Abstract
To curb the increasing demand for nitrogenous fertilizers, it is imperative to develop new cultivars with comparatively greater nitrogen use efficiency (NUE). Nonetheless, so far very meager information is available concerning the variances among barley (Hordeum vulgare L.) varieties for their response to nitrogen deprivation. The current study was carried out to explore the potential of barley genotypes for higher NUE. A hydroponic experiment was conducted at seedling stage to compare the performance of four barley genotypes, ZD9 and XZ149 (with higher NUE) and HXRL and XZ56 (with lower NUE) in response to low (0.1 mM) and normal nitrogen (2 mM) levels. Under low N, all the genotypes expressed less number of tillers, decreased soluble proteins, chlorophyll and N concentrations in both roots and shoots, in comparison with normal N supply. However, significant differences were found among the genotypes. The genotypes with high NUE (ZD9 and XZ149) showed higher N concentration, increased number of tillers, improved chlorophyll and soluble proteins in both roots and shoots as compared to the inefficient ones (HXRL and XZ56). Furthermore, nitrate transporter gene (NRT2.1) showed higher expression under low N, both in roots and leaves of N efficient genotypes, as compared to the N inefficient ones. However, N assimilatory genes (GS1 and GS2) showed higher expression under normal and low N level, in leaves and roots respectively. The outcome of the study revealed that genotypes with higher NUE (ZD9 and XZ149) performed better under reduced N supply, and may require relatively less N fertilizer for normal growth and development, as compared to those with lower NUE. The study also revealed a time-specific expression pattern of studied genes, indicating the duration of low N stress. The current study suggested that future work must involve the time course as a key factor while studying expression patterns of these genes to better understand the genetic basis of low-N tolerance.
Keywords: Barley, Nitrogen metabolism, Gene expression
Introduction
Nitrogen (N) is a vital element in plant nutrition and is known to be a key factor in limiting crop productivity (Kraiser et al. 2011). Being a major component of RNA, DNA, chlorophyll, ATPs, cytokinins, auxin and enzymes, N plays a pivotal role in growth and development of plants (Raven et al. 2004; Hawkesford et al. 2012). In 2016, the world total use of nitrogenous fertilizer was 110 million tones, representing a 34% increase with respect to 2002 (FAO 2018). However, it has substantial impacts on the quality of environment throughout the globe. Only 30–50% of the applied N is taken up by crops, depending upon the crop species and cultivars, as well as management practices, with the rest fertilizer amount being lost and ultimately polluting the local agro-ecology (Garnett et al. 2009).
Within soil, most of the nitrogen is absorbed by plants as nitrate (NO3−) (Crawford and Forde 2002), which is reduced to ammonium by nitrate reductase (NR) and nitrite reductase (NiR), followed by amino acid and protein biosynthesis. Little et al. (2005) has narrated an important function of NRT2.1, a high-affinity nitrate transporter, in controlling branching pattern of roots, which implies that the NUE may directly be influenced by genes involved in nitrogen absorption (Lea and Azevedo 2007). Owing to its pivotal role in assimilation of inorganic nitrogen, glutamine synthetase (GS; EC 6.3.1.2) fascinated the researchers for several decades (Mc Nally et al. 1983; Edwards et al. 1990). The studies regarding expression and knockout behavior of individual GS isogenes revealed their cell and tissue specific localization and their pivotal role in growth and development of cereals. The expression pattern of NRT2.1 gene has thoroughly been investigated at mRNA level. Its expression is stimulated by low NO3− availability which controls its uptake by roots in response to plant’s nitrogen status (Okamoto et al. 2003; Filleur and Daniel-Vedele 1999). The expression of NRT2.1 is suppressed by NO3− itself, through a mechanism independent of the feedback repression, employed by nitrogen metabolites, but specifically prompted by the dual-affinity NRT1.1 NO3− transporter (Munos et al. 2004; Krouk et al. 2006). The NO3− high affinity nitrate transporters (HATS) are regulated by the same factors, showing a robust correlations of changes in NRT2.1 transcript level and the activity of HATS, indicating a central role of NRT2.1 transcriptional regulation in NO3− absorption.
The genetic diversity of cultivated barley becomes narrower due to intensive breeding and cultivation, posing a bottleneck for genetic improvement. Contrarily, wild barley has a highly rich source of genetic variability, and can serve as a valuable gene reservoir for future breeding programs (Dai et al. 2012). Due to immense genetic diversity, it is hypothesized that wild barley can perform better under low N availability. To curb the ever rising global N demands, it is imperative to identify the barley genotypes showing tolerance to N starvation, which could certainly serve as a genetic resource for developing high yielding crops, with comparatively low N requirement, ultimately giving a healthy environment to the future generations.
Genetic differences in physiological responses to applied N have already been pointed out in various species of plants like wheat, maize, barley, rice and sorghum (Ortiz-Monasterio et al. 1997; Le Gouis et al. 2000; Muchow 1998; Presterl et al. 2003; Namai et al. 2009; Anbessa et al. 2009). However, the information regarding intra-specific genetic diversity is very limited. A hydroponic experiment was designed to investigate the genotypic differences between Tibetan wild and cultivated barley under reduced N availability.
