Significance
Photon-induced near-field electron microscopy (PINEM) enables the visualization of nanoscale structures and interfaces with enhanced contrast due to the energy gain of electrons from light-excited evanescent electromagnetic fields. Here we optimized a PINEM approach to directly image and quantify these fields at the surface of T cells as a function of various stimulation conditions. We found that while PINEM is sensitive to multiple biophysical changes associated with T cell activation, it strongly correlates with binding of specific antigen major histocompatibility complex, even without accompanying functional T cell activation. Our results suggest that PINEM can find use as a cellular imaging tool, with exquisite surface selectivity and high sensitivity to structural changes on the length scale of cell surface protein aggregates.
Keywords: T cell activation, PINEM, electron microscopy, electron-photon coupling, T cell receptor
Abstract
T cells can be controllably stimulated through antigen-specific or nonspecific protocols. Accompanying functional hallmarks of T cell activation can include cytoskeletal reorganization, cell size increase, and cytokine secretion. Photon-induced near-field electron microscopy (PINEM) is used to image and quantify evanescent electric fields at the surface of T cells as a function of various stimulation conditions. While PINEM signal strength scales with multiple of the biophysical changes associated with T cell functional activation, it mostly strongly correlates with antigen-engagement of the T cell receptors, even under conditions that do not lead to functional T cell activation. PINEM image analysis suggests that a stimulation-induced reorganization of T cell surface structure, especially over length scales of a few hundred nanometers, is the dominant contributor to these PINEM signal changes. These experiments reveal that PINEM can provide a sensitive label-free probe of nanoscale cellular surface structures.
Photon-induced near-field electron microscopy (PINEM) integrates the ultra-high spatial resolution of transmission electron microscopy with the extreme temporal resolution of femtosecond (fs) light spectroscopy (1). The basic concept is that, under certain conditions, transmitted electrons can absorb or emit photons. In free space, such a process is forbidden due to energy–momentum conservation. However, when the electron–photon interaction is both spatially and temporally confined, a material structure of sufficiently small dimensions can alter the dispersion relation of the photons at the surface, so as to eliminate this energy–momentum mismatch (2). In other words, the structure and composition of the interface can play a role in the generation of a PINEM signal. A formal theory of PINEM was initially developed and then tested using model nanostructures (2), such as plasmonic silver nanowires (metals) (3), carbon nanotubes (1), or protein vesicles (dielectrics) (4), each with critical dimensions below the wavelength of the incident radiation. Cancer cells with diameters substantially larger than the incident radiation wavelength, have surprisingly been shown to yield a weak, but measurable, PINEM signal, and that signal could be influenced through growth factor stimulation of the cells (5). However, the specific physical cellular properties that contributed to the PINEM signal, or the growth-factor induced PINEM signal changes, were unresolved.
Here, we use PINEM for the analysis of human T lymphocyte activation. PINEM can potentially provide a view of T cell activation, but perhaps surprisingly, T cell activation can also help provide insight into the physical parameters that affect the PINEM signal. Selective triggers for T cell activation include the use of peptide-major histocompatibility complexes (p-MHCs) for peptide antigen-specific engagement of the T cell receptor (6). T cell activation can also proceed via nonselective triggers, such as combining phorbol 12-myristate 13-acetate (PMA) to activate protein kinase C, with ionomycin, which is a calcium ionophore (7). We tap the known biology of different T cell stimulation protocols, coupled with biophysical properties measurements, to extract the physical parameters that influence the PINEM signal. We used a CD8-negative Jurkat T cell line transduced with T cell receptors (TCRs) specific for a melanoma-associated antigen (MART-1) (8). Controlled stimulation of these cells can be used to dial up a variety of biological or biochemical signals. We explored stimulation protocols that were limited to alterations in the cell surface biochemical composition, or which, at a second extreme, led to full biological activation. In both cases, the PINEM signal was altered by a much greater extent than was reported for EGF-stimulated cancer cells (5). This increased signal to noise allowed for correlations of PINEM signal strength, with PINEM imaging of cell/vacuum interfacial structure, with distinct biophysical and biochemical properties of the T cells, as stimulation protocols were titrated. We find that cellular surface structure is altered by both partial and full stimulation protocols. These measurements reveal that PINEM imaging can provide a highly sensitive tool for interrogating changes in cell surface structure. They also demonstrate that a nonactivating, but antigen-specific stimulation of CD8-negative T cells can lead to significant cell surface reorganization.
