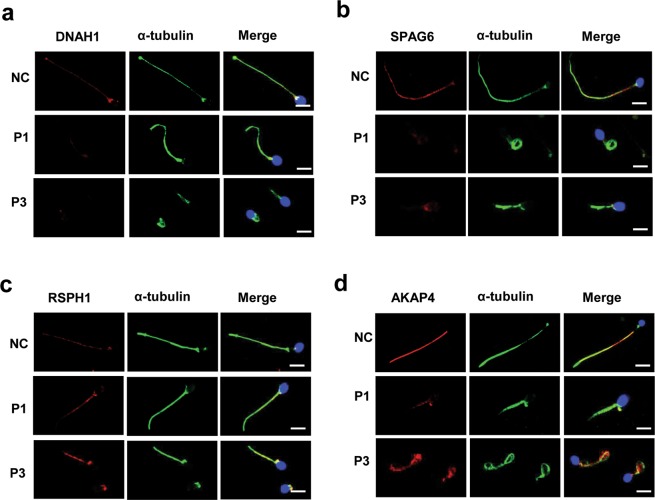
To characterize the ultrastructural defects observed by TEM at the molecular level, we performed immunostaining using antibodies targeting various axonemal proteins. In spermatozoa from DNAH6-mutant patients, expression of DNAH1 and SPAG6 (markers for the inner dynein arm and CPC, respectively) were remarkably reduced relative to the control (Fig. 5a,b). In patients with DNAH6 mutations, expression levels of the radial spoke marker RSPH1 were comparable to that in the control (Fig. 5c), suggesting that the mutations do not affect the radial spokes of flagella. However, in DNAH6-mutant patients, the fibrous sheath marker AKAP4 showed abnormally diffuse expression all along the midpiece and principle piece (Fig. 5d), whereas a restricted staining in the principle piece was in the normal control, suggesting the affected location and disorganized assembly of fibrous sheath due to mutations in DNAH6. Taken together, these results suggest that DNAH6 mutations cause severe axonemal or peri-axonemal disorganization, which might lead to the impair of sperm motility.
Figure 5.
DNAH6-mutant patients have abnormal, disorganized sperm axonemes. (a,b) Spermatozoa from normal control (NC) and DNAH6-mutant patients (P1 and P3) were double-labeled with antibodies against acetylated α-tubulin to identify axonemes and basal bodies (green), DNAH1 as a marker of the inner dynein arm (red), and SPAG6 as marker of CPC (red). Both α-tubulin and DNAH1 or SPAG6 were colocalized along the full length of axonemes in sperm flagellum from the NC (upper panel). In contrast, in MMAF patients (P1 and P3) (lower panels), DNAH1 or SPAG6 expression was markedly reduced in the axonemes compared to NC. (c,d) Spermatozoa from control and DNAH6-mutant patients (P1 and P3) was double-labeled with antibodies against acetylated α-tubulin to identify axonemes and basal bodies (green), RSPH1 as a marker of the radial spoke (red), and AKAP4 as a marker of the fibrous sheath (red). Scale bars = 5 μm.
Discussion
Infertility is a global medical and social problem with both physical and psychosocial consequences. Asthenozoospermia is particularly prevalent, occurring in ~19% of infertile men21; Generally, sever asthenozoospermia cases represent a heterogeneous group, and genetically-associated asthenozoospermia mainly includes PCD and MMAF6,7,22. In this study, we identified novel compound heterozygous mutations in the DNAH6 gene in three patients with MMAF from two unrelated Chinese families. This is the first study reporting DNAH6 as a recessive causative gene for this condition.
