Significance
Pregnancy is one of the most dynamic periods in a woman’s life, involving a remarkable potential for brain plasticity that promotes cognitive and emotional adjustments to the newborn. We provide evidence for a relationship between number of childbirths and brain aging in 12,021 middle-aged women, suggesting that potential parity-related brain changes may endure beyond the postpartum period and influence the course of neurobiological aging.
Keywords: pregnancy and childbirth, brain imaging, genetics, machine learning
Abstract
Maternal brain adaptations have been found across pregnancy and postpartum, but little is known about the long-term effects of parity on the maternal brain. Using neuroimaging and machine learning, we investigated structural brain characteristics in 12,021 middle-aged women from the UK Biobank, demonstrating that parous women showed less evidence of brain aging compared to their nulliparous peers. The relationship between childbirths and a “younger-looking” brain could not be explained by common genetic variation or relevant confounders. Although prospective longitudinal studies are needed, the results suggest that parity may involve neural changes that could influence women’s brain aging later in life.
During pregnancy and postpartum, fundamental biological processes are instigated to support maternal adaptation and ensure protection of the offspring (1). In rodents, brain adaptations across pregnancy and postpartum include reduced neurogenesis in the dentate gyrus (2, 3) and changes in volume, dendritic morphology, and cell proliferation in the hippocampus (1, 4). In humans, reduction in total brain volume has been observed during pregnancy, with reversion occurring within 6 mo of parturition (5). In the postpartum period, regional gray matter increases have been found in the amygdala, hypothalamus, and prefrontal cortex (6). Postpartum regional reductions have also been reported (7–9), some of which are positively related to maternal attachment (8). While some maternal brain changes revert during the postpartum period, others extend well beyond this phase (1, 7, 8, 10) and may influence the course of neurobiological aging later in life. Regional reductions in brain volume have been found to endure for at least 2 y postpregnancy in humans (8), and reproductive history has been linked to cortical thickness later in life (10). Parous rats have increased hippocampal neurogenesis in middle age (3) and show fewer signs of brain aging relative to nulliparous rats (1, 11). Such long-lasting brain adaptations could also reflect genetic pleiotropy. Reproductive behaviors are complex, heritable traits, and their polygenic architecture is likely to overlap with other traits that influence brain-aging trajectories (12, 13). Parsing the effects of common genetic variation is thus important to delineate potential effects of parity on the brain.
We investigated structural brain characteristics in 12,021 women from the UK Biobank, hypothesizing that parity would be associated with apparent brain aging. Machine learning and brain age prediction was used to test 1) whether a classifier could identify women as parous or nulliparous based on morphometric brain characteristics and 2) whether brain age gap (estimated brain age minus chronological age) differed between parous (n = 9,568) and nulliparous (n = 2,453) women. Mean age SD was 54.72 7.29 y for the full sample and 55.23 7.22 y for parous and 52.79 7.23 y for nulliparous women. To investigate the impact of number of childbirths, we tested for associations between number of births and the probabilistic scores from the group classification and brain age gap, in addition to comparing women who had given 1 birth, 2 births, 3 births, 4 births, and 5 to 8 births to nulliparous women.
To parse the effects of common genetic variation, we performed a genome-wide association study (GWAS) on the phenotype number of births in 271,312 healthy women in the UK Biobank (excluding our MRI subsample). We then computed polygenic score for each European individual in our MRI subsample (n = 10,289; Materials and Methods) and tested for associations between polygenic scores and 1) the probability score from the group classification and 2) brain age gap. Next, the main analyses were rerun using polygenic scores as a covariate.