Results
PINEM Provides a Quantitative View of T Cells with High Spatiotemporal Resolution.
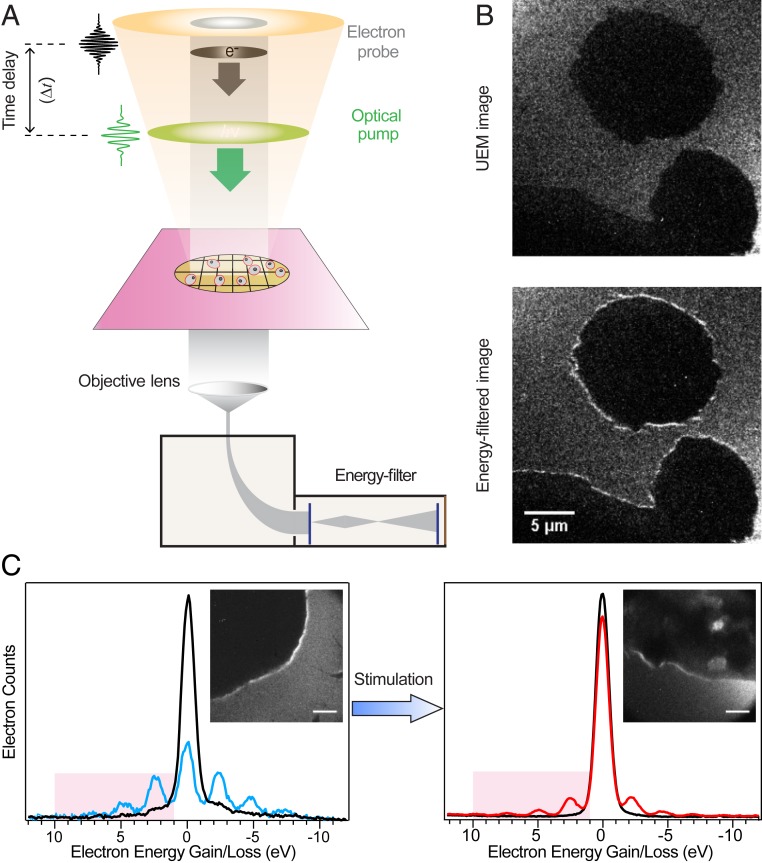
The basic instrumentation for PINEM imaging of Jurkat T cells is illustrated in Fig. 1. Modification of a conventional TEM allows for the production of ultrashort packets of imaging electrons, which are transmitted through the cells supported on a silicon nitride-covered TEM grid (Fig. 1A). The same cells are excited with a fs optical pulse that can be delayed to overlap in space and time with the electron pulse. The optical excitation creates an evanescent electric field at the surface of the cells, and the longitudinal components of this near-field inelastically scatters the electron pulse. This results in quantized energy exchange, yielding distinct gain and loss peaks in the electron energy spectrum that can be gated for imaging. PINEM imaging selects only the energy-gained electrons (1 to 11 eV) to form an image, enabling the visualization of the evanescent field at the cell boundaries. Contrast enhancement from the PINEM signal is clearly shown in Fig. 1B, in which unstimulated Jurkat cells appear to glow at the edges. The average excitation fluence (2.5 mJ⋅cm−2) of the pump pulse is optimized to yield a measurable electron energy gain/loss spectra, but without inducing thermal damage to the cells. This contrast enhancement from the evanescent photon-induced near-field effect is at a maximum when the 2 beams are coincident, and gradually disappears and is absent at a 1 picosecond (ps) delay (SI Appendix, Fig. S1, quantified using pixel-by-pixel analysis with 10 nm × 10 nm pixel size). The PINEM effect also exhibits a dependence on the optical field polarization with respect to the surface normal of the cells (SI Appendix, Fig. S2), consistent with previous investigations. Importantly, when Jurkat cells are stimulated with a standard T cell activation procedure that cross-links CD2, CD3, and CD28 cell surface ligands, the PINEM intensity was sharply attenuated relative to unstimulated cells (Fig. 1C).
Fig. 1.