DNAH6 encodes axonemal dynein heavy chain that functions as an inner dynein arm component13. Dyneins are ATP-fueled motor proteins that generate force and movement on microtubules in a range of biological processes including ciliary beating, cell division, and intracellular transport15. Mutations in DNAH6 have been shown to cause PCD as well as heterotaxy by disrupting the CPC of motile cilia16. DNAH6 can act both recessively and possibly through trans-heterozygous interactions with other PCD genes such as DNAH1 or DNAH516. Another member of the dynein heavy chain family, DNAH1, is required in spermatozoa for the formation of inner dynein arms and is important for the assembly and biogenesis of the flagellar axoneme6,23. Mutations in DNAH1 can cause MMAF without any other PCD-associated symptoms6,23. Considering their structural and functional similarities, we speculate that DNAH6 may also participate in axoneme assembly and flagellar motility. In the present study, we confirmed that DNAH6 was highly expressed in tissues harboring cilia in both adult humans and mice such as testis, ovary, brain, and lung. Moreover, both TEM analysis and immunostaining revealed that DNAH6 mutations were associated with severe axonemal disorganization and markedly reduced DNAH1 expression. Additionally, similar to DNAH1 mutation phenotypes, patients with DNAH6 mutations in our study presented MMAF without other PCD-related symptoms as well as aberrant axonemal structure of sperm flagella, suggesting that DNAH6 plays distinct roles in axoneme regulation in somatic and germ cells. Dynein is involved in the process of spermatogenesis24; patients with a rare homozygous missense mutation in DNAH6 exhibited azoospermia and oligozoospermic (one patient)17. These findings provide evidence that DNAH6 contributes to flagellar axoneme assembly during spermatogenesis.
Recently, compound heterozygous missense mutations (c.2454 A > T, p.E818D; c.7706 G > A, p.R2569H) in DNAH6 were shown to cause sperm head anomalies (30% headless spermatozoa and 69% globozoospermia)19. These investigators found that DNAH6 was localized in the neck region of normal spermatozoa, and DNAH6 mRNA and protein were completely absent in carriers of the mutations. We carefully considered their findings, however, in our study, DNAH6 was detected in the sperm tail and ultrastructural analysis did not find obvious abnormality in sperm head or the neck region in patients with DNAH6 mutations. We found that compound heterozygous missense mutations in DNAH6 were associated with MMAF.
We identified novel compound heterozygous mutations of DNAH6 in both patients with MMAF that were predicted to be pathogenic by in silico analysis. The mutated amino acids (Asp2194Glu and p.Gly3753Asp) in P1—located in the third P-loop and dynein heavy chain domains, respectively, of DNAH6—were shown to be highly conserved across species. The P-loop has nucleoside triphosphate hydrolase activity and binds and hydrolyzes ATP25, and is the main functional domain of the dynein protein family. The dynein heavy chain domain located in the C terminus of DNAH6 has ATPase activity and binds microtubules, and functions as a motor for the movement of organelles and vesicles along microtubule tracks26,27. The three-dimensional model of the protein suggested that the identified compound heterozygous mutations in DNAH6 alters part of the protein structure, resulting in protein misfolding; this could reduce the ATPase activity and microtubule-binding ability of DNAH6. These results suggest that the amino acids at these positions are critical for the process of spermatogenesis. Although our identification of DNAH6 mutations in unrelated patients with MMAF suggests a causative relationship, functional validation of these mutations and their effects on spermatogenesis require further investigation.
Intracytoplasmic sperm injection (ICSI) is an assisted reproductive technology that is an effective way to achieve pregnancy in cases where the male exhibits grossly abnormal semen parameters, including MMAF10,28–31. Patients with MMAF harboring DNAH1 mutations have a good prognosis following ICSI, with 70.8% overall fertilization, 50.0% pregnancy, and 37.5% delivery rates32. In our study, ICSI using ejaculated live spermatozoa from the proband (P1) with MMAF resulted in the fertilization of 14/17 oocytes, with eight transferable embryos. Clinical pregnancy was confirmed after three embryos were transferred to his wife. However, no fetal heartbeat was detected by ultrasound and the pregnancy ended in abortion (data not shown). Therefore, whether successful ICSI outcomes can be achieved in MMAF patients with DNAH6 mutations requires further investigation.