Results
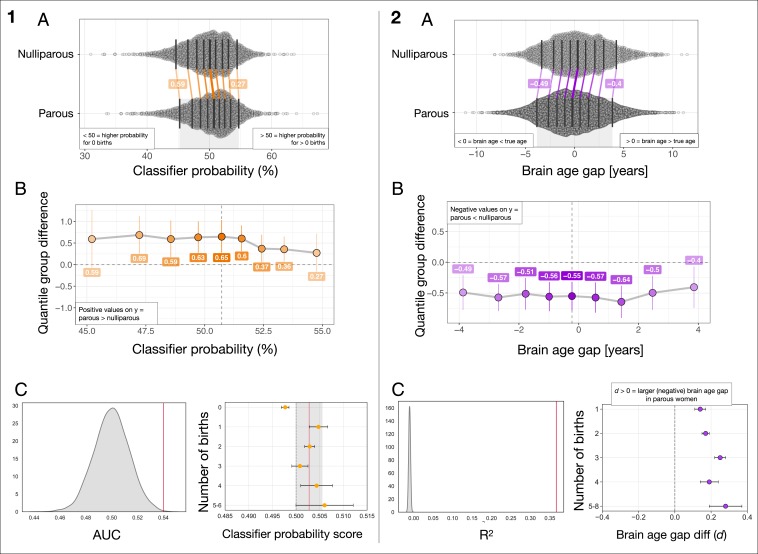
Fig. 1 and Tables 1 and 2 show the results from the group classification and the brain age prediction. For the classification, the average area under the receiver operating characteristic curve (AUC) was 0.54 (SD = 0.02), (Materials and Methods). The probability of being classified as parous was higher for the parous group than for the nulliparous group (mean difference SD , , Cohen’s ). Within the parous group, the classifier probability score was not related to number of births (, , CI = [−0.05, 0.02]).
Fig. 1.
(Left, A) The distributions of classifier probability scores in nulliparous and parous women. The x axis refers to the estimated percentage probability of having given birth. The black vertical lines mark the deciles of the distributions. The matching deciles in the 2 groups are joined by colored lines, showing a uniform, positive shift in the group of parous women. (Left, B) The portion of the x axis in A marked by the gray shaded area at the bottom of the plot. The y axis shows the group differences between deciles (parous group minus nulliparous group), while the x axis shows the deciles of the parous group. (Left, C) Left plot shows the mean SD AUC for the classifier was 0.54 0.02, based on a 10-fold cross-validation (red vertical line). The null distribution calculated from 10,000 permutations is shown in gray, with a mean SD AUC of 0.50 0.01. The number of permuted results from the null distribution exceeding the mean from the cross-validation was 6 (). Right plot shows mean classifier probability for each of the subgroups of women based on number of childbirths. The red vertical line shows the mean classifier probability in the groups of parous women, while the lighter gray area illustrates the SD. The plot is displayed with balanced group samples (n nulliparous women = 2,453, 1 birth = 442, 2 births = 1,331, 3 births = 523, 4 births = 122, 5 to 6 births = 35; see Materials and Methods for details). The error bars represent the SE on the means. The dashed line indicates 0.5 on the x axis. (Right, A) The distributions of bias-corrected, estimated brain age gap in nulliparous and parous women. Negative values indicate a predicted brain age that is lower than chronological age, i.e., a “younger-looking” brain. The plot shows a uniform, negative shift in the group of parous women. (Right, B) The y axis shows the group differences between deciles (parous group minus nulliparous group), while the x axis shows the deciles of the parous group. (Right, C) Left plot shows the mean SD for the XGboost regressor model was , based on a 10-fold cross-validation with 10 repetitions per fold (red vertical line). The null distribution calculated from 10,000 permutations is shown in gray, with a mean SD of (). Right plot shows the difference in brain age gap between each of the subgroups and nulliparous women as measured by Cohen’s . The error bars represent the SD of the effect size. Higher values on the x axis indicate a larger effect size. The dashed line indicates 0 on the x axis. Number of subjects in each group: 1 birth = 1,630, 2 births = 5,315, 3 births = 2,021, 4 births = 476, 5 to 8 births = 126, nulliparous women = 2,453.
Table 1.
Results from the group classification and brain age prediction, including correlation analyses and logistic regression
| Pearson’s r | Logistic regression | ||||||
| r | p | 95% CI | β | SE | z | p | |
| Classifier probability score and no. of childbirths | −0.02* | 0.45* | −0.05, 0.02* | 2.19 | 0.73 | 3.01 | |
| Brain age gap and no. of childbirths | −0.07 | −0.09, −0.06 | −0.06 | 0.01 | −8.22 | ||
SE, standard error. Number of women with >1 birth = 9,568, nulliparous women = 2,453.
within parous group
Table 2.