Time-resolved PINEM on T cells. (A) Scheme depicting the basic components of PINEM imaging. A packet of probing electrons (gray) and a fs laser pulse (green) are directed to the cells. At the cell surface, some of the electrons gain or lose integer number of photon energy quanta. An energy filter is used to retain the electrons that gained energy, which are used to generate a PINEM image. (B) Representative UEM (Top) and PINEM (Bottom) images of Jurkat T cells. (C) Electron energy spectra of Jurkat T cell without (blue) or with (red) T cell activation (CD3/CD28/CD2) treatment and their corresponding PINEM images (Inset), showing the relative difference in contrast. Both spectra are measured at zero time delay, while spectra with zero loss peaks (black) are measured at +1 ps time delay. Electron energy gain equivalent to 1 to 4 photon quanta is highlighted (2.4 to 9.6 eV). (Scale bars: 1 μm.)
To systematically assess the PINEM signal of different stimulation conditions, we developed a standard procedure to measure and calculate the PINEM intensity. In a typical PINEM measurement, the second condenser aperture (9, 10) was adjusted to image an isolated cell on a TEM grid. Depending on the cell orientation, the PINEM intensity was maximized by tuning the optical polarization. Then, the electron energy spectra was collected so that the edge of the cell appears with highest the contrast enhancement relative to the interior of the cell. A given image covers an area of about 50 μm2 at 8,000× magnification. A background measurement was also collected either without optical excitation or with optical excitation at 1 ps time delay. The PINEM intensity is defined as the integral under the electron energy spectrum from +1 to +11 eV at zero time delay, normalized to the integration of the background spectrum at the same interval. When reporting on the PINEM intensity and SEM for a given experimental condition, the PINEM measurements are averaged over 4 to 12 individual cells.
PINEM Is Sensitive to T Cell Activation.
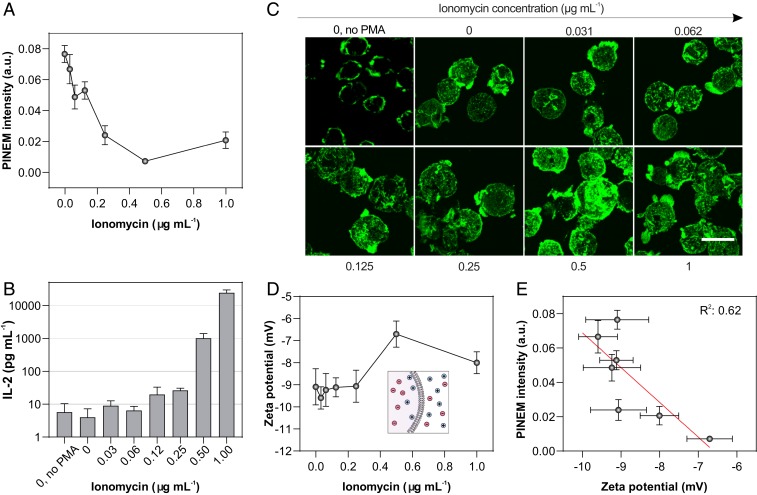
We correlated the strength of the PINEM intensity with the biological and biophysical changes associated with strong, non-antigen-specific T cell activation by titrating the Jurkat cells with ionomycin over the range 0 to 1.0 μg⋅mL−1 in the presence of 50 ng⋅mL−1 PMA. PMA diffuses into the cytoplasm to activate protein kinase C. Ionomycin triggers the release of Ca2+ from intracellular storage compartments. For a given treatment condition, the cells were stimulated for a period of up to 24 h (Methods), after which the thoroughly washed cells were casted on TEM grids for PINEM analysis. The PINEM intensity as a function of ionomycin concentration drops more than 5-fold relative to unstimulated cells (Fig. 2A), reaching a minimum at around 0.5 μg⋅mL−1 ionomycin. T cell activation as assessed by immunoassay quantitation of secreted interleukin (IL)-2 (Fig. 2B) and C-C motif chemokine ligand 4 (CCL4) (SI Appendix, Fig. S3), reveals that functional activation is well established at 0.5 μg⋅mL−1 ionomycin, and increases up to at least 1.0 μg⋅mL−1 ionomycin. T cell cytoskeletal reorganization and an increase in T cell size also evolved up to about 0.5 μg⋅mL−1 ionomycin (Fig. 2C). The surface charge of the T cells also varied with ionomycin treatment (Fig. 2D), as reflecting in an increasing zeta potential. The zeta potential moderately correlates (R2 = 0.62) with the PINEM intensity (Fig. 2E), but also exhibits no significant changes above 0.5 μg⋅mL−1 ionomycin. These results suggest that multiple biophysical properties including surface composition, surface charge, cytoskeleton rearrangement, and cell volume could potentially contribute to the attenuation of PINEM signal, but no specific property is implied as dominant.
