Significance
APOBEC3B (A3B) is overexpressed in multiple tumor types and a major source of mutation by converting DNA cytosines to uracils. A3B-positive tumors are often more heterogenous and result in poor clinical outcomes. We show that a proportion of A3B-catalyzed uracil lesions is repaired by uracil DNA glycosylase 2 (UNG)-driven base excision repair and that rendering cells UNG-defective leads to cytotoxicity. This cell death mechanism requires uracil processing by mismatch repair proteins and functional p53. Our results indicate that UNG inhibition could be a strategy to kill a subset of A3B-positive tumors.
Keywords: APOBEC3B mutagenesis, DNA deamination, mismatch repair, synthetic lethality, uracil base excision repair
Abstract
Human cells express up to 9 active DNA cytosine deaminases with functions in adaptive and innate immunity. Many cancers manifest an APOBEC mutation signature and APOBEC3B (A3B) is likely the main enzyme responsible. Although significant numbers of APOBEC signature mutations accumulate in tumor genomes, the majority of APOBEC-catalyzed uracil lesions are probably counteracted in an error-free manner by the uracil base excision repair pathway. Here, we show that A3B-expressing cells can be selectively killed by inhibiting uracil DNA glycosylase 2 (UNG) and that this synthetic lethal phenotype requires functional mismatch repair (MMR) proteins and p53. UNG knockout human 293 and MCF10A cells elicit an A3B-dependent death. This synthetic lethal phenotype is dependent on A3B catalytic activity and reversible by UNG complementation. A3B expression in UNG-null cells causes a buildup of genomic uracil, and the ensuing lethality requires processing of uracil lesions (likely U/G mispairs) by MSH2 and MLH1 (likely noncanonical MMR). Cancer cells expressing high levels of endogenous A3B and functional p53 can also be killed by expressing an UNG inhibitor. Taken together, UNG-initiated base excision repair is a major mechanism counteracting genomic mutagenesis by A3B, and blocking UNG is a potential strategy for inducing the selective death of tumors.
APOBEC signature mutations are common in several cancer types including breast, head/neck, cervical, bladder, and lung (1–4). This signature is comprised of C-to-T transitions and C-to-G transversions within TC(A/T) trinucleotide motifs resulting in TT(A/T) and TG(A/T) mutations (1, 2, 4). Multiple lines of evidence indicate that APOBEC3B (A3B), the only constitutively nuclear single-stranded (ss)DNA deaminase (5), is the primary source of these mutations (1–4). First, A3B is overexpressed in primary tumors compared to normal tissues (e.g., >50% of breast tumors and >90% of head/neck tumors) (2, 6). Second, increased A3B levels correspond positively with overall cytosine mutational loads (6). Third, A3B expression associates with worse clinical outcomes including hormone therapy resistance in breast cancer (7–10). APOBEC3A and APOBEC3H have also been implicated in cancer mutation, although to lesser extents (11–13).
APOBEC-catalyzed C-to-U lesions must escape multiple DNA repair processes before becoming immortalized as mutations in tumor genomes. Most DNA uracil lesions are counteracted by base excision repair (BER), which excises the uracil base and choreographs its faithful replacement by another cytosine (reviewed by refs. 14–16). Uracil BER is primarily initiated by uracil DNA glycosylase 2 (hereafter UNG), which catalyzes uracil excision, resulting in an abasic site that is further processed by endonucleolytic cleavage, local DNA synthesis, and ligation. Intermediates in UNG-initiated BER (abasic sites and ssDNA nicks) need to be processed efficiently to avoid becoming more detrimental lesions such as double-stranded (ds)DNA breaks, which can precipitate translocations, amplifications, deletions, and/or duplications. As a backup to BER, mismatch repair (MMR) can also mediate the repair of uracil lesions, specifically U/G mispairs (reviewed by refs. 17 and 18). Moreover, unrepaired uracils template the insertion of adenines during DNA replication, which leads to C-to-T transition mutations (19–21). The combination of APOBEC-catalyzed DNA cytosine deamination, incomplete processing by BER, and erroneous insertion by REV1 (resulting in C-to-G transversions) accounts for the APOBEC signature in cancer (22). It is therefore possible that inhibiting the repair of A3B-catalyzed uracil lesions will result in toxic accumulation of genomic uracils and genome instability, which may be an effective way to selectively kill A3B-expressing tumor cells. This strategy parallels the classical example of synthetic lethality in which PARP inhibitors suppress DNA repair in homologous recombination-deficient tumors (e.g., BRCA1 mutant cells; reviewed by ref. 23).
Here, we test the hypothesis that UNG inhibition will be toxic to A3B-overexpressing cells. In support of this idea, 2 different UNG knockout (KO) cell lines showed hypersensitivity to A3B. This synthetic lethal phenotype required functional MMR proteins as both UNG KO/MSH2 KO and UNG KO/MLH1 knockdown cells became resistant to A3B. Several lines of evidence indicated that U/G mispairs and ssDNA tracts are molecular intermediates in this process. Finally, a role for p53 was shown by restoring p53 function in a breast cancer cell line and causing a synthetic lethal interaction between A3B and UNG inhibition. These results suggest that UNG inhibition may be a strategy to selectively kill A3B-positive tumors.