Materials and methods
Plant materials, growth conditions and N doses
This experiment was performed under a controlled environment at Zijingang Campus of Zhejiang University, Hangzhou (30.29°N and 120.16°E), P.R. China. On the basis of previous investigation (Shah et al. 2017a), two accessions (XZ56 and XZ149) of Tibetan annual wild barley, and two cultivars (HXRL and ZD9), varying in NUE, were chosen for the experiment. Seeds of barley cultivars were obtained from key Laboratory of Crop Germplasm Resource of Zhejiang Province, Department of Agronomy, Zhejiang University, Hangzhou, China. Two Tibetan annual wild barley accessions (XZ56 and XZ149) were obtained from Huazhong Agricultural University, China. Seeds were treated with 2% H2O2 for 30 min, for surface sterilization (Quan et al. 2016). After washing, the seeds were germinated on moist Whatman grade-1 filter papers. 12 days after germination (second leaf stage), uniformly healthy and vigorous seedlings were transplanted to containers filled with 5 L basic nutrition solution. The concentration of different nutrients in the solution was: 2 mmol NaNO3−, 0.63 mmol MgSO4, 0.18 mmol K2SO4, 0.18 mmol KH2PO4, 0.36 mmol CaCl2, 20.9 µmol Fe-citrate, 4.5 µmol MnCl2, 0.38 µmol ZnSO4, 0.16 µmol CuSO4, 46.9 µmol H3BO3 and 0.062 µmol H2MoO4. The solution pH was managed at 5.8 ± 0.1, by using HCl or NaOH. Prior to treatment, the plants were grown normally for 7 days for acclimatization in basic nutrient solution. Based on the previous studies (Quan et al. 2016), two N concentrations, i.e. 0.1 (low) and 2 mM (normal), were maintained in the basic nutrition solution for the study. Completely randomized block design was employed with four independent replicates. The containers were continuously supplied with air through pumps and the nutrition solution was replaced after every 5 days. After 22 days exposure to treatments, the plants were harvested for measuring morphological, physiological and biochemical characteristics of roots and shoots. However, the samples for molecular study were harvested after 24 h, 10 days and 15 days exposure to N treatments and immediately stored at − 80 °C.
Measurement of N concentration
Total N concentration in plant tissues was measured by the Kjeldahl method (Jones 1991). After drying, the material was ground into fine powder. Around 0.2 g sample powder was weighed, digested in sulfuric acid, and analyzed for N content according to Li et al. (2006).
Measurement of soluble protein
Bradford (1976) method was employed to determine the soluble protein contents. Fresh leaf samples (2 g each) were homogenized with 0.05 M Tris buffer (1 mL pH 8.5) using pestle and mortar, followed by centrifugation at 9000 for 10 min. The supernatant (100 µL) was mixed with 3 mL Bradford reagent (Sigma, prepared using 10 mL reagent + 50 mL distilled water), followed by incubation for 5 min. The absorbance was recorded at 595 nm using spectrophotometer. Standard calibration curve was generated by using bovine serum (Sigma). The concentration of protein in leaf samples was demonstrated in terms of mg/g fresh weight.
Chlorophyll content
On fresh weight basis, total chlorophyll content was determined as mentioned by Farida et al. (2017). The leaf disks, excised from the youngest fully expanded leaves, were embedded in acetone (20 mL) under dark conditions for approximately 24 h at 4 °C till the complete disappearance of green color. The resultant solution was used to measure the absorbance at 664 and 647 nm using a spectrometer (UV-3101P, Labomed Inc., USA).
qRT-PCR and gene expression analysis
By using the TRIzol reagent, total leaf and root RNA was extracted according to manufacturers’ protocols (Invitrogen, Karlsruhe, Germany). cDNA was synthesized using 1 μg of each RNA sample with 0.5 of oligo (dT) 12–18 and 200 units of Superscript II (Invitrogen, Karlsruhe, Germany). cDNA samples were analyzed by quantitative real time PCR (qRT-PCR) in the iCycler iQTM Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA) using the SYBR Green PCR Master Mix (Applied Biosystems). The PCR outlines were as follows: pre-denaturation at 95 °C for 30 s, 40 cycles of denaturation at 95 °C for 5 s and annealing at 60 °C for 30 s, followed by steps for Melt-Curve analysis (60–95 °C, 0.5 °C increment for 5 s per step). Each sample was replicated three times. Primer express soft-ware (Applied Biosystems) was used to design gene-specific primers. The primer used for HvGS1; (accession no JX878489 forward-5′-GAGCGTCACCATTGCTCTCT-3′, reverse-5′-CTGCCTTCCTTCTCGGTGTC-3′) for HvGS2; (accession no AK360336 forward-5′-GACACCTACACACCACAGGG-3′, reverse -5′-GCTATGTGCAAGTCATGGCG-3′) for HvNRT2.1; (accession no U34198 forward-5′-CTGACCTTGGTGCCCGTTAT-3′, reverse-5′-GGAGACCCTTGGCTTTCTCC-3′). Barley GAPDH (glyceralde-hyde-3-phosphate dehydrogenase) gene (accession no. M36650, forward-5′-AAGCATGAAGATACAGGGAGTGTG-3′, reverse-5′- AATTTATTCTCGGAAGAGGTTGTACA-3′) was used as control. The relative expression level was determined by using 2−ΔΔCT method, described by Livak and Schmittgen (2001).
Statistical analysis
The data are the means of four independent replicates. The significance of the differences between two barley genotypes, under varying N supply, was evaluated by two-way analysis of variance (ANOVA), followed by the LSD (least significant difference) multiple range test (P < 0.05), employing DPS 9.50 (Data Processing System) (Bukhari et al. 2016).