Fig. 2.
Non-antigen-specific T cell activation. (A) Plot of PINEM intensity of ionomycin-treated Jurkat T cells with increasing concentration of ionomycin. Error bars represent ± SEM (n = 8 to 12 cells). (B) Bar plot of IL-2 secretion by ionomycin-treated Jurkat cells measured by ELISA. Top error bars represent s.d. (n = 3). (C) Corresponding confocal fluorescence micrographs of filamentous actin. (Scale bar: 20 μm.) (D) Plot of zeta potential of ionomycin-treated Jurkat cells. Error bars represent ± SEM (n = 3). (E) Correlation analysis of PINEM intensity (A) and zeta potential (D) (R2 = 0.62). Unless otherwise specified, PMA concentration is fixed at 50 ng⋅mL−1.
Antigen-specific Surface Binding Leads to Attenuation of PINEM Signal.
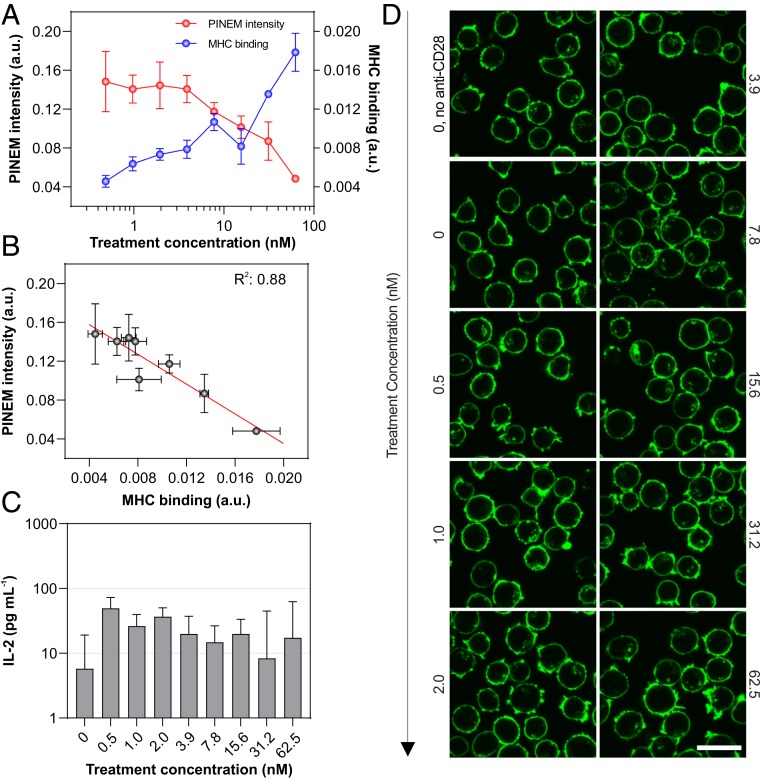
To single out an individual component in the T cell activation process, we used a nonactivating stimulation protocol that modifies the cell surface via antigen-specific interaction without triggering downstream signaling. These CD8− Jurkat cells express MART-1 TCRs (8) that selectively bind to MART-1 pMHC tetramers independent of CD8 coreceptor binding (11), and so do not initiate T cell activation (12). Here, we titrated the concentrations MART-1 pMHC tetramers (0 to 62.5 nM) while monitoring the PINEM intensity, surface binding of the tetramer, and biophysical changes of the cells. The surface binding of the tetramer (Fig. 3A) strongly correlates (R2 = 0.88) with an ∼4-fold PINEM intensity drop (Fig. 3B), with no accompanying signs of T cell activation. As shown in Figs. 3 C and D and SI Appendix, Fig. S4, no changes of cytokine secretion or cytoskeletal restructuring were observed. Finally, as shown in SI Appendix, Fig. S5, essentially no correlation (R2 = 0.03) was observed between membrane zeta potential and PINEM intensity. These results demonstrate that TCR binding to pMHC tetramers is sufficient to attenuate the PINEM intensity.
Fig. 3.