Results
UNG Knockout Is Synthetic Lethal with Enforced A3B Overexpression.
To test the hypothesis that inhibition of uracil BER would result in a synthetic lethal combination with A3B overexpression, CRISPR was used to disrupt the UNG gene in a system that allows for doxycycline (Dox)-inducible expression of an A3B-eGFP construct (293-TREx-A3Bi-eGFP [293-A3B]). Maximal A3B-eGFP induction elicits a strong DNA damage response (DDR) and cytotoxicity in this system (6, 24). However, here we wanted to use the lowest Dox concentration for A3B induction and minimal toxicity, which in titration experiments was determined to be 1 ng/mL with A3B-eGFP fluorescence still approaching 100% (SI Appendix, Fig. S1A). UNG KO clones were validated by uracil excision activity assays and immunoblotting (SI Appendix, Fig. S1B). Knockout specificity was demonstrated by restoring both uracil excision activity and immunoreactivity upon complementation with a WT UNG cDNA (SI Appendix, Fig. S1B).
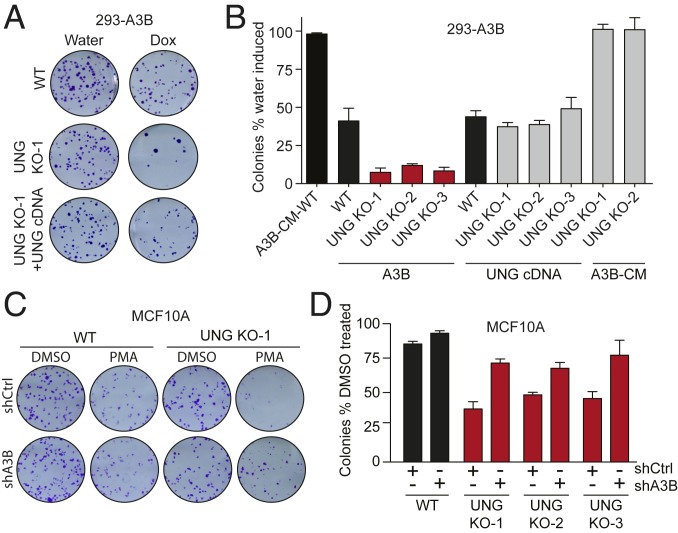
Colony formation assays were done to directly compare the viability (colony formation efficiency) of parental (WT) cells and UNG-null derivatives in the absence or presence of 1 ng/mL Dox for A3B-eGFP induction. Under control conditions, WT, UNG KO clones, and UNG-complemented derivatives showed similar colony formation efficiencies and doubling times (SI Appendix, Fig. S1 C and D). In contrast, A3B-eGFP induction caused a 50% decline in parental cell viability and a synergistic >90% reduction in UNG-null cell viability (Fig. 1 A and B). This synthetic lethal phenotype could be reversed by complementation with a retroviral construct expressing WT UNG. Furthermore, synthetic lethality was dependent on A3B catalytic activity, because induced expression of an A3B-eGFP catalytic mutant (CM) caused no change in survival with or without UNG (Fig. 1B).
Fig. 1.
UNG KO exacerbates A3B-induced cytotoxicity. (A) Scans of representative 293-A3B colony formation assays. (B) Quantitative summary of 293-A3B colony formation assay results for cell lines with indicated genotypes. Each histogram bar represents the colony formation efficiency following A3B-eGFP expression (1 ng/mL Dox) relative to controls (n = 3 biological replicates ± SEM). Key results such as A3B WT vs. A3B UNG KO and A3B UNG KO vs. A3B UNG KO+UNG cDNA are significant by paired 2-sided t test (P < 0.05). (C) Scans of representative MCF10A colony formation assays. (D) Colony formation assay quantification of PMA-treated (25 ng/mL, 6 h) MCF10A cells relative to control (DMSO). Bars represent the average of 3 biological replicates ± SEM. Key results (WT vs. UNG KO and in shCTRL vs. shA3B) are significant by paired 2-sided t test (P < 0.01).
UNG Knockout Is Synthetic Lethal with Endogenous A3B Up-Regulation.
To further investigate the synthetic lethal phenotype between A3B and UNG ablation, we used the breast epithelial cell line MCF10A where endogenous A3B can be induced by phorbol 12-myristate 13-acetate (PMA) treatment and signal transduction through the PKC and noncanonical NF-κB pathways (25). PMA-induced A3B mRNA levels are similar to those reported in many cancer cell lines and tumors (6, 25). As above, MCF10A cells were engineered to lack UNG (SI Appendix, Fig. S1E). MCF10A parental and UNG KO clones were then transduced with lentiviral constructs expressing either a nontargeting shRNA (shCTRL) or a previously validated A3B-specific shRNA (shA3B) to be able to determine whether observed phenotypes are due to endogenous A3B (6, 25). A3B knockdown was confirmed by treating with PMA and quantifying mRNA levels by RT-qPCR (SI Appendix, Fig. S1F). PMA treatment had a modest effect on parental (WT) MCF10A viability, but this was independent of A3B as no significant difference was observed between shCTRL and shA3B conditions (Fig. 1 C and D). In contrast, 3 independent UNG KO clones showed a 40–50% reduction in viability following A3B induction, with the majority of this synthetic lethal phenotype suppressible by endogenous A3B knockdown (Fig. 1 C and D).