Results
N concentration in shoots and roots
Shoot N concentration (SNC) was significantly reduced in low N treatment relative to control in all barley genotypes (Table 1). However, a considerable variation among the four genotypes was observed, with ZD9 (3.74 and 4.95% at 0.1 and 2.0 mM N respectively) and XZ149 (3.76 and 5.79% at 0.1 and 2.0 mM N respectively) showing significantly higher SNC at both N levels than other two genotypes. Similarly root N concentration (RNC) was also significantly reduced in low N level relative to control (Table 1). Again ZD9 (1.44 and 3.10% at 0.1 and 2.0 mM N respectively) and XZ149 (1.66 and 2.87% at 0.1 and 2.0 mM N respectively) depicted the higher RNC than HXRL (1.12 and 2.51% at 0.1 and 2.0 mM N respectively) and XZ56 (1.43 and 2.73% at 0.1 and 2.0 mM N respectively). Moreover, wild genotype XZ149 contained the highest SNC and RNC at both N levels, indicating its high capability of N uptake and transportation.
Table 1.
Effect of N levels on chlorophyll content, N concentration (roots and shoots) and number of tillers per plant of four barley genotypes
| N level (mM) | Genotype | Chl a mg g−1 FW | Chl b mg g−1 FW | Chl a + b | SNC (%) | RNC (%) | Tillers per plant |
|---|---|---|---|---|---|---|---|
| 0.1 | ZD9 | 38.86 ab | 60.44 c | 99.83 b | 3.74 c | 1.44 d | 2.39 cd |
| XZ149 | 38.26 ab | 64.81 abc | 103.19 ab | 3.76 c | 1.66 d | 2.53 c | |
| HXRL | 38.38 ab | 50.54 d | 89.82 c | 2.95 de | 1.12 e | 1.41 e | |
| XZ56 | 38.75 ab | 61.56 bc | 100.31 b | 2.81 e | 1.43 d | 2.21 d | |
| 2.00 | ZD9 | 39.39 a | 70.25 a | 109.11 a | 4.95 b | 3.10 a | 3.25 a |
| XZ149 | 39.28 a | 67.95 ab | 106.20 ab | 5.79 a | 2.87 ab | 3.30 a | |
| HXRL | 38.08 b | 67.43 abc | 105.51 ab | 4.78 b | 2.51 c | 2.17 d | |
| XZ56 | 38.95 ab | 62.23 bc | 101.18 b | 3.60 cd | 2.74 bc | 2.95 b | |
| Interaction (G × N) | ns | ** | * | * | ns | ** |
Means sharing the common letter(s) are statistically at par with each other
ns non-significant, RNC root nitrogen concentration, SNC shoot nitrogen concentration
*Significant at P < 0.05, **significant at P < 0.01
Chlorophyll content and soluble proteins
As compared with the control, a declining trend was observed for chlorophyll content in the low N treatment. Chlorophyll a + b significantly differed among all genotypes at both N levels. However, chl a showed variation at higher N dose only. ZD9 (38.86, 39.39 and 99.83, 109.11 mg g−1 FW Chl a and Chl a + b at 0.1 and 2.0 mM N respectively) and XZ149 (64.81, 67.95 and 103.19, 106.20 mg g−1 FW Cha a and Chl a + b at 0.1 and 2.0 mM N respectively) had higher chlorophyll a and a + b contents at both N treatments (Table 1).
Like N concentration, the soluble protein contents were also significantly reduced in low N treatment relative to control (Fig. 1) in both shoots and roots of all four barley genotypes. Meanwhile, ZD9 and XZ149 had significantly higher leaf soluble protein contents than other two genotypes. However for root soluble protein contents, no considerable variation was noticed among the genotypes at low nitrogen level, except XZ56 which showed significantly lower root soluble protein content than other three genotypes. At normal nitrogen level, ZD9 had the highest soluble protein content than other three genotypes while HXRL exhibited the lowest value.
Fig. 1.
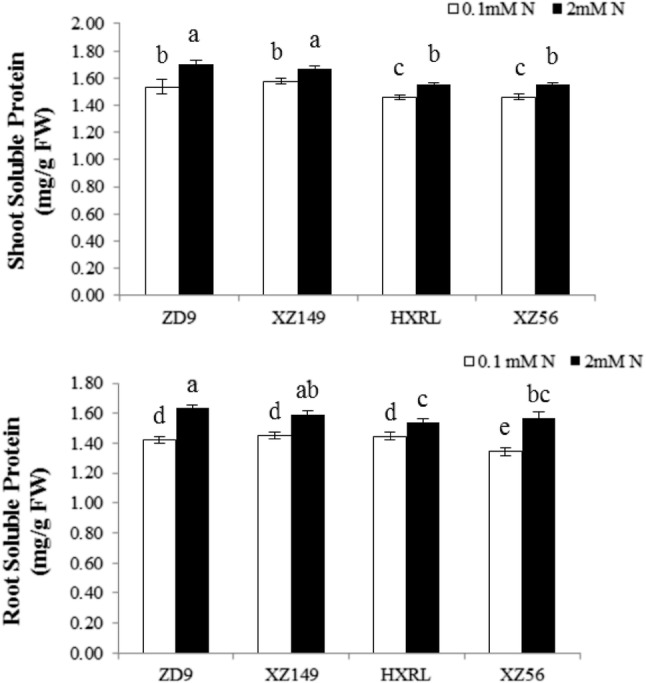
Effect of N levels on soluble proteins in shoot and root of four barley genotypes. Different letters indicate significant difference among the genotypes at P < 0.05
Number of tillers per plant
The tillering capacity of plants was significantly reduced under low N availability, as compared with the control. However, considerable differences were found among different barley genotypes for this trait, in response to low N supply. Both ZD9 (2.39 and 3.25 tillers at 0.1 and 2.0 mM N respectively) and XZ149 (2.53 and 3.30 tillers at 0.1 and 2.0 mM N respectively) produced more tillers per plant than other two genotypes at both N levels (Table 1).