Antigen-specific T cell stimulation. (A) Plot of PINEM intensity (red) and MHC binding (blue) of Jurkat cells treated with increasing concentration of pMHC tetramer (nanomolar concentrations). PINEM intensity is shown as mean ± SEM, collected from 4 to 10 individual cells. MHC binding (surface fluorescence intensity per cell) is shown as mean±SEM (n = 2); each measurement is from 28,750 to 90,000 cells in individual wells. (B) Correlation analysis of A shows that PINEM intensity vs. MHC binding represents a strong correlation (R2 = 0.88). (C) Bar plot of IL-2 secretion by pMHC-treated Jurkat cells measured by ELISA. Top error bars represent SD (n = 3). (D) Corresponding confocal fluorescence micrographs of filamentous actin. (Scale bar: 20 μm.) Unless otherwise specified, anti-CD28 antibody concentration is fixed at 1 μg⋅mL−1.
PINEM Intensity Drop Is Associated with the Loss of Cell Surface Structural Features in the Nano- to Microregime.
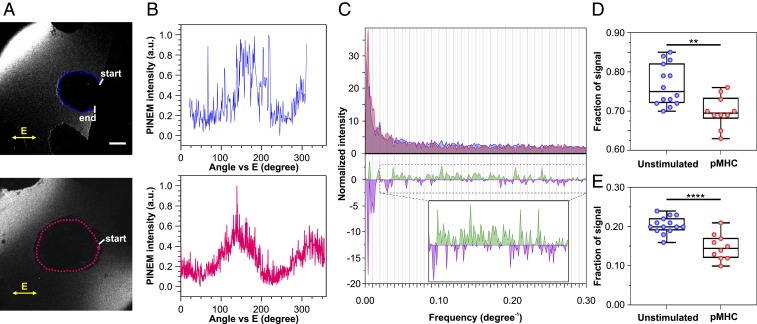
The PINEM signal originates at the T cell/vacuum interface, with nanoscale structures on the T cell surface playing the critical role of energy–momentum matching (2). Thus, we looked to the imaging aspect of PINEM to try to resolve whether nanoscale changes in the T cell/vacuum interface structure might account for the drop in PINEM signal that accompanies TCR–pMHC binding. To this end, we traced the PINEM intensity around the circumference of a cell (Fig. 4 A and B). The Fourier transform of this line analysis revealed a low frequency component arising from the polarization dependence (0 to 0.02 degree−1 frequency range), plus higher frequency fluctuations (0.02 to 0.3 degree−1 frequency range), presumably arising from nanoscale structural variations of the T cell surface (Fig. 4 C, Top). A statistically significant decrease in the fraction of higher frequency components was observed for cells treated with pMHC (Fig. 4D; P = 0.0026). By subtracting the average Fourier spectrum for pMHC-treated cells from that of untreated cells (Fig. 4 C, Bottom), we identified that the most significant changes occurred within the range of 0.10 to 0.18 degree−1 (Fig. 4 C, Inset and Fig. 4E; P < 0.0001), corresponding to a length scale of around 500 to 900 nm for a 10-μm diameter T cell. This analysis implies that the stimulations explored here led to significant nanoscale structural rearrangements of the T cell surface, even when those stimulations do not lead to T cell activation.
Fig. 4.
Analysis of PINEM micrographs of unstimulated or pMHC tetramer-treated Jurkat T cells. Representative analysis of PINEM micrographs (A and B) for an unstimulated (Top) or a pMHC tetramer-treated (Bottom) cell. The images are generated with the fs laser light linearly polarized in a plane indicated by the double-headed arrows. Dotted lines around the cells indicate the regions where a python script traces and records the PINEM intensity from start to end. (Scale bar: 5 μm.) (B) Plots showing pixel-by-pixel tracing of the normalized intensity around the circumference of the cells. (C, Top) Overlaid area plots of the average Fourier spectrum for unstimulated (blue) and pMHC tetramer-treated (red) cells for the full frequency range (0 to 0.3 degree−1). The spectrum from each cell is normalized so that the area under the curve equals 1. (C, Bottom) Spectral subtraction plot of the average Fourier spectrum corresponding to unstimulated cells minus treated cells. The positive spectrum (green) and negative spectrum (purple) arise from the contribution of the unstimulated and treated cells, respectively. (Inset) Zoom of the same plots in the 0.02 to 0.3 degree−1 frequency range. (D) Box plots of the fraction of signal from the 0.02 to 0.3 degree−1 frequency range. Unpaired 2-tailed t test, P = 0.0026. (E) Box plots of the fraction of signal from the 0.10 to 0.18 degree−1 frequency range. Unpaired 2-tailed t test, P < 0.0001. The whiskers on the box plots show the minimum to maximum range. Analysis was performed on 5 (treated) to 7 (unstimulated) unique cells, and additional analysis was performed on the same set of cells, using an orthogonal optical polarization.