In conclusion, we identified novel compound heterozygous mutations in DNAH6 that are likely responsible for the disrupted flagellation of spermatozoa in patients with MMAF. Our findings expand the spectrum of known DNAH6 mutations and phenotypes and provide useful information for genetic and reproductive counseling of patients with MMAF.
Methods
Patients
Ten male patients aged 26–46 years (mean age: 35.0 years) of the Hand ethnic group from Hunan province in China were treated in the Reproductive and Genetic Hospital of CITIC-Xiangya for primary infertility. The proband P1 (41 years old) and P2 (34 years old) from one family (family 1) were brothers whose parents were not consanguineous (Fig. 1a). The sister of the proband (29 years old) lacked any observable symptoms but had not attempted to procreate; therefore, her fertility status was unclear. The other eight unrelated patients (P3–P10), whose parents were not consanguineous either, were not related to one another. All patients had a normal 46, XY karyotype, and no abnormalities were observed by Y chromosome microdeletion analysis. Other causes of infertility such as drugs and exposure to gonadotoxic factors were excluded. None of the patients reported any additional symptoms associated with PCD. The patients’ parents were all healthy and fertile.
The study protocol was approved by the Institutional Ethics Committees of Central South University and Reproductive Genetic Hospital of CITIC-Xiangya, China. Written, informed consent was obtained from all participants. All procedures were performed in accordance with approved guidelines.
Semen and sperm morphological analyses
Semen samples were collected from each patient through masturbation after 3–5 days of sexual abstinence. Each semen parameter was measured at least three times according to the World Health Organization (WHO) 2010 guidelines33. Morphological abnormalities of the flagella were classified as (1) absent, (2) short, (3) coiled, (4) bent, or (5) irregular width6 as determined by Papanicolaou staining. The percentages of morphologically normal and abnormal spermatozoa were evaluated according to WHO guidelines.
Transmission electron microscopy (TEM)
Seminal fluid samples from normal control subjects (fertile adult donors who provided written informed consent to participate in this study) and patients were fixed overnight with 2.5% glutaraldehyde (Sigma-Aldrich, St. Louis, MO, USA) in 0.1 M phosphate buffer (pH 7.4) followed by 1% osmium tetroxide for 2 h for TEM analysis, which was carried out as previously described34. The 1-μm-thick sections were stained with toluidine blue for light microscopy, and 70- to 90-nm-thick sections were contrasted with uranyl acetate and lead citrate and examined with an H7700 electron microscope (Hitachi, Tokyo, Japan). Digital images were captured using a MegaView III digital camera (EMSIS GmbH, Münster, Germany).
WES and bioinformatic analysis
WES analysis was performed for samples from nine patients (P1 and P3–P10) and targeted Sanger sequencing was performed for P2, whose sample had not been available in time for inclusion in WES. Sequences were captured with the SureSelect v.4 platform (Agilent Technologies, Santa Clara, CA, USA) and the enriched library was sequenced with the Illumina HiSeq. 2000 system as previously described35. Raw reads after removing the adaptors were aligned to NCBI GRCh37 (Human reference genome Hg19) using the Burrows–Wheeler Aligner36, followed by the removal of PCR duplicates and sorting using Picard (http://broadinstitute.github.io/picard/). Variant identification was performed using GATK package37 according to the recommended best practices, including base recalibration variant calling with Haplotype Caller, quality score recalibration, and annotation using ANNOVAR software38.
We filtered the variants obtained by WES with minor allele frequencies of 5% in dbSNP, 1000 Genomes Project, or ExAC databases according to the following criteria: predicted to be deleterious; homozygous or compound heterozygous for the alternate allele; relevant phenotype based on comprehensive expression data (high expression in testis)39; the Gene Ontology term “biological process associated with spermatogenesis40”; and model organism data (male infertility phenotype observed in animal models)41.