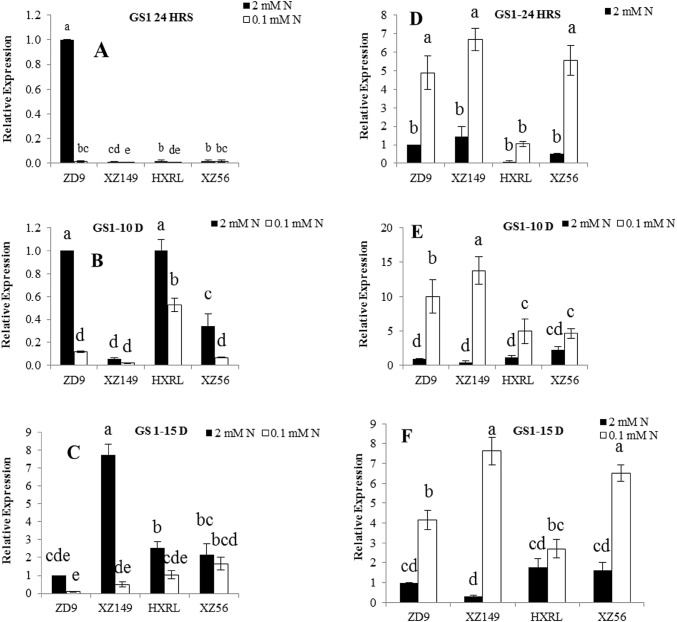
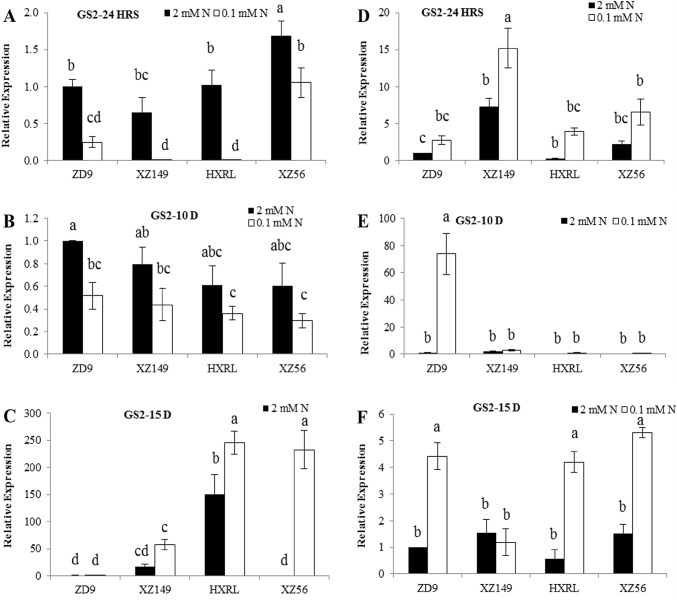
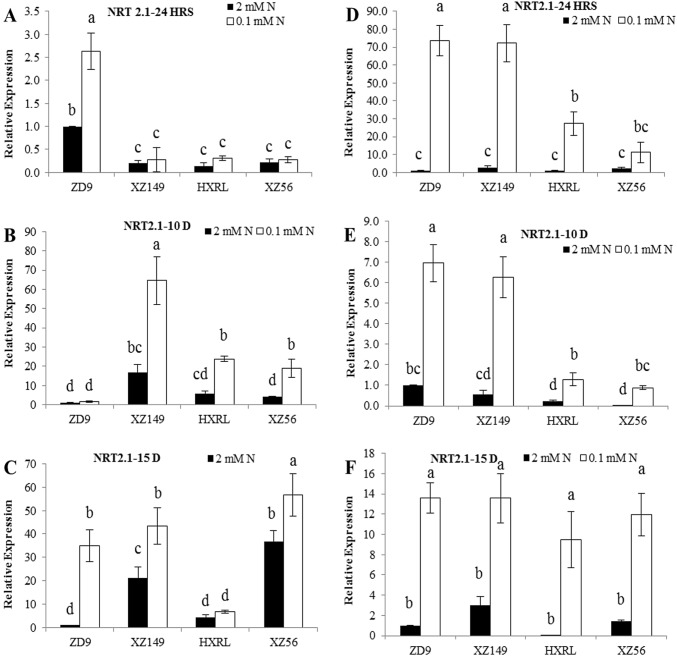
GS1, GS2 and NRT2.1 expression
The results indicated a significant influence of time interval on the expression pattern of genes in both leaves and roots. The interaction between genotype and time interval was also significant for all three genes (Figs. 2, 3, 4). The expression of GS1 and GS2 in leaves was dramatically reduced in low N treatment relative to control for all the genotypes except GS2 at 15 days after N treatment, where its expression was increased at low N in all four genotypes, while in roots, it was up-regulated under low N at all three sampling times. In contrast to N assimilatory genes, an increasing trend in expression of NRT2.1 (nitrate transporter) gene was observed, both in roots and leaves of the four genotypes under low N level during the whole stress treatment, with a marked genotypic difference. The expression of GS1 in ZD9 leaves was significantly increased from 24 h to 10 days, and then decreased at 15 days. In HXRL and XZ149, the expression of GS1 in leaves was increased at 10 days and 15 days, respectively (Fig. 2a–c). The expression level of GS1 in ZD9 was not different from that of HXRL at 10 days, but became significantly lower than other genotypes at 15 days (Fig. 2b, c). In roots, significant difference was found for expression level of GS1 among genotypes at 10 days, with HXRL being considerably lower at all sampling times than other genotypes. In addition, a considerable difference in GS1 expression was found between the two efficient genotypes at 10 and 15 days, with the wild barley (XZ149) exhibiting higher GS1 expression level than the cultivar ZD9.
Fig. 2.