Discussion
PINEM signal has been reported from systems ranging from plasmonic nanostructures (2) to cells (4, 5). PINEM scattering theory (2) teaches that polarizable nanoscale structures at the vacuum interface give rise to momentum spread, and so permit energy–momentum matching between the electron and light pulses. We find that the PINEM signal negatively correlates with the binding of pMHC tetramers to TCRs (Fig. 3B). TCRs are abundant surface proteins (13) with up to 105 copies per T cell (14). These TCRs appear to play a dominant role in generating the PINEM signal in unstimulated T cells, and reorganization of those TCRs (and likely other associated subcellular structures) after pMHC binding greatly reduces that role. Our observations thus suggest that there is a significant spatial reorganization of the T cell receptors on the cell surface (15, 16). Such spatial reorganization has been previously reported for stimulations that functionally activate T cells. Our findings indicate that it can happen after just pMHC tetramer binding to TCRs, without accompanying activation. In fact, the surface binding of pMHC to TCR was strongly correlated (R2 = 0.88) with the PINEM intensity drop, which, in turn, is associated with the loss of surface structural features in the few hundred nanometers size range. A second significant finding is that PINEM appears to provide a highly sensitive (label-free) probe of nanoscale cellular surface structure, in a manner that was not anticipated by previous PINEM studies of dielectric spheres (4) and cells (5). In particular, the spectroscopy module is useful for detecting small changes in the PINEM intensity, and the imaging module is useful for extracting nanoscale features of the biological specimen.
The findings reported here represent an initial effort toward a quantitative understanding of biological imaging with PINEM. As a label-free high-resolution method, PINEM imaging can provide insights into cell biology, but the imaging method itself needs to be better understood. One challenge will be to bridge PINEM’s imaging module with its spectroscopy module by establishing a mathematic relationship between these 2. However, PINEM imaging effectively provides a 1-dimensional spatial view of the cell, which may not provide resolution of those structural features that are most responsible for the loss or gain of PINEM signal strength. One option might be to integrate a second label-free imaging method such as scanning probe microscopy to provide an independent, surface-sensitive view of those subcellular structures that influence PINEM signal (17). A second exciting avenue will be to study living cell activities, which can be done by equipping PINEM with liquid cell (18, 19).
Materials and Methods
Materials.
RPMI 1640 Medium (22400-071), FITC-conjugated streptavidin (SA1001), Neutravidin (31000), APC-conjugated FITC monoclonal antibody (17-7691-80), Alexa Fluor 488 Phalloidin (A12379), formaldehyde (28906), and PBS were purchased from ThermoFisher Scientific. Poly-L-lysine solution (P8920), Ionomycin calcium salt (I0634), and PMA (P8139) were purchased from Sigma-Aldrich. CD28 monoclonal antibody (MAB342), human CCL4 ELISA kit (DMB00), and human IL-2 ELISA kit (D2050) were purchased from R & D Systems. FBS (30-2020) was purchased from ATCC. Penicillin-streptomycin mixture (17-602E) was purchased from Lonza (Basel, Switzerland). BSA solution was purchased from Miltenyi Biotec (Bergisch Gladbach, Germany). ImmunoCult Human CD3/CD28/CD2 T Cell Activator was purchased from STEMCELL Technologies Inc.
Cell Culture.
Jurkat T cell line transduced with the F5 MART-1 TCR is a gift from David Baltimore. These MART-1-specific Jurkat cells were cultured in growth media (RPMI 1640 containing 10% FBS, 100 U⋅mL−1 penicillin, and 100 µg⋅mL−1 streptomycin) in a humidified incubator at 37 °C with 5% CO2.
Preparation of PINEM Samples.
For PINEM experiments, silicon nitride or silicon oxide TEM grids (Ted Pella Inc., 200 mesh) were used. To increase the hydrophilicity of the surface of the grid, it was passivated with a plasma for 2 min. Thereafter, a droplet (2 to 3 µl) from Jurkat cells, with given treatments, was loaded onto the grid. To ensure the transfer, cells were counted through optical microscope and compared with the numbers estimated at low magnification in EM. The sample was allowed to dry briefly in the air before loading it in to the microscope.
PINEM Methodology.