Differences in brain age gap between nulliparous women and the subgroups of parous women
| Group differences in brain age gap | ||||
| Group | Mean diff (SD) | t | p | Cohen’s d |
| 1 birth | 0.56 (3.76) | 8.27 | 0.15 | |
| 1 birth (n = 1,630) | 0.43 (2.98) | 4.49 | 0.14 | |
| 2 births (n = 5,315) | 0.52 (2.98) | 7.14 | 0.17 | |
| 3 births (n = 2,021) | 0.75 (2.99) | 8.33 | 0.25 | |
| 4 births (n = 476) | 0.58 (2.98) | 3.88 | 0.19 | |
| 5 to 8 births (n = 126) | 0.82 (2.96) | 3.02 | 0.28 | |
Number of women with >1 birth = 9,568, nulliparous women = 2,453.
In the brain age analysis, the correlation between predicted and chronological age was , , CI = [0.6, 0.62], and root-mean-square error (rmse) = 5.78 (SD = 0.10), . To account for age-related bias in the predicted age (14, 15), we employed a quadratic regression to the data (Eq. 1, Materials and Methods). Bias-corrected brain age gap correlated negatively with number of births (, , CI = [−0.09, −0.06]), indicating a younger-looking brain in multiparous women. The correlation remained significant when including only parous women (, , CI = [−0.05, −0.01]). To assess the robustness of these effects, the brain age analysis was rerun using predicted brain age estimates based on an independent approach and training set (16, 17). In brief, the results were consistent with the main findings (see Materials and Methods for full description). The results are provided in SI Appendix, page 2. As a follow-up, first- and second-degree polynomial fits were performed. The results are shown in SI Appendix, page 3.
To investigate relevant confound variables, we performed additional analyses testing the associations between brain age gap and number of childbirths when accounting for age at first birth, ethnic background, education, and BMI. None of these variables fully explained the differences in brain age gap between parous and nulliparous women. The results are provided in SI Appendix, Tables S2–S5. To test for additional confound variables, partial correlation analyses were performed with 1) age at menarche and age at menopause and 2) number of incomplete pregnancies as covariates. The correlation between number of births and brain age gap persisted when including age at menarche and age at menopause (, , CI = [−0.07, −0.02], n = 6007) and when including number of incomplete pregnancies (, , CI = [−0.1, −0.03], n = 3,760). To test whether parity-related complications or disease influenced the results, we excluded women with reported diagnoses in ICD10 (18, 19) chap. XV, “Pregnancy, childbirth, and the puerperium” and reran the analysis. For the remaining sample (n = 9,064), the relationship between brain age gap and number of births was , , CI = [−0.08, −0.03]. Next, we excluded women with reported diagnoses in ICD10 chap. V, “Mental and behavioral disorders” and/or chap. VI, “Diseases of the nervous system.” For the remaining sample (n = 9,207), the relationship between brain age gap and number of births remained significant (, , CI = [−0.08, −0.04]). See http://biobank.ndph.ox.ac.uk/showcase/field.cgi?id=41202 for an overview of the ICD10 categories.