A3B-Induced DNA Damage Responses Are UNG Dependent.
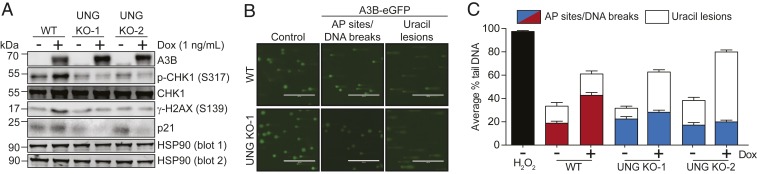
We next assessed DDR markers starting with the 293-A3B system. In WT cells, A3B induction caused increased CHK1 phosphorylation, γ-H2AX accumulation, and p21 expression (Fig. 2A), consistent with prior reports (6, 24, 26). Interestingly, induction of these DDR markers by A3B-eGFP was fully dependent upon UNG (Fig. 2A). Levels of p21 and, to a lesser extent, phosphorylated CHK1 even appeared to decline below uninduced levels in UNG-null clones overexpressing A3B-eGFP (potentially related to leaky A3B-eGFP expression in this system; refs. 6 and 24). Nevertheless, these results clearly showed that UNG is required for A3B-induced activation of the DDR.
Fig. 2.
A3B expression induces UNG-dependent DNA breaks. (A) Immunoblots of DDR proteins in control or A3B-eGFP induced 293-A3B cells (Dox treated, 48 h). HSP90 blot 1 is the loading control for anti-A3B, p-CHK1 (S317), and γ-H2AX. HSP90 blot 2 is the loading control for anti-CHK1 and p21. (B) Images of representative alkaline COMETs. 293-A3B cells with the indicated genotypes were water or Dox (1 ng/mL) treated 48 h prior to alkaline or modified alkaline COMET assays. (Scale bars, 400 µm.) The former assay provides a sum measure of AP sites and ss/dsDNA breaks and the latter with hUNG pretreatment additionally measures genomic uracil lesions (see text, SI Appendix, Fig. S2, and Materials and Methods for additional information). (C) Quantification of 293-A3B COMET experiments. Hydrogen peroxide was a positive control. Each histogram bar represents the average % tail DNA ± SEM (3 biological replicates, >100 comets each). Red/blue shaded bars show data for standard alkaline COMET assays, and open bars data for modified alkaline COMET assays. Key results for the standard COMET (WT vs. WT with induced A3B-eGFP, WT induced vs. UNG KO induced) and for the modified COMET (uninduced vs. induced for all genotypes) are significant by 1-way ANOVA (P < 0.0001).
To further investigate the mechanism by which A3B mediates death of UNG-null cells, alkaline COMET assays were done to quantify AP sites and ss/dsDNA breaks (i.e., intermediates in uracil excision repair). 293-A3B cells with and without UNG were treated 48 h with Dox to induce A3B-eGFP and then harvested for COMET assays. A3B-eGFP induction in WT cells caused a 2-fold increase in the amount of genomic DNA in COMET tails, which was completely dependent upon uracil excision activity because UNG KO clones did not show similar increases (representative images in Fig. 2B and quantification by red/blue bars in Fig. 2C; larger number of images from this and another independent experiment in SI Appendix, Fig. S2). Moreover, neutral COMET assays, which only detect dsDNA breaks, showed that A3B can also cause this type of lesion through an UNG-dependent mechanism (SI Appendix, Fig. S3 A and B). As an independent measure of DNA damage, A3B induction also increased γ-H2AX foci through UNG (SI Appendix, Fig. S3 C and D). Taken together, these results showed that UNG is required for both A3B-induced activation of the DDR and accumulation of AP sites and ssDNA breaks, and also for an increase in dsDNA breaks.
A3B-Induced Uracil Lesions (Not DNA Breaks nor DDR) Are Molecular Intermediates in the Synthetic Lethal Phenotype.