The experiments described in this report were performed using the second-generation ultrafast electron microscope (UEM) located in the Physical Biology Center for Ultrafast Science and Technology at Caltech. The experimental details of the UEM are described elsewhere. The microscope is combined with Yb-fiber oscillator/amplifier laser system (1,038 nm, IMPULSE; Clark-MXR, Inc.). PINEM images can be obtained either at 519 or 1,038 nm in the laser system used here. The higher-energy (2.4 eV) excitation wavelength was chosen because it renders more readily filtered PINEM peaks with energy of 200 keV +2.4 eV. The laser system generates 2 separate pulses: the frequency-doubled green (519 nm) one for the specimen excitation and the quadrupled UV (259 nm) one for the generation of a single pulse of electrons to create time-resolved images and spectra. A tunable optical delay line was used to provide a well-defined temporal delay between 2 pulses. Before gathering PINEM images from specimen, it was prerequisite to determine time 0, when the ultrafast electrons and photons are overlapped in space and time, using silver nanowires. The angle of polarization of the excitation pulse was determined by changing the half-wave plate at the entrance of the microscope. PINEM data were only obtained when the energy spread of the zero-loss peak (ZLP) is narrower than the amount of the energy gained. For this purpose, the UV intensity on the cathode was reduced using aperture as well, resulting in ideal narrow ZLP with FWHM of ∼0.9 eV. The repetition rates of the laser used in the described experiments were 100 to 500 kHz. PINEM images were typically acquired with an excitation fluence of 2.5 mJ⋅cm−2, which was focused on the specimen with a diameter (FWHM of ∼50 μm). The selected electrons that had gained energy between 1 and 11 eV were used to produce PINEM images by adjusting the energy slit with a width of 10 eV and by being centered at +6 eV at the energy gain side of the ZLP. The electron energy gain/loss spectra (SI Appendix, Fig. S6) of a Jurkat T cell after 1 (blue) or 2 (red) PINEM measurements, as well as inspection of the UEM and PINEM images, indicated that samples remained undamaged during PINEM imaging.
T Cell Activation.
MART-1-specific Jurkat cells (∼400,000) were cultured for 18 to 24 h, using CD3/CD28/CD2 T cell activator (1:40 in growth media). In titration experiments, cells were cultured for 18 to 24 h in 200 µL growth media containing varying concentrations of ionomycin (0 to 1 µg⋅mL−1) and 0 or 50 ng⋅mL−1 of PMA (phorbol 12-myristate 13-acetate; Sigma).
Secretion Characterization.
ELISA kits were used according to manufacturer’s instructions to measure IL-2 and CCL4 in the supernatant of cells treated with ionomycin-PMA or MART-1 tetramer. In these experiments, 3 replicates were evaluated for each sample by absorbance, measured at 450 nm with plate reader (Synergy H4).
Actin Imaging.
To image actin cytoskeleton of Jurkat cells, around 50,000 cells were adhered in 96-well glass-bottomed plates modified with poly-l-lysine. The cells were fixed with 4% formaldehyde in PBS, washed with PBS, and stained with phalloidin-488 according to manufacturer’s protocol. Fluorescent images were acquired with a Nikon C2 confocal microscope (Ti) using Plan Apo Lambda 20× or 60× oil objective (Nikon Inc., Melville, NY), controlled by NIS elements AR (4.51.00).
Zeta Potential Measurement.
To measure the membrane zeta potential of Jurkat cells, cells were thoroughly washed with and resuspended in PBS. The zeta potential was determined with a Malvern Zetasizer Nano ZS90 (Malvern Instruments).
MART-1 MHC Tetramer Preparation.
MART-1 MHC was first folded from MHC heavy chain inclusion body (HLA-A*02:01), Beta-2 microglobulin inclusion body, and MART-1 peptide (ELAGIGILTVI), according to literature protocol. The MART-1 MHC was then biotinylated using the BirA biotin ligase. MART-1 MHC tetramers were formed by incubating biotinylated MART-1 MHC with neutravidin or FITC-streptavidin at 4 to 1 molar ratio for 1 h at room temperature.
MART-1 MHC Tetramer Binding and Characterization.