Expression patterns of GS1 gene in four barley genotypes at different time intervals. a, b and c in leaves; d, e and f in roots at 24 h, 10 days and 15 days after N treatment, respectively. Different letters indicate significant difference among the genotypes at P < 0.05
Fig. 3.
Expression patterns of GS2 gene in four barley genotypes at different time intervals. a, b and c in leaves; d, e and f in roots at 24 h, 10 days and 15 days, respectively. Different letters indicate significant difference among the genotypes at P < 0.05
Fig. 4.
Expression patterns of NRT2.1 gene in four barley genotypes at different time intervals. a, b and c in leaves; d, e and f in roots at 24 h, 10 days and 15 days, respectively. Different letters indicate significant difference among the genotypes at P < 0.05
The GS2, a main isoform of GS in leaves, depicted a similarly clear reduction in its expression under low N availability relative to control. For GS2 expression level in leaves, only XZ56 was significantly higher at 24 h while no significant difference was found among the genotypes at 10 days after N treatment (Fig. 3a, b). At 15 days, GS2 expression reversed where HXRL showed increase in leaves (Fig. 3c). In roots, all the genotypes showed the similar expression. ZD9, XZ149 and XZ56 showed higher NRT2.1 expression in leaves at 24 h, 10 and 15 days after N treatment, respectively (Fig. 4a–c). XZ149 showed significantly higher expression level than ZD9 and HXRL, but lower than XZ56 at 15 days. No significant difference was found for root NRT2.1 expression level at 24 h and 15 days. However at 10 days, ZD9 (high N efficient genotype) showed significantly higher expression than the two genotypes with low NUE.
Low N decreased and increased the overall expression of GS1 and GS2 in leaves and roots, respectively, as compared to normal N supply. However slight difference was observed among the genotypes as well as sampling times. For GS1 expression, XZ149 and ZD9 showed the lowest value at 24 h, while HXRL exhibited the highest figure at 10 days treatment at low N (Fig. 2a–c). In roots, N efficient genotypes showed enhanced GS1 expression at low N as compared to the inefficient genotypes (Fig. 2d–f). Interestingly, the two N efficient genotypes differed in root GS1 expression at 10 and 15 days, XZ149 with higher expression than ZD9 (Fig. 2e, f). Similar trend was observed for GS2 in leaves where its expression was reduced under low N supply, but increased when the stress prolonged, i.e. at 15 days. Contrasting results were obtained at 15 days in low N level and genotypes differed significantly, where HXRL and XZ56 showed higher values than ZD9 and XZ149 in GS2 expression (Fig. 3c). In roots, N efficient genotypes XZ149 and ZD9 showed significant increase in GS2 expression at 24 h and 10 days, while XZ149 showed marked decrease at 15 days. NRT2.1 was highly expressed in ZD9 and XZ149 at 24 h and 10 days, respectively, in leaves at low N, while at 15 days, XZ56 had higher expression.
Discussion
The N concentrations often fluctuate under field condition, thus affecting the normal growth and development of plants. Therefore, adaption to low N availability is an important mechanism for the existence of plants in changing environments, and in crops, it is vital to maximize the yields (Kant et al. 2011). Plants’ adaptation to N deficiency consists of composite morphological, physiological, and developmental responses (Yang et al. 2011). Wide-range of alterations in primary and secondary metabolism, protein synthesis and cellular growth processes can be observed in plants under low N environments (Peng et al. 2008). Various crops, including maize, rice, oilseed rapes have been extensively investigated for their response to varying N doses (Kessel et al. 2012; Ikram et al. 2012; Wei et al. 2012; Abdel-Ghani et al. 2013). However, comparatively less information is available regarding the physiological and molecular variations in barley under reduced N availability. The traits that contribute to NUE comprise of a wider range of genetic variations (Kessel et al. 2012; Dawson et al. 2008). The emphasis of this experiment was to observe the expression of nitrogen transporter and assimilation genes in roots and leaves under different N supply. The plants growing under high nitrate supply showed down-regulation of leading GS1 and GS2 genes in the roots and a relative up-regulation in leaves, particularly HvGS1 and HvGS2 (Fig. 4). This response shows that HvGS1 gene responds to the shift from roots to shoot nitrate assimilation, owing to enhanced movement of nitrate to leaves as indicated by previous researchers (Lewis et al. 1982; Goodall et al. 2013). HvGS1, therefore has an important role to assimilate ammonia generated as a result of nitrate reduction in leaves. Similarly improved GS expression in leaves enhanced GS2 protein in the mesophyll and GS1 protein in vascular tissues (bundle sheath and vascular bundle) of plants growing under increased level of ammonium or nitrate (Tobin and Yamaya 2001; Goodall et al. 2013). The expression of GS2 under low nitrogen in leaves was higher at 15 days after N treatment (Fig. 3c). It has also been reported that in leaves, GS2 activity is usually greater, but it decreases with progressing senescence as chloroplasts are degraded (Mc Nally et al. 1983; Bernard et al. 2008). Thus, obvious increase was observed in N inefficient genotypes (Shah et al. 2017a, b). The up-regulation of HvGS1 and HvGS2 in plant roots, grown under reduced N supply is interesting (Figs. 2, 3). The higher expression of these genes under N scarcity might be the consequence of general stress response. Previous studies reported the highest expression of HvGS1 at 0.1 mM NH4NO3, which endorses this possibility, as ammonia is known to limit nitrate absorption and as a result aggravates the N deficiency in barley (Kronzucker et al. 1999; Lewis et al. 1982; Goodall et al. 2013). In contrast to that, increasing N level decreases expression of HvGS1 isoforms and HvGS2 in all tissues, further showing its role in re-assimilation of ammonia, synthesized during N remobilization. Likewise, in rice, Zhao and Shi (2006) observed an up-regulation of root GS isoforms under high N tension, while under normal N, GS1 had high expression.