Although DNA breaks are established cytotoxic lesions, the results above indicated counterintuitively that these may not be the primary reason for A3B-induced synthetic lethality (i.e., UNG ablation promoted A3B-dependent death but suppressed DNA break accumulation). Moreover, standard alkaline COMET assays were negative with the MCF10A system, which expresses lower A3B levels (SI Appendix, Fig. S4). Therefore, to further investigate the lesion responsible for the synthetic lethal phenotype, an additional step was added to the COMET protocol to quantify genomic uracil (27). Permeabilized cell nucleoids were treated with recombinant human UNG (hUNG) to excise DNA uracil. The resulting abasic sites were then converted to ssDNA breaks under the alkaline conditions of the assay. Thus, through quantification of these additional tail DNA signals, over and above those described above, this modified COMET protocol provided a readout of genomic uracil levels. As expected, hUNG treatment caused an increase in COMET tail DNA in every condition with larger increases evident in UNG-null cells (representative images in Fig. 2B; quantification with open bars in Fig. 2C; larger number of images from this and another independent experiment in SI Appendix, Fig. S2). Complementary results were also obtained by applying this modified assay to MCF10A, where UNG-null cells showed 2-fold more uracil COMET signal and nearly all of this increase could be suppressed by depleting endogenous A3B (SI Appendix, Fig. S4). Altogether, these COMET experiments showed that A3B inflicts uracil lesions in genomic DNA that are processed in an UNG-dependent manner into detectable DNA breaks (AP sites/ssDNA breaks and dsDNA breaks). Moreover, when considered with the colony formation data in Fig. 1, these results indicated that A3B-induced genomic uracils (not DNA breaks) are responsible for the observed cell death by either leading directly to toxicity and/or being processed by another pathway into lesions with greater cytotoxicity.
Mismatch Recognition Proteins MSH2 and MLH1 Are Required for A3B-Mediated Toxicity in UNG-Null Cell Lines.
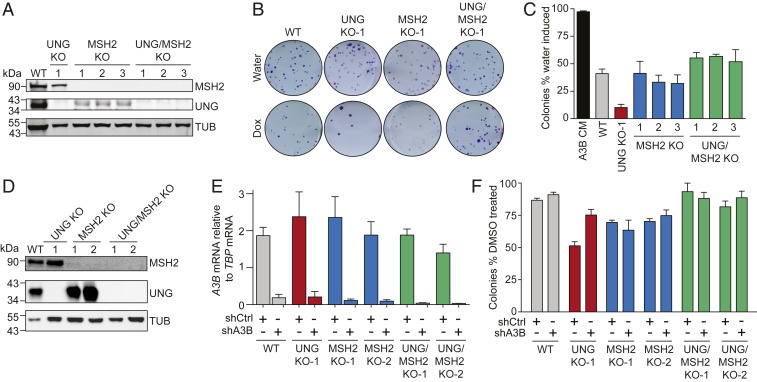
Considering the likely stochastic nature of A3B-catalyzed DNA deamination, any resulting mutations would have to accumulate in essential genes to develop into a lethal phenotype in UNG-null cells. A more likely mechanism based on biochemical insights and precedents for activation induced deaminase (AID)-mediated antibody gene diversification is uracil processing by MMR (17, 18, 28). MMR is capable of recognizing U/G mispairs and creating long patches of ssDNA that could become substrates for additional A3B deamination events and/or templates for error-prone DNA synthesis. To determine whether MMR may be involved in A3B-induced cytotoxicity, CRISPR was used to knock out MSH2 in 293-A3B cells. MSH2 KO and UNG/MSH2 double-KO clones were generated and confirmed by immunoblotting (Fig. 3A). The MSH2 KO alone had no additional impact on colony formation efficiency with or without A3B induction in 293-A3B cells. However, the KO of MSH2 in 3 independent UNG-null clones resulted in complete suppression of the synthetic lethal phenotype (Fig. 3 B and C). Suppression was specific to MSH2 disruption because complementation with WT MSH2 cDNA restored lethality (SI Appendix, Fig. S5 A–C). A similar MSH2 dependence was evident in the MCF10A system (Fig. 3 D–F). Additionally, shRNA-mediated knockdown of MLH1 in 293-A3B UNG KO cells was also able to revert the synthetic lethal phenotype (SI Appendix, Fig. S5 D–F). These results strongly indicated that a MSH2- and MLH1-dependent process provides an alternative route for genomic uracil processing and, importantly, that this process mediates A3B-induced toxicity in UNG-null cells.
Fig. 3.
MMR is required for A3B-induced lethality of UNG KO cells. (A) Immunoblots of 293-A3B UNG, MSH2, and UNG/MSH2 KO clones. (B) Scans of representative 293-A3B colony formation assays. (C) Quantitative summary of 293-A3B colony formation assay results for cells with indicated genotypes. Each histogram bar represents the colony formation efficiency following A3B-eGFP induction relative to water-treated controls (n = 3 biological replicates ± SEM). Key results (WT vs. UNG KO and UNG KO vs. UNG/MSH2 KO) are significant by paired 2-sided t test (P < 0.05). (D) Immunoblots of MCF10A UNG, MSH2, and UNG/MSH2 KO clones. (E) RT-qPCR data confirming A3B knockdown in the indicated PMA-treated cell lines. (F) Quantitative summary of MCF10A colony formation assay results for cells with indicated genotypes. Each histogram bar represents the colony formation efficiency following PMA treatment relative to DMSO-treated controls (n = 3 biological replicates ± SEM). Key results (WT shCtrl vs. UNG KO shCtrl and UNG KO shCtrl vs. UNG/MSH2 KO shCtrl) are significant by paired 2-sided t test (P < 0.01).