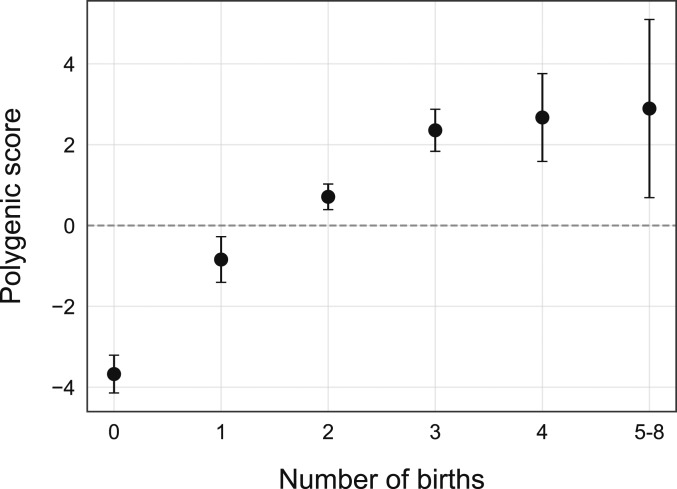
The mean polygenic scores for number of births in each of the subgroups are shown in Fig. 2. Within the group of parous women, a positive correlation was found between polygenic scores and number of births (, , CI = [0.03, 0.07]). Polygenic scores and classifier probability scores showed a correlation of (, CI = [−0.0, 0.08]) for parous women and (, CI = [−0.04, 0.04]) for nulliparous women (full sample: , , CI = [−0.0, 0.06]). Polygenic scores and brain age gap showed a correlation of (, CI = [−0.0, 0.04]) for parous women and (, CI = [−0.01, 0.08]) for nulliparous women (full sample: , , CI = [−0.0, 0.06]). The correlation between number of births and brain age gap persisted when partialling out polygenic scores (, , CI = [−0.1, −0.06] for the full sample and , , CI = [−0.05, −0.01] within the group of parous women).
Fig. 2.
The black circles show the mean polygenic score in each of the subgroups based on number of births. The error bars represent the SE on the means.
Discussion
Summarized, the results show that parity can be linked to women’s brain age in midlife, in line with a recent analysis that tested for associations between brain age and a range of phenotypes in the UK Biobank (14). We found no evidence that common polygenetic variation or confound variables could fully explain the differences in brain age gap between parous and nulliparous women. Although prospective longitudinal studies are needed to conclude, the findings suggest that parity may involve long-lasting neural changes (8, 20–24) that could influence brain aging later in life (10, 25) and that such effects may be more prominent following multiple childbirths. While the present results demonstrated a negative linear relationship between parity and brain age gap, follow-up analyses also showed evidence for a moderate quadratic effect (SI Appendix, Fig. S2), suggesting that any protective effects of parity may level off and be less pronounced in grand-parous (5 births) women. In line with this observation, parity has been linked to risk of Alzheimer’s disease (AD) (26, 27), with a higher risk in women with 5 or more completed pregnancies (28). Recent studies have also shown a J-shaped relationship between parity and mortality, with longevity peaking at 3 to 4 births (29). Nulliparity has been associated with increased risk of autoimmune conditions (30), while grand multiparity has been linked to cardiovascular diseases including stroke and associated risk factors such as adiposity and diabetes (26, 28). Although it is possible that moderate-level parity could be more beneficial for brain aging relative to nulliparity and grand multiparity, such effects could also be driven by other variables not considered, for example differences in socioeconomic factors or stress levels (29). More studies controlling for relevant confounding factors are needed to fully understand the nature of the relationship between parity and brain aging.