The expression of the N assimilation genes HvGS1 and HvGS2 in the roots of N-efficient barley genotype ZD9 and XZ149 were greater than that in HXRL and XZ56 (inefficient) genotypes at early stage of N stress, i.e. 24 h and 10 days, suggesting that these genes may play important role in manipulating the NUEs of crops by controlling N uptake during N stress. The higher activity of GS and protein synthesis, in response to reduced N supply and stressful environment, is in agreement with the up-regulation of HvGS (Mack 1995; Peat and Tobin 1996; Goodall et al. 2013). In this study, the increased expression of GS in barley leaves at normal N condition in N efficient genotypes resulted in increased synthesis of soluble proteins (Fig. 1), higher N concentration and chlorophyll contents which improved the vegetative growth of plants, as reflected by number of tillers per plant (Table 1). Previous reports indicated that soluble protein synthesis and GS activity were directly linked to GS expression and that root GS expression increased under low N environment and other abiotic stresses, when rapid re-assimilation of ammonia, produced by protein degradation, was required (Mack 1995; Peat and Tobin 1996; Goodall et al. 2013). Furthermore, in view of the low N availability for protein synthesis, two scenarios might also be advocated for explaining the lower protein concentration in N-starved plants: (1) NO3−–N at comparatively high concentration would lead to an obvious increase in transcripts encoding various proteins (Stitt 1999); and (2) greater activity of proteases in N-starved plants which would lead to a rise in protein degradation (Galangau et al. 1988). Whatever the case, the capacity of maintaining a large proportion of N in a soluble form and divert it to the formation of soluble proteins might help plants to tolerate low N stress. Similar results were also observed previously in barley (Robredo et al. 2011) and in cassava (Gleadow et al. 2009).
Plants absorb N from the soil by nitrate transporters. The studies revealed a preferential expression of NRT2 genes in roots. However, no clear information is reported regarding the organ specificity in a wide range of plant species. In general, the expression of high affinity nitrate transport system increases under N starvation (Crawford and Glass 1998). It was also reported earlier that high affinity nitrate transporter AtNRT2.1 was induced in N-deprived Arabidopsis roots (Kiba et al. 2012; Lezhneva et al. 2014). In this study, nitrate transporter NRT2.1 was up-regulated at low N only in the roots and leaves of N efficient genotypes XZ149 and ZD9 (Fig. 4), suggesting enhanced uptake and translocation of NO3− from roots to leaves. While in inefficient genotypes, it showed a slower response. This distinctive up-regulation of nitrate transporters in XZ149 and ZD9 may contribute to enhanced NO3− absorption, generating higher amount of N-containing metabolites essential for their endurance under N scarcity. Thus, it could be deduced that better performance of ZD9 and XZ149 genotypes under low N availability is attributed to higher N absorption and accumulation.
Conclusion
By improving NUE of the crops, application of fertilizer and pollution in the environment could be decreased. The present study illustrated a huge variation among 4 barley genotypes in terms of NUE, providing an opportunity in developing new cultivars with high NUE, which will ultimately reduce the fertilizer application and its loss into environment. The plants, supplied with limited N revealed a negative correlation with total chlorophyll content, soluble proteins and tissue N concentration. The studied genes showed time specific expression patterns which indicates that the time of the stress is an important factor while manipulating variances among genotypes. So the present study suggests that the future work must involve the time course as a key factor while studying expression patterns of these genes which would enable us to better understand the genetic basis of low-N tolerance. The present study also supports the genes HvNRT2.1, HvGS1 and HvGS2 as possible targets to improve N uptake and assimilation in barley that could improve NUE.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jawad Munawar Shah, Email: shah8712@yahoo.com.
Sidra tul Muntaha, Email: smsidra@ymail.com.
Essa Ali, Email: essaali@upr.edu.pk.
Azhar Abbas Khan, Email: drkupchani@bzu.edu.pk.
Syed Hassan Raza Zaidi, Email: hasanzaidi72@yahoo.com.
Ahmad Naeem Shahzad, Email: anaeems@gmail.com.
Zeshan Hassan, Email: zeshan.hassan@bzu.edu.pk.
Ahmad Nawaz, Email: ahmadnawaz2006@gmail.com.
Muhammad Rashid, Email: muhammad.rashid@luawma.edu.pk.
Syed Asad Hussain Bukhari, Phone: +923018445080, Email: bukhariasad@yahoo.com.