A3B Induction in UNG Knockout Cells Leads to MSH2-Dependent ssDNA Tracts and PCNA Monoubiquitylation.
The MMR excision process results in ssDNA tracts up to several kilobase pairs long that can serve as templates for synthesis by replicative and error-prone DNA polymerases (17, 29, 30). To probe for ssDNA accumulation, a series of BrdU immunofluorescence experiments was performed under nondenaturing conditions (31). These experiments showed that A3B induction causes a modest but significant increase in ssDNA in WT cells (SI Appendix, Fig. S6 A and B). In comparison, A3B induction in UNG KO cells caused 3-fold higher levels of ssDNA and this increase was fully dependent upon MSH2 (SI Appendix, Fig. S6 A and B). A3B-induced UNG KO cells also showed elevated RPA staining consistent with an accumulation of more of ssDNA (SI Appendix, Fig. S6 A and B).
To estimate the capacity for translesion DNA synthesis, WT, UNG KO, MSH2 KO, and UNG/MSH2 KO 293-A3B cells were blotted for PCNA. PCNA monoubiquitylation (PCNA-Ub) is required for translesion DNA synthesis (30). These immunoblots indicated that there is no obvious difference in PCNA-Ub levels when A3B is induced in WT cells (SI Appendix, Fig. S6C). However, A3B induction in UNG KO 293-A3B cells caused an increase in PCNA-Ub, which could also be suppressed by ablating MSH2 (SI Appendix, Fig. S6C). Altogether, these results indicated that in the absence of UNG, A3B-catalyzed uracil lesions (likely U/G mispairs) trigger a MSH2-dependent generation of ssDNA tracts and PCNA-Ub indicative of translesion DNA synthesis.
Synthetic Lethality of A3B-High, UNG-Inhibited Cancer Cells Requires p53.
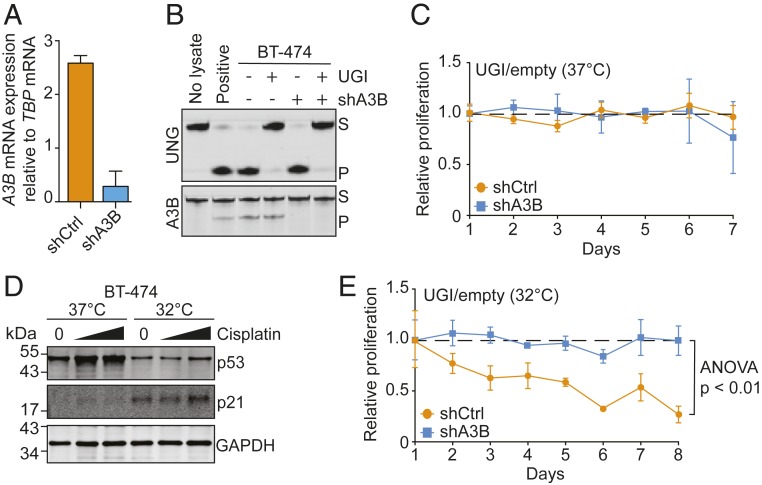
We next investigated the role of UNG in preserving the viability of cancer cells with elevated levels of endogenous A3B. We chose the breast cancer cell line, BT-474, because it expresses tumor-like levels of A3B (Fig. 4A) and has a strong APOBEC mutation signature (32). We reasoned that a knockout approach might not be successful here because targeted cells might die before analysis. We therefore first used lentiviral transduction to create A3B-high (shCTRL) and A3B-low (shA3B) pools, as above, and then used another round of transduction to create UNG-proficient (vector control) and UNG-inhibited (UGI expressing) subpools (i.e., 4 conditions total). The bacteriophage-derived uracil DNA glycosylase inhibitor (UGI) protein is a universal UNG antagonist with proven potency in vertebrate cells (33). RT-qPCR confirmed A3B knockdown, and uracil excision assays confirmed UNG inhibition (89% and 94% efficient, respectively; Fig. 4 A and B). The viability of each pool was compared daily by MTS assays and, curiously, all 4 conditions maintained similarly high viabilities over 7 d (i.e., relative proliferation rates of 1; Fig. 4C).
Fig. 4.
Functional restoration of p53 sensitizes BT-474 to UNG inhibition. (A) A3B knockdown confirmation by RT-qPCR. (B) UNG activity assays (Upper) and A3B activity assays (Lower) using whole-cell extracts from BT-474 cells with the indicated genotypes (S, substrate oligo; P, product oligo). (C) Relative 37 °C proliferation rates (MTS assay) for BT-474 cells with the indicated genotypes. Relative proliferation is represented as MTS absorbance of cells expressing UGI over empty vector relative to day 1 (each data point is the mean of 3 biological replications ± SEM). (D) Immunoblots of BT-474 cells grown at 37 °C or 32 °C following 24-h treatment with vehicle (DMF) or cisplatin (10 and 50 µM). (E) Relative 32 °C proliferation rates (MTS assay) for BT-474 cells with the indicated genotypes. Relative proliferation is represented as MTS absorbance of cells expressing UGI over empty vector relative to day 1 (each data point is the mean of 3 biological replications ± SEM; significance determined by 2-way ANOVA).