Sanger sequencing
Forward and reverse PCR primers targeting mutation regions in the DNAH6 gene had the following sequences: c.6582 C > A: 5′-CCATCTTGTGCCCTGACAGT-3′ and 5′-GTCTGTCCACCCTCTGAAGC-3′; c.11258 G > A: 5′-GGAAGGAAATTCTGTTGTGTGCT-3′ and 5′-TGGGGATAGGGTGTGGCTATAA-3′; c.10025 G > A: 5′-CTGAATGCGAACAAGGGAGAC-3′ and 5′-AGATAGGGTTCCTTGCTGACG-3′; and c.2823dupT: 5′-TGAAAGGAATAAACAGGTGATGCT-3′ and 5′-ACTCTGAATGCCCCTCCTAAC-3′. These regions were amplified by PCR using Ex Taq DNA polymerase (Bio-Rad, Hercules, CA, USA) in all patients and their family members. PCR products were sequenced on a 3730XL system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions.
Evolutionary conservation and in silico analyses
Evolutionary conservation analysis was performed by aligning the amino acid sequences of DNAH6 proteins from different vertebrate species obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/homologene/). The potential pathogenicity of DNAH6 mutations was predicted by in silico analysis using MutationTaster (http://www.mutationtaster.org/), Sorting Intolerant From Tolerant (http://sift.jcvi.org/), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2), and Combined Annotation Dependent Depletion (cadd.gs.washington.edu/) programs. Structural analysis of DNAH6 (NP_001361.1) variants in P1 was carried out using SWISS-MODEL software (https://swissmodel.expasy.org) based on the template of dynein heavy chain, cytoplasmic (3vkh.pdb). PyMol (http://www.pymol.org) was used to visualize protein structure.
Expression analysis
Total RNA was extracted from tissues of fertile adult donors (who provided written informed consent for their participation in this study) and wild-type C57BL/6 adult mice using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and 1 μg was reverse transcribed into cDNA using a the GoScript Reverse Transcription System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. To evaluate the expression analysis of human DNAH6 and mouse Dnah6 genes, PCR amplification was performed with 1 μl of cDNA. The mRNA expression levels of human DNAH6 and mouse Dnah6 were normalized to those of the actin gene (ACTB and Actb, respectively) using the following forward and reverse primers: DNAH6, 5′-CTAGAGCCTTTGCCAGTGCTA-3′ and 5′-AGGAGAGGGAGAGAACACCTC-3′; ACTB, 5′-CCTGGCACCCAGCACAAT-3′ and 5′-GGGCCGGACTCGTCATAC-3′; Dnah6, 5′-CTGGAGCCTCTGCCAGTGCTA-3′ and 5′-CGGAGAAGGAGAGAACACATC-3′; and Actb, 5′-AGATCAAGATCATTGCTCCTCC-3′ and 5′-AGCTCAGTAACAGTCCGCCT-3′. PCR conditions were as follows: 95 °C for 10 min; 34 cycles of 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s; and 72 °C for 10 min. The assay was performed three times.
Immunofluorescence analysis
Sperm specimens were fixed in 4% paraformaldehyde for 30 min and permeabilized with 0.5% Triton X-100 for 10 min. Non-specific sites were blocked with 10% normal goat serum and 3% bovine serum albumin in phosphate-buffered saline, and the samples were incubated overnight at 4 °C with monoclonal anti-acetylated α-tubulin (T5168, 1:800), SPAG6 (HPA038440, 1:400), polyclonal anti- RSPH1 (HPA017382, 1:400), and polyclonal anti-AKAP4 (HPA020046, 1:200) antibodies (all from Sigma-Aldrich, St. Louis, MO, USA); and polyclonal antibodies against DNAH6 (ab122333, 1:50) and DNAH1 (ab122367, 1:100) (both from Abcam, Cambridge, UK). Alexa Fluor 488 anti-mouse (A-21121, 1:300) and Alexa Fluor 555 anti-rabbit (A31572, 1:300 dilution) IgG (both from Life Technologies, Carlsbad, CA, USA) were used as secondary antibodies. Specimens were counterstained with 4′,6-diamidino-2-phenylindole for 5 min, and fluorescence signals were visualized with a BX-51 fluorescence microscope (Olympus, Tokyo, Japan). Images were captured using VideoTesT-FISH v.2.0 software (VideoTesT, St. Petersburg, Russia).