MART-1-specific Jurkat cells (∼100,000) were cultured for 14 to 16 h in 100 µL growth media containing varying concentrations of MART-1 MHC tetramers (0 to 500 nM) and 1 µg/mL anti-CD28 antibody. Cell surface binding of the MHC tetramers was characterized using tetramers formed from FITC-streptavidin and subsequent antibody staining of FITC. Briefly, after MHC tetramer binding, cells were fixed with 4% formaldehyde in PBS for 10 min, washed twice in PBS (no permeabilization), and stained with APC-conjugated FITC antibody (10 µg⋅mL−1) in PBS containing 0.5% BSA for 1 h. After washing with PBS, the cells were counted; distributed in wells of a low-volume, 384-well plate; and analyzed using a plate reader (Synergy H4, excitation: 594/20.0, emission: 667/20.0, gain: 150, optics: top, read height: 10 mm). The fluorescent intensity of each well was normalized to cell count (28,750 to 90,000 cells were analyzed per well). PINEM intensity tracing and Fourier analysis was performed on unstimulated cells or cells treated with 500 nM MART-1 MHC and 1 µg/mL of anti-CD28 antibody.
Data and Materials Availability.
All data needed to evaluate the conclusions in the paper are present in the paper and/or the SI Appendix.
Supplementary Material
Acknowledgments
We thank D. Baltimore for providing MART-1-specific Jurkat cells and K. A. Woodrow for providing the Zetasizer. Funding: This work was supported by the National Cancer Institute of the National Institutes of Health (U01CA217655). The authors thank the Parker Institute for Cancer Immunotherapy (PICI) for support. A.H.C.N. is supported by a Banting Postdoctoral Fellowship from the Government of Canada. M.M.L. acknowledges the Welch Foundation for support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914078116/-/DCSupplemental.
References
- 1.Barwick B., Flannigan D. J., Zewail A. H., Photon-induced near-field electron microscopy. Nature 462, 902–906 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Park S. T., Lin M., Zewail A. H., Photon-induced near-field electron microscopy (PINEM): Theoretical and experimental. New J. Phys. 12, 123028 (2010). [Google Scholar]
- 3.Piazza L., et al. , Simultaneous observation of the quantization and the interference pattern of a plasmonic near-field. Nat. Commun. 6, 6407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flannigan D. J., Barwick B., Zewail A. H., Biological imaging with 4D ultrafast electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 9933–9937 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan M., et al. , Photon-induced near-field electron microscopy of Eukaryotic cells. Angew. Chem. Int. Ed. Engl. 56, 11498–11501 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Germain R. N., MHC-dependent antigen processing and peptide presentation: Providing ligands for T lymphocyte activation. Cell 76, 287–299 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Chatila T., Silverman L., Miller R., Geha R., Mechanisms of T cell activation by the calcium ionophore ionomycin. J. Immunol. 143, 1283–1289 (1989). [PubMed] [Google Scholar]
- 8.Johnson L. A., et al. , Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J. Immunol. 177, 6548–6559 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park H. S., Baskin J. S., Kwon O.-H., Zewail A. H., Atomic-scale imaging in real and energy space developed in ultrafast electron microscopy. Nano Lett. 7, 2545–2551 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Barwick B., Park H. S., Kwon O.-H., Baskin J. S., Zewail A. H., 4D imaging of transient structures and morphologies in ultrafast electron microscopy. Science 322, 1227–1231 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Choi E. M.-L., et al. , High avidity antigen-specific CTL identified by CD8-independent tetramer staining. J. Immunol. 171, 5116–5123 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Wooldridge L., et al. , Tricks with tetramers: How to get the most from multimeric peptide-MHC. Immunology 126, 147–164 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans E. J., et al. , The T cell surface–How well do we know it? Immunity 19, 213–223 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Sykulev Y., et al. , Kinetics and affinity of reactions between an antigen-specific T cell receptor and peptide-MHC complexes. Immunity 1, 15–22 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Hu Y. S., Cang H., Lillemeier B. F., Superresolution imaging reveals nanometer- and micrometer-scale spatial distributions of T-cell receptors in lymph nodes. Proc. Natl. Acad. Sci. U.S.A. 113, 7201–7206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pageon S. V., et al. , Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. U.S.A. 113, E5454–E5463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zewail A. H., Four-dimensional electron microscopy. Science 328, 187–193 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Fu X., Chen B., Tang J., Hassan M. T., Zewail A. H., Imaging rotational dynamics of nanoparticles in liquid by 4D electron microscopy. Science 355, 494–498 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Fu X., Chen B., Tang J., Zewail A. H., Photoinduced nanobubble-driven superfast diffusion of nanoparticles imaged by 4D electron microscopy. Sci. Adv. 3, e1701160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.