However, a key difference between the systems above (293T and MCF10A) and the cancer cell line used here is functional versus mutant p53, respectively. Together with our prior demonstration that TP53-null 293-A3B cells become resistant to A3B-induced cell death (24), we hypothesized that the synthetic lethality observed between A3B overexpression and UNG ablation requires functional p53. Fortunately, BT-474 cells express a temperature-sensitive p53 mutant (E285K) that misfolds at 37 °C but folds properly and maintains WT function at 32 °C (34). Indeed, as shown previously, this p53 mutant is misfolded, resilient to MDM2-mediated degradation, and accordingly expressed at higher levels at 37 °C, whereas the same mutant is folded properly, degraded by MDM2, and expressed at lower levels at 32 °C (Fig. 4D). Moreover, the p53-responsive gene CDKN1a (p21) was expressed at higher levels at 32 °C and could be further up-regulated by cisplatin (Fig. 4D). The MTS experiment described above was repeated at 32 °C and, as anticipated, the restoration of p53 activity resulted in a concomitant emergence of the synthetic lethal phenotype between A3B overexpression and UNG inhibition (Fig. 4E).
Discussion
These studies implicate uracil BER as a target to selectively kill tumor cells overexpressing A3B. We show that disruption of uracil BER by deleting UNG sensitizes cells to A3B expression. Synthetic lethality was quantified by colony formation assays, which are a gold standard for viability measurements. Furthermore, A3B expression in UNG-null cells did not activate a DDR or induce DNA breaks but, instead, it induced the accumulation of uracil lesions in chromosomal DNA. Epistasis analysis revealed that the synthetic lethal phenotype is dependent on MMR proteins, MSH2 and MLH1, as well as associated with ssDNA accumulation. Finally, UNG inhibition by UGI caused A3B-dependent proliferation defects in the breast cancer cell line BT-474 but only when p53 function was restored.
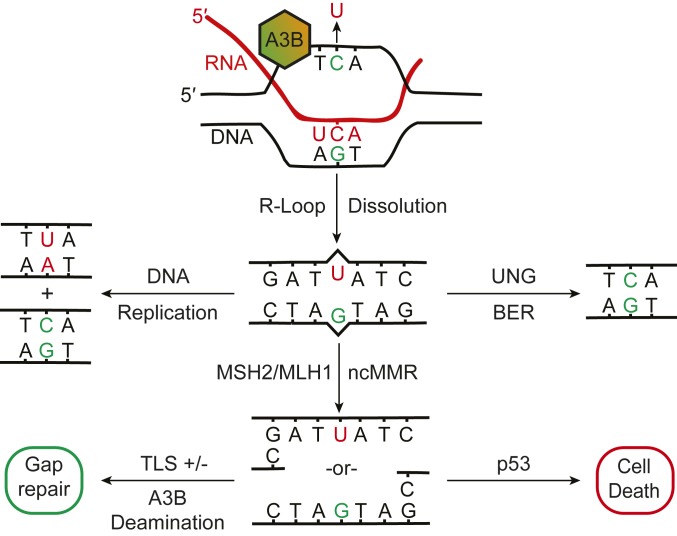
Our results support a model in which A3B-induced uracil lesions are substrates for both uracil BER and MMR proteins (Fig. 5; see next paragraph for precedents). A key inference from MSH2 and MLH1 involvement is that A3B-catalyzed deamination is likely to lead to U/G mispairs. These lesions probably emerge from transcription-associated events, such as deamination of cytosines in single-stranded R loops followed by R-loop dissolution to leave a mismatch-containing DNA duplex. U/G mispairs are less likely to arise during DNA replication, where U/A pairs are more common (due to U misincorporation) and an additional proportion of APOBEC mutagenesis occurs by deamination of ssDNA replication intermediates and mismatch-free resolution into C-to-T and C-to-G mutations (19–21). Our results indicate that uracil lesions in U/G mispairs are repaired in most instances in a nontoxic manner by the highly efficient uracil BER pathway (reviewed by refs. 14–16). Alternatively, U/G lesions can be processed in a MSH2- and MLH-1– dependent manner, and greatly exacerbated by UNG deficiency. Taken together with the observed increases in ssDNA accumulation in A3B-expressing UNG-null cells, these genetic dependencies strongly implicate noncanonical MMR (17, 18, 29, 30). This process is equally likely to excise either DNA strand, leading to exposure of ssDNA tracts and potentially also to additional A3B-catalyzed deamination events prior to gap repair to restore DNA duplex integrity. We further postulate that intermediates in the MSH2-dependent processing of U/G lesions, such as ssDNA tracts or downstream lesions, may then be capable of triggering cell death in a p53-dependent manner.
Fig. 5.