References
- Abdel-Ghani AH, Kumar B, Reyes-Matamoros J, Gonzalez-Portilla PJ, Jansen C, Martin JPS, Lee M, Lübberstedt T. Genotypic variation and relationships between seedling and adult plant traits in maize (Zea mays L.) inbred lines grown under contrasting nitrogen levels. Euphytica. 2013;189:123–133. [Google Scholar]
- Anbessa Y, Juskiw P, Good A, Nyachiro J, Helm J. Genetic variability in nitrogen use efficiency of spring barley. Crop Sci. 2009;49:1259–1269. [Google Scholar]
- Bernard SM, Moller AL, Dionisio G, Kichey T, Jahn TP, Dubois F, Baudo M, Lopes MS, Tercé-Laforgue T, Foyer CH, Parry MA, Forde BG, Araus JL, Hirel B, Schjoerring JK, Habash DZ. Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivumL.) Plant Mol Biol. 2008;67:89–105. doi: 10.1007/s11103-008-9303-y. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bukhari SAH, Wang R, Wang W, Ahmed IA, Zheng W, Cao F. Genotype-dependent effect of exogenous 24-epibrassinolide on chromium-induced changes in ultrastructure and physicochemical traits in tobacco seedlings. Environ Sci Pollut Res. 2016;23:18229–18238. doi: 10.1007/s11356-016-7017-2. [DOI] [PubMed] [Google Scholar]
- Crawford NM, Forde BG. Molecular and developmental biology of inorganic nitrogen nutrition. In: Meyerowitz E, Somerville C, editors. The arabidopsis book. Rockville: American Soci Plant Biologists; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. [Google Scholar]
- Dai F, Nevo E, Wu D, Comadran J, Zhou M, Qiu L, Chen Z, Beiles A, Chen G, Zhang G. Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci USA. 2012;109:16969–16973. doi: 10.1073/pnas.1215265109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson JC, Huggins DR, Jones SS. Characterizing nitrogen use efficiency to improve crop performance in organic and sustainable agricultural systems. Field Crops Res. 2008;107:89–101. [Google Scholar]
- Edwards JW, Walker EL, Coruzzi GM. Cell-specific expression in transgenic plants reveals nonoverlapping roles for chloroplast and cytosolic glutamine synthetase. Proc Natl Acad Sci USA. 1990;87:3459–3463. doi: 10.1073/pnas.87.9.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2018) World Food and Agriculture—statistical pocketbook 2018. Rome. pp 8. http://www.fao.org/economic/ess/ess-publications/ess-yearbook/en/#.XTZhXK_lYdU
- Farida M, Ali S, Rizwan M, Ali Q, Abbas F, Bukhari SAH, Saeed R, Wu L. Citric acid assisted phytoextraction of chromium by sunflower; morphophysiological and biochemical alterations in plants. Ecotoxicol Environ Saf. 2017;145:90–102. doi: 10.1016/j.ecoenv.2017.07.016. [DOI] [PubMed] [Google Scholar]
- Filleur S, Daniel-Vedele F. Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta. 1999;207:461–469. doi: 10.1007/s004250050505. [DOI] [PubMed] [Google Scholar]
- Galangau F, Danielvedele F, Moureaux T, Dorbe MF, Leydecker MT, Caboche M. Expression of leaf nitrate reductase from tomato and tobacco in relation to light-dark regimes and nitrate supply. Plant Physiol. 1988;88:383–388. doi: 10.1104/pp.88.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett T, Conn V, Kaiser BN. Root based approaches to improving nitrogen use efficiency in plants. Plant Cell Environ. 2009;32:1272–1283. doi: 10.1111/j.1365-3040.2009.02011.x. [DOI] [PubMed] [Google Scholar]
- Gleadow RM, Evans JR, McCaffery S, Cavagnaro TR. Growth and nutritive value of cassava (Manihot esculenta Cranz) are reduced when grown in elevated CO2. Plant Biol. 2009;11:76–82. doi: 10.1111/j.1438-8677.2009.00238.x. [DOI] [PubMed] [Google Scholar]
- Goodall AJ, Kumar P, Tobin AK. Identification and expression analyses of cytosolic glutamine synthetase genes in barley (Hordeum vulgare L.) Plant Cell Physiol. 2013;54:492–505. doi: 10.1093/pcp/pct006. [DOI] [PubMed] [Google Scholar]
- Hawkesford M, Horst W, Kichey T, Lambers H, Schjoerring J, Skrumsager Møller I, White P. Functions of macronutrients. In: Marschner P, editor. Marschner’s mineral nutrition of higher plants. 3. Amsterdam: Elsevier; 2012. pp. 135–189. [Google Scholar]
- Ikram S, Bedu M, Daniel-Vedele F, Chaillou S, Chardon F. Natural variation of Arabidopsis response to nitrogen availability. J Exp Bot. 2012;63:91–105. doi: 10.1093/jxb/err244. [DOI] [PubMed] [Google Scholar]
- Jones JB., Jr . Kjeldahl method for nitrogen (N) determination. Athens: Micro-Macro Publishing; 1991. [Google Scholar]
- Kant S, Bi YM, Rothstein SJ. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J Exp Bot. 2011;62:1499–1509. doi: 10.1093/jxb/erq297. [DOI] [PubMed] [Google Scholar]
- Kessel B, Schierholt A, Becker HC. Nitrogen use efficiency in a genetically diverse set of winter oilseed rape (Brassica napus L.) Crop Sci. 2012;52:2546–2554. [Google Scholar]
- Kiba T, Feria-Bourrellier AB, Lafouge F, Lezhneva L, Boutet-Mercey S, Orsel M, Bréhaut V, Miller A, Daniel-Vedele F, Sakakibara H, Krapp A. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen starved plants. Plant Cell. 2012;24:245–258. doi: 10.1105/tpc.111.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiser T, Gras DE, Gutiérrez AG, González B, Gutiérrez RA. A holistic view of nitrogen acquisition in plants. J Exp Bot. 2011;62:1455–1466. doi: 10.1093/jxb/erq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker HJ, Glass ADM, Siddiqi MY. Inhibition of nitrate uptake by ammonium in barley. Analysis of component fluxes. Plant Physiol. 1999;120:283–291. doi: 10.1104/pp.120.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Tillard P, Gojon A. Regulation of the high-affinity NO3 uptake system by NRT1.1-mediated NO3 demand signaling in Arabidopsis. Plant Physiol. 2006;142:1075–1086. doi: 10.1104/pp.106.087510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gouis J, Beghin D, Heumez E, Pluchard P. Genetic differences for nitrogen uptake and nitrogen utilization efficiency in winter wheat. Eur J Agron. 2000;12:163–173. [Google Scholar]