Endocrinological modulations play an important role in the altered brain plasticity that occurs during and after pregnancy (22, 24). Changes in sex steroid hormones are known to influence human brain structure through regulation of neuronal morphology (31), and hormones such as estradiol, progesterone, prolactin, oxytocin, and cortisol are known to regulate brain plasticity (22, 31). Hormonal profiles are thus likely to contribute to maternal brain adaptations during pregnancy and postpartum, and their fluctuations may have implications for brain aging later in life. However, the long-term effects are not fully understood, and while endogenous exposure to estrogen has been suggested to be neuroprotective (32), a recent metaanalysis found no evidence of an association between endogenous estrogen exposure and incident dementia (33). Another proposed mechanism for enduring effects is the long-lasting presence of fetal cells in the maternal body (34–36), and such fetal microchimerism provides an avenue for biological interactions between fetal and maternal cells long after delivery. In an evolutionary framework, this has been conceptualized as a mother–offspring negotiation (35), providing an intriguing link to the maternal immune system. There is strong evidence for a crucial role of immune factors in pregnancy (37), which represents a state of low-level inflammation characterized by a balance between anti-inflammatory and proinflammatory cytokines (1, 38). Pregnancy is known to influence and modify inflammatory disease activity and symptomology in conditions such as multiple sclerosis, asthma, and rheumatoid arthritis (39), and the pregnancy-induced increase in concentration of regulatory T cells may have implications for inflammatory susceptibility later in life. A higher cumulative time spent pregnant in the first trimester, which is when the proliferation of regulatory T cells is highest, has been shown to protect against AD (39), of which the pathogenesis is known to involve inflammatory processes (40). Genetic differences have also been shown to interact with age and parity to influence cognitive function, expression of proteins related to synaptic plasticity, and AD neuropathology in mice (41) and humans (42), indicating differential genotype-dependent effects of parity on the brain across life. In humans, the polygenetic architecture of reproductive behavior is known to overlap with other complex traits that affect brain-aging trajectories, such as biological fecundity and education (12, 13). Parity is also associated with societal factors such as economy, culture, and availability of contraception and childcare, emphasizing that complex social–environmental interactions represent potentially important confounders in genome-wide association studies (43). It should also be noted that the presented results may not apply to populations beyond those represented in the UK biobank. The use of large datasets does, however, enable the identification of subtle effects that could go undetected in smaller samples. Although parity may explain only a small portion of the variance in brain aging, the current findings represent an important part of the wider picture of women’s brain aging, as well as sex differences in risk factors and disease (9, 30).
In conclusion, our results provide evidence that parity is linked to women’s brain aging in midlife and that this association cannot be fully explained by common genetic variation or relevant confounding variables. Potential parity-related neural changes may thus extend beyond the postpartum period and influence the course of women’s brain aging later in life.
Materials and Methods
The sample was drawn from the UK Biobank (http://www.ukbiobank.ac.uk) and included 12,021 women. The data are available for researchers through the UK Biobank application procedure (http://www.ukbiobank.ac.uk/researchers). Code can be provided upon request. Sample demographics are provided in Table 3 and SI Appendix, Table S1. Due to the low number of grand-multiparous women (n for 5 births = 85, 6 births = 31, 7 births = 6, and 8 births = 4), these subgroups were merged.
Table 3.
Demographics for each group
| Births | N | Age (M SD) | Ethnic background, % | Educational qualification, % |
| 0 | 2,453 | 52.73 (7.21) | W97.02 B0.78 M0.73 A0.65 C0.37 O0.37 | U51.90 A13.58 O20.42 C3.67 N2.41 P4.81 Noa3.22 |
| 1 | 9,568 | 55.23 (7.22) | W97.55 B0.52 M0.46 A0.68 C0.36 O0.40 | U40.52 A14.14 O23.06 C4.54 N3.58 P6.26 Noa7.90 |
| 1 | 1,630 | 53.82 (7.19) | W96.87 B0.67 M0.74 A0.68 C0.55 O0.43 | U40.67 A14.60 O23.74 C4.72 N4.48 P5.40 Noa6.38 |
| 2 | 5,315 | 55.23 (7.13) | W97.89 B0.49 M0.38 A0.56 C0.38 O0.26 | U40.67 A14.60 O23.44 C4.72 N5.40 P7.17 Noa8.03 |
| 3 | 2,021 | 55.94 (7.26) | W97.33 B0.54 M0.49 A0.74 C0.25 O0.64 | U40.72 A14.30 O22.61 C3.66 N3.66 P6.73 Noa8.31 |
| 4 | 476 | 56.80 (7.45) | W97.06 B0.42 M0.42 A1.68 C0.00 O0.42 | U39.28 A11.97 O21.43 C4.41 N4.41 P9.03 Noa9.45 |
| 5 to 8 | 126 | 56.33 (6.96) | W97.62 B0.00 M0.00 A0.79 C0.00 O1.59 | U38.89 A12.70 O22.22 C2.38 N3.97 P10.32 Noa9.52 |
M ± SD, mean ± SD. Ethnic background: A, Asian; B, black; C, Chinese; M, mixed; O, other; W, white. Educational qualification: A, A levels or equivalent; C, CSE or equivalent; N, NVQ/HNS/HNS or equivalent; Noa, none of the above; O, O levels/GCSE or equivalent; P, professional qualification, e.g., nursing/teaching; U, university/college degree. For the categories, see http://biobank.ctsu.ox.ac.uk/crystal/coding.cgi?id=100305 and http://biobank.ctsu.ox.ac.uk/crystal/coding.cgi?id=1001.