Working model for processing of A3B-induced uracil lesions in genomic DNA. The Upper Center schematic depicts A3B-mediated deamination of a ssDNA cytosine in an R loop. U/G mispairs may arise following R-loop dissolution. A majority of these uracil lesions would be repaired in an error-free manner by UNG-mediated BER (Upper Right schematic). A minority may serve as templates for DNA replication and, following UNG-mediated BER, become immortalized as C-to-T mutations (Upper Left schematic). However, as shown here, UNG KO or inhibition leads to increased processing of U/G lesions by an MSH2/MLH1-dependent process (potentially noncanonical [nc]MMR). These events are likely to orchestrate the generation of exposed ssDNA tracts without strand discrimination (Lower Center schematic). These ssDNA tracts may be subject to translesion DNA synthesis (TLS) and additional A3B mutagenesis (Lower Left) or, with functional p53, lead to the accumulation of toxic intermediates that result in cell death.
Existing literature also provides support for many aspects of this model. First, UNG/BER and MSH2/MLH1/MMR are established DNA repair mechanisms with well-defined biochemical activities consistent with this model (reviewed by refs. 14–18). Second, the initial step of the model resembles somatic hypermutation and class switching of Ig genes in B cells, where transcription is required, R loops occur, and AID-catalyzed uracils become substrates for both uracil BER and MMR (35–39). Third, for mismatches not arising during DNA replication, MMR has been documented to lack directionality and create extensive ssDNA excision tracts by incision- or nick-primed exonucleolytic degradation of either DNA strand (30, 40). Reminiscent to our work here, MMR was found to be responsible for DDR activation and mediating cytotoxicity in response to 5-fluorouracil treatment of Caenorhabditis elegans (41). Fourth, p53 inactivation renders cells more resistant to A3B overexpression (24). Fifth, multiple types of DNA damage are recognized by MMR including uracil lesions and, in some instances, are similarly able to trigger p53-dependent cell death (42–44). Last, RNA-sequencing analysis of patient breast tumors has revealed a positive correlation between A3B expression and TP53 inactivation (6), further supporting the idea that cells cannot tolerate A3B-catalyzed DNA damage without first suppressing a p53-dependent death mechanism.
Our results indicate that UNG is likely to be critical for the survival of A3B-positive tumors. This provides a molecular explanation for why UNG is rarely (if ever) defective in human cancer (45) and why a positive correlation exists between A3B and UNG expression levels in tumors (SI Appendix, Fig. S7). Thus, we suggest that UNG inhibition may be an effective therapeutic strategy for A3B-overexpressing tumors with WT MMR and p53 functions. UNG inhibition may also be effective in a subset of p53 mutant tumors, where p53 function can be restored pharmacologically (reviewed by ref. 46). Overall, our studies reveal key intermediates in the APOBEC mutation process and indicate that efforts to target this process in cancer should not only consider the enzyme that initiates genomic DNA deamination (A3B) but also downstream DNA repair and cell death pathways.
Materials and Methods
UNG and MSH2 KO Generation.
Guide RNAs (gRNA) were designed for UNG exon 2 using http://crispor.tefor.net. Annealed gRNA sequences were cloned into the BsmBI site of pLentiCRISPR-1000 (47). Transducing viral stocks were used to create puromycin-resistant UNG KO pools, and the CRISPR/Cas9-puroR cassette was removed using Cre recombinase (47). KO pools were then cloned by limiting dilution and validated. UNG KO clones 1 and 2 were made using gRNA 1, and UNG KO clone 3 was made using gRNA 2. MSH2 exon 1 KO clones were generated similarly. UNG KO pools were also transduced with MSH2 KO virus to create UNG/MSH2 double KO pools that were subcloned prior to analysis. All oligos including gRNA sequences are listed in SI Appendix, Table S1.
Colony Formation Assays.
293-A3B cells were seeded into 6-well plates with media containing either water (vehicle) or Dox (1 ng/mL) (500 and 750 cells per well, respectively). After 24 h, this media was replaced with fresh media. MCF10A cells were first plated into a 6-well plate (300,000 cells per well) and treated 6 h with dimethyl sulfoxide (vehicle) or PMA (25 ng/mL). After treatment, cells were harvested and reseeded in a 6-well plate (400 cells per well) containing 25% conditioned mammary epithelial growth medium media. Once vehicle-treated wells reached >50 cells per colony (7–9 d), plates were washed with phosphate-buffered saline, stained with 50% methanol/0.5% crystal violet, and counted. Colony formation efficiencies (number of visible colonies divided by number of cells plated) were used to measure survival between wells with different seeding densities. Statistical analyses were done with GraphPad Prism 6.
Supplementary Material
Acknowledgments
We thank Salvatore Caradonna for providing rabbit anti-UNG polyclonal sera, Rick Fishel and Alan Ashworth for thoughtful comments, and James Stivers for providing a human UNG cDNA. These studies were supported by National Cancer Institute Grant P01-CA234228 (to R.S.H.), Norwegian Cancer Society Grant 182793 (to H.N.), and a Norwegian Centennial Chair Program grant (to R.S.H. and H.N.). R.S.H. is the Margaret Harvey Schering Land Grant Chair for Cancer Research, a Distinguished University McKnight Professor, and an Investigator of Howard Hughes Medical Institute.