Supplementary information
Acknowledgements
The authors thank the individuals who participated in this study as well as their families. This study was supported by grants from the National Natural Science Foundation of China (81771645 and 81471432 to Y.T.); Teacher Research Fund of the Central South University (905010042 to J.D.), and Graduate Research and Innovation Projects of Central South University (Grant 2017zzts071 to CF. T).
Author contributions
Yue-Qiu Tan designed the research; Chaofeng Tu, Hongchuan Nie, Lanlan Meng and Shimin Yuan performed the experiments; Wenbin He, Aixiang Luo and Haiyu Li carried out the bioinformatics analysis; Wen Li, Juan Du, Guangxiu Lu, Ge Lin, Yue-Qiu Tan, Chaofeng Tu and Hongchuan Nie analyzed the data; and Yue-Qiu Tan, Chaofeng Tu and Hongchuan Nie wrote the paper. All authors reviewed the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chaofeng Tu and Hongchuan Nie.
Supplementary information
is available for this paper at 10.1038/s41598-019-52436-7.
References
- 1.Boivin J, Bunting L, Collins JA, Nygren KG. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum Reprod. 2007;22:1506–1512. doi: 10.1093/humrep/dem046. [DOI] [PubMed] [Google Scholar]
- 2.Storm van’s Gravesande K, Omran H. Primary ciliary dyskinesia: clinical presentation, diagnosis and genetics. Ann Med. 2005;37:439–449. doi: 10.1080/07853890510011985. [DOI] [PubMed] [Google Scholar]
- 3.Escalier D, David G. Pathology of the cytoskeleton of the human sperm flagellum: axonemal and peri-axonemal anomalies. Biol Cell. 1984;50:37–52. doi: 10.1111/j.1768-322X.1984.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 4.Ray PF, et al. Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin Genet. 2017;91:217–232. doi: 10.1111/cge.12905. [DOI] [PubMed] [Google Scholar]
- 5.Amiri-Yekta A, et al. Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum Reprod. 2016;31:2872–2880. doi: 10.1093/humrep/dew262. [DOI] [PubMed] [Google Scholar]
- 6.Ben Khelifa M, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang SM, et al. Morphological characteristics and initial genetic study of multiple morphological anomalies of the flagella in China. Asian J Androl. 2015;17:513–515. doi: 10.4103/1008-682X.138190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang S, et al. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am J Hum genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutton C, Escoffier J, Martinez G, Arnoult C, Ray PF. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update. 2015;21:455–485. doi: 10.1093/humupd/dmv020. [DOI] [PubMed] [Google Scholar]
- 10.Chemes HE, Alvarez Sedo C. Tales of the tail and sperm head aches: changing concepts on the prognostic significance of sperm pathologies affecting the head, neck and tail. Asian J Androl. 2012;14:14–23. doi: 10.1038/aja.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baccetti B, et al. Gene deletions in an infertile man with sperm fibrous sheath dysplasia. Hum Reprod. 2005;20:2790–2794. doi: 10.1093/humrep/dei126. [DOI] [PubMed] [Google Scholar]
- 12.Coutton C, et al. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun. 2018;9:686. doi: 10.1038/s41467-017-02792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hom EF, et al. A unified taxonomy for ciliary dyneins. Cytoskeleton (Hoboken) 2011;68:555–565. doi: 10.1002/cm.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaughan KT, et al. Multiple mouse chromosomal loci for dynein-based motility. Genomics. 1996;36:29–38. doi: 10.1006/geno.1996.0422. [DOI] [PubMed] [Google Scholar]
- 15.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, et al. DNAH6 and Its Interactions with PCD Genes in Heterotaxy and Primary Ciliary Dyskinesia. PLoS Genet. 2016;12:e1005821. doi: 10.1371/journal.pgen.1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gershoni M, et al. A familial study of azoospermic men identifies three novel causative mutations in three new human azoospermia genes. Genet Med. 2017;19:998–1006. doi: 10.1038/gim.2016.225. [DOI] [PubMed] [Google Scholar]