MRI Processing.
Raw T1-weighted MRI data for all participants were processed using a harmonized analysis pipeline, including automated surface-based morphometry and subcortical segmentation as implemented in FreeSurfer 5.3 (44). In line with a recent large-scale implementation (45), we utilized a fine-grained cortical parcellation scheme (46) to extract cortical thickness, area, and volume for 180 regions of interest per hemisphere, in addition to the classic set of subcortical and cortical summary statistics from FreeSurfer (44). This yielded a total set of 1,118 structural brain imaging features (360/360/360/38 for cortical thickness/area/volume, as well as cerebellar/subcortical and cortical summary statistics, respectively). To remove outliers, the Euler numbers (47) were extracted from FreeSurfer and averaged across the left and right hemispheres. The average values were then residualized with respect to age and scanning site using linear models, before subjects with average Euler numbers of SD 4 were identified and excluded (n = 109). In addition, subjects with SD 4 on the global MRI measures mean cortical or subcortical gray matter volume were excluded (n = 10 and n = 12, respectively), yielding a total of 12,021 subjects for the main analyses. As a data quality cross-check, the main analyses (binary classification and brain age prediction) were rerun using MRI data that were first residualized with respect to the average Euler numbers in addition to the other covariates. In brief, the results were consistent with the main findings (see SI Appendix, Table S2 for full results). For the binary classification, we residualized all variables with respect to age, scanning site, ethnic background, education, and intracranial volume (ICV) using linear models. For the brain age prediction, we residualized all variables with respect to scanning site, ethnic background, education, and ICV using linear models.
Principal Component Analysis.
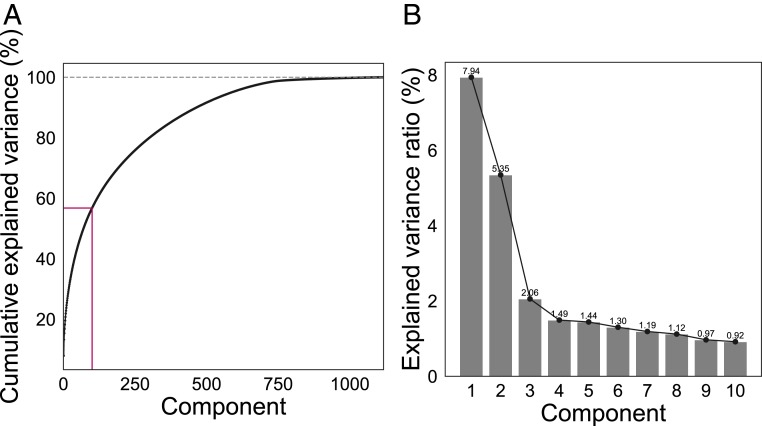
Principal component analyses (PCA) were run with z-transformed MRI variables , where is an MRI variable of mean and SD ). The top 100 components were used in the subsequent analyses, explaining 56.77% of the total variance for the classifier variables and 56.78% for the brain age prediction variables, as shown in Fig. 3. As a cross-check, the correlations between number of births and 1) classifier prediction value and 2) brain age gap were rerun with 200 components, explaining 71.62% and 70.98% of the total variance, respectively. With 200 components included, the correlation between number of births and classifier prediction value was , , CI = [−0.06, 0.02], while the correlation between number of births and brain age gap showed , , CI = [−0.09, −0.05]. As the results were consistent, 100 components were chosen to reduce computational time.
Fig. 3.
(A) Cumulative explained variance for the PCA components based on 1,118 z-transformed MRI variables used in the brain age analysis. (B) Explained variance ratio shown for the top 10 PCA components used in the brain age analysis.