Footnotes
Competing interest statement: R.S.H. is a co-founder, shareholder, and consultant for ApoGen Biotechnologies Inc. The other authors have no conflicts of interest to declare.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904024116/-/DCSupplemental.
References
- 1.Alexandrov L. B., et al. ; Australian Pancreatic Cancer Genome Initiative ; ICGC Breast Cancer Consortium ; ICGC MMML-Seq Consortium ; ICGC PedBrain , Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burns M. B., Temiz N. A., Harris R. S., Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45, 977–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bruin E. C., et al. , Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts S. A., et al. , An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lackey L., Law E. K., Brown W. L., Harris R. S., Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle 12, 762–772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns M. B., et al. , APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Law E. K., et al. , The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2, e1601737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sieuwerts A. M., et al. , Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm. Cancer 5, 405–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan S., et al. , Increased APOBEC3B predicts worse outcomes in lung cancer: A comprehensive retrospective study. J. Cancer 7, 618–625 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L., et al. , High APOBEC3B expression is a predictor of recurrence in patients with low-risk clear cell renal cell carcinoma. Urol. Oncol. 33, 340.e1–340.e8 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Chan K., et al. , An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 47, 1067–1072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nik-Zainal S., et al. , Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 46, 487–491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starrett G. J., et al. , The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat. Commun. 7, 12918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krokan H. E., et al. , Error-free versus mutagenic processing of genomic uracil–relevance to cancer. DNA Repair (Amst.) 19, 38–47 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Lindahl T., Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res. 462, 129–135 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Nilsen H., Krokan H. E., Base excision repair in a network of defence and tolerance. Carcinogenesis 22, 987–998 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Crouse G. F., Non-canonical actions of mismatch repair. DNA Repair (Amst.) 38, 102–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Modrich P., Strand-specific mismatch repair in mammalian cells. J. Biol. Chem. 272, 24727–24730 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Bhagwat A. S., et al. , Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 113, 2176–2181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoopes J. I., et al. , APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 14, 1273–1282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seplyarskiy V. B., et al. , APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res. 26, 174–182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan K., Resnick M. A., Gordenin D. A., The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst.) 12, 878–889 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lord C. J., Ashworth A., PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikkilä J., et al. , Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br. J. Cancer 117, 113–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonard B., et al. , The PKC/NF-κB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res. 75, 4538–4547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor B. J., et al. , DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2, e00534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsen H., et al. , Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell 5, 1059–1065 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Rada C., Di Noia J. M., Neuberger M. S., Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16, 163–171 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Jeon Y., et al. , Dynamic control of strand excision during human DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 113, 3281–3286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peña-Diaz J., et al. , Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell 47, 669–680 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Toledo L. I., et al. , ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Jarvis M. C., Ebrahimi D., Temiz N. A., Harris R. S., Mutation signatures including APOBEC in cancer cell lines. JNCI Cancer Spectr. 2, pky002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Noia J., Neuberger M. S., Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419, 43–48 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Müller P., Ceskova P., Vojtesek B., Hsp90 is essential for restoring cellular functions of temperature-sensitive p53 mutant protein but not for stabilization and activation of wild-type p53: Implications for cancer therapy. J. Biol. Chem. 280, 6682–6691 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Basu U., et al. , The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144, 353–363 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rada C., González-Fernández A., Jarvis J. M., Milstein C., The 5′ boundary of somatic hypermutation in a V kappa gene is in the leader intron. Eur. J. Immunol. 24, 1453–1457 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Sohail A., Klapacz J., Samaranayake M., Ullah A., Bhagwat A. S., Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31, 2990–2994 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiedemann E. M., Peycheva M., Pavri R., DNA replication origins in immunoglobulin switch regions regulate class switch recombination in an R-loop-dependent manner. Cell Rep. 17, 2927–2942 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Di Noia J. M., Neuberger M. S., Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76, 1–22 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez G. P., et al. , Mismatch repair-dependent mutagenesis in nondividing cells. Proc. Natl. Acad. Sci. U.S.A. 109, 6153–6158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.SenGupta T., et al. , Base excision repair AP endonucleases and mismatch repair act together to induce checkpoint-mediated autophagy. Nat. Commun. 4, 2674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Atri S., et al. , Involvement of the mismatch repair system in temozolomide-induced apoptosis. Mol. Pharmacol. 54, 334–341 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Duckett D. R., Bronstein S. M., Taya Y., Modrich P., hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl. Acad. Sci. U.S.A. 96, 12384–12388 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu L., Wu J., Qiu L., Jennings C. D., Li G. M., Involvement of DNA mismatch repair in folate deficiency-induced apoptosis small star, filled. J. Nutr. Biochem. 13, 355–363 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez-Perez A., et al. , IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods 10, 1081–1082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bykov V. J. N., Eriksson S. E., Bianchi J., Wiman K. G., Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 18, 89–102 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Carpenter M. A., Law E. K., Serebrenik A., Brown W. L., Harris R. S., A lentivirus-based system for Cas9/gRNA expression and subsequent removal by Cre-mediated recombination. Methods 156, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.