- 18.Norling A, et al. Identification of a duplication within the GDF9 gene and novel candidate genes for primary ovarian insufficiency (POI) by a customized high-resolution array comparative genomic hybridization platform. Hum Reprod. 2014;29:1818–1827. doi: 10.1093/humrep/deu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, L. et al. DNAH6 is a novel candidate gene associated with sperm head anomaly. Andrologia (2018). [DOI] [PubMed]
- 20.Blue E, et al. Variation in Cilia Protein Genes and Progression of Lung Disease in Cystic Fibrosis. Ann Am Thorac Soc. 2018;15:440–448. doi: 10.1513/AnnalsATS.201706-451OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curi SM, et al. Asthenozoospermia: analysis of a large population. Arch Androl. 2003;49:343–349. doi: 10.1080/01485010390219656. [DOI] [PubMed] [Google Scholar]
- 22.Chemes HE. Phenotypes of sperm pathology: genetic and acquired forms in infertile men. J Androl. 2000;21:799–808. [PubMed] [Google Scholar]
- 23.Sha Y, et al. DNAH1 gene mutations and their potential association with dysplasia of the sperm fibrous sheath and infertility in the Han Chinese population. Fertil Steril. 2017;107:1312–1318 e1312. doi: 10.1016/j.fertnstert.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 24.McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olbrich H, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 26.Koonce MP, Grissom PM, Lyon M, Pope T, McIntosh JR. Molecular characterization of a cytoplasmic dynein from Dictyostelium. J Eukaryot Microbiol. 1994;41:645–651. doi: 10.1111/j.1550-7408.1994.tb01528.x. [DOI] [PubMed] [Google Scholar]
- 27.Mocz G, Gibbons IR. Model for the motor component of dynein heavy chain based on homology to the AAA family of oligomeric ATPases. Structure. 2001;9:93–103. doi: 10.1016/S0969-2126(00)00557-8. [DOI] [PubMed] [Google Scholar]
- 28.Payne D, Flaherty SP, Jeffrey R, Warnes GM, Matthews CD. Successful treatment of severe male factor infertility in 100 consecutive cycles using intracytoplasmic sperm injection. Hum Reprod. 1994;9:2051–2057. doi: 10.1093/oxfordjournals.humrep.a138392. [DOI] [PubMed] [Google Scholar]
- 29.Yang S, et al. Successful intracytoplasmic sperm injection with testicular spermatozoa from a man with multiple morphological abnormalities of the sperm flagella: a case report. J Assist Reprod Genet. 2018;35:247–250. doi: 10.1007/s10815-017-1057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chemes EH, Rawe YV. Sperm pathology: a step beyond descriptive morphology. Origin, characterization and fertility potential of abnormal sperm phenotypes in infertile men. Hum Reprod Update. 2003;9:405–428. doi: 10.1093/humupd/dmg034. [DOI] [PubMed] [Google Scholar]
- 31.Yang SM, et al. Intracytoplasmic sperm injection outcomes in Chinese men with multiple morphological abnormalities of sperm flagella. Asian J Androl. 2016;18:809–811. doi: 10.4103/1008-682X.150038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wambergue C, et al. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 33.Cooper TG, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 34.Tan YQ, et al. Loss-of-function mutations in TDRD7 lead to a rare novel syndrome combining congenital cataract and nonobstructive azoospermia in humans. Genet Med. 2019;21:1209–1217. doi: 10.1038/gim.2017.130. [DOI] [PubMed] [Google Scholar]
- 35.Zhu F, et al. Biallelic SUN5 Mutations Cause Autosomal-Recessive Acephalic Spermatozoa Syndrome. Am J Hum genet. 2016;99:942–949. doi: 10.1016/j.ajhg.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eppig JT, et al. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.