- Lea PJ, Azevedo RA. Nitrogen use efficiency. 2. amino acid metabolism. Ann Appl Biol. 2007;151:269–275. [Google Scholar]
- Lewis OAM, James DM, Hewitt EJ. Nitrogen assimilation in barley (Hordeum vulgare L Cv. Mazurka) in response to nitrate and ammonium nutrition. Ann Bot. 1982;49:39–49. [Google Scholar]
- Lezhneva L, Kiba T, Feria-Bourrellier AB, Lafouge F, Boutet-Mercey S, Orsel M, Bréhaut V, Miller A, Daniel-Vedele F, Sakakibara H, Krapp A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2014;80:230–241. doi: 10.1111/tpj.12626. [DOI] [PubMed] [Google Scholar]
- Li BZ, Xin WJ, Sun SB, Shen QR, Xu GH. Physiological and molecular responses of nitrogen-starved rice plants to re-supply of different nitrogen sources. Plant Soil. 2006;287:145–159. [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mack G. Organ -specific changes in the activity and subunit composition of glutamine-synthetase isoforms of barley (Hordeum vulgare L) after growth on different levels of NH4. Planta. 1995;196:231–238. [Google Scholar]
- Mc Nally SF, Hirel B, Gadal P, Mann AF, Stewart GR. Glutamine synthetases of higher plants evidence for a specific isoform content related to their possible physiological role and their compartmentation within the leaf. Plant Physiol. 1983;72:22–25. doi: 10.1104/pp.72.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchow RC. Nitrogen utilization efficiency in maize and grain sorghum. Field Crops Res. 1998;56:209–216. [Google Scholar]
- Munos S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell. 2004;16:2433–2447. doi: 10.1105/tpc.104.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namai S, Toriyama K, Fukuta Y. Genetic variations in dry matter production and physiological nitrogen use efficiency in rice (Oryza sativa L.) varieties. Breed Sci. 2009;59:269–276. [Google Scholar]
- Okamoto M, Vidmar JJ, Glass AD. Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol. 2003;44:304–317. doi: 10.1093/pcp/pcg036. [DOI] [PubMed] [Google Scholar]
- Ortiz-Monasterio RJI, Sayre KD, Rajaram S, McMa-hon M. Genetic progress in wheat yield and nitrogen use efficiency under four nitrogen rates. Crop Sci. 1997;37:898–904. [Google Scholar]
- Peat LJ, Tobin AK. The effect of nitrogen nutrition on the cellular localization of glutamine synthetase isoforms in barley roots. Plant Physiol. 1996;111:1109–1117. doi: 10.1104/pp.111.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Hudson D, Schofield A, Tsao R, Yang R, Gu H, Bi Y-M, Rothstein SJ. Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J Exp Bot. 2008;59(11):2933–2944. doi: 10.1093/jxb/ern148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presterl T, Seitz G, Landbeck M, Thiemt EM, Schmidt W, Geiger HH. Improving nitrogen use efficiency in European maize: estimation of quantitative genetic parameters. Crop Sci. 2003;43:1259–1265. [Google Scholar]
- Quan X, Zeng J, Ye L, Chen G, Han Z, Shah JM, Zhang G. Transcriptome profiling analysis for two Tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol. 2016;16:30. doi: 10.1186/s12870-016-0721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Handley LL, Andrews M. Global aspects of C/N interactions determining plant environment interactions. J Exp Bot. 2004;55:11–25. doi: 10.1093/jxb/erh011. [DOI] [PubMed] [Google Scholar]
- Robredo A, Pérez-López U, Miranda-Apodaca J, Lacuesta M, Mena-Petite A, Muñoz-Rueda A. Elevated CO2 reduces the drought effect on nitrogen metabolism in barley plants during drought and subsequent recovery. Environ Exp Bot. 2011;71:399–408. [Google Scholar]
- Shah JM, Bukhari SAH, Jianbin Z, Xiaoyan Q, Ali E, Noor M, Zhang G. Nitrogen (N) metabolism related enzyme activities, cell ultrastructure and nutrient contents as affected by N level and barley genotype. J Integr Agric. 2017;16:190–198. [Google Scholar]
- Shah JM, Jianbin Z, Xiaoyan Q, Essa A, Imran HS, Zhang G. Growth and physiological characterization of low nitrogen responses in Tibetan wild and cultivated barleys. J Plant Nutr. 2017;40:861–868. [Google Scholar]
- Stitt M. Nitrate regulation of metabolism and growth. Curr Opin Plant Biol. 1999;2:178–186. doi: 10.1016/S1369-5266(99)80033-8. [DOI] [PubMed] [Google Scholar]
- Tobin AK, Yamaya T. Cellular compartmentation of ammonium assimilation in rice and barley. J Exp Bot. 2001;52:591–604. [PubMed] [Google Scholar]
- Wei D, Cui K, Ye G, Pan J, Xiang J, Huang J, Nie L, Estavillo JM. QTL mapping for nitrogen use efficiency and nitrogen deficiency tolerance traits in rice. Plant Soil. 2012;359:281–295. [Google Scholar]
- Yang X, Wu J, Ziegler TE, Yang X, Zayed A, Rajani MS, Zhou D, Basra AS, Schachtman DP, Peng M, Armstrong CL, Caldo RA, Morrell JA, Lacy M, Staub JM. Gene expression biomarkers provide sensitive indicators of in planta nitrogen status in maize. Plant Physiol. 2011;157:1841–1852. doi: 10.1104/pp.111.187898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XQ, Shi WM. Expression analysis of the glutamine synthetase and glutamate synthase gene families in young rice (Oryza sativa) seedlings. Plant Sci. 2006;170:748–754. [Google Scholar]