Binary Classification.
Gradient boosting classification was performed using Scikit-learn (https://scikit-learn.org). Parameters were set to max depth = 1, number of estimators = 100, and learning rate = 0.1 (defaults). To account for differences in group size between nulliparous and parous women, undersampling was performed using Imbalanced-learn (https://imbalanced-learn.readthedocs.io/en/stable/user_guide.html), randomly selecting samples (without replacement). The balanced sample included 2,453 nulliparous women and 2,453 parous women with a maximum of 6 births (n 1 birth = 442, 2 births = 1,331, 3 births = 523, 4 births = 122, 5 to 6 births = 35). The classifier probability score was estimated based on a 10-fold cross-validation, assigning a probability of being labeled as parous (having given birth) to each of the subjects.
Brain Age Prediction.
Brain age prediction estimates an individual’s apparent brain aging based on structural brain characteristics derived from MRI. Subtracting chronological age from estimated brain age provides a measure of individuals’ brain age gap: the difference between their estimated brain age and their chronological age. For instance, if a 60-y-old individual shows a brain age gap of −5 y, the typical aging pattern resembles the brain structure of a 55-y-old individual; i.e., the brain is younger looking than what is expected for the chronological age (48). The XGBRegressor model from XGBoost (https://xgboost.readthedocs.io/en/latest/python/index.html) was used to run the brain age prediction analysis with an algorithm that has been used in recent large-scale brain age studies (14, 45). Parameters were set to max depth = 3, number of estimators = 100, and learning rate = 0.1 (defaults). The predicted age based on the PCA components was estimated in a 10-fold cross-validation with 10 repetitions per fold, assigning an estimated brain age to each individual. Brain age gap was calculated using estimated brain age minus true age. Average rmse SD = 5.78 based on a 10-fold cross-validation with 10 repetitions per fold. The null distribution calculated from 10,000 permutations showed an average rmse of 7.33 0.01, and the number of permuted results from the null distribution that exceeded the mean from the cross-validation was 0 ().
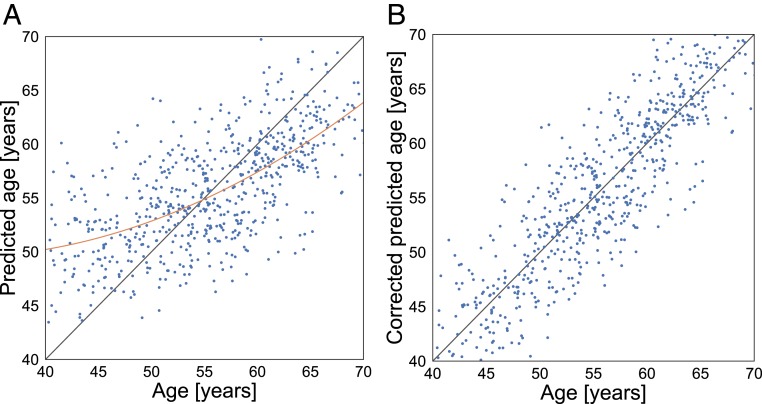
To adjust for a frequently observed bias leading to generally overestimated age predictions at low age and underestimated predictions at high age (14, 15), we employed the regression
| [1] |
where the coefficients , , and parameterize the relationship between the true and predicted age. These coefficients were then used to remove the effect of the bias, to achieve a linear dependence with slope = 1 between the true and predicted age values, as illustrated in Fig. 4.
Fig. 4.
(A) Machine performance is biased toward the mean age, resulting in overestimated predictions at low age and underestimated predictions at high age. (B) After bias correction using Eq. 1, the predictions follow the expected dependence.
To ensure that the bias correction was employed successfully, we tested the association between bias-corrected brain age delta and number of births while controlling for chronological age. The test showed results consistent with the main findings: , CI = [−0.09, −0.06].
To assess the robustness of the effects, the brain age analysis was rerun using predicted brain age estimates based on an independent approach and training set from the brainageR software (https://github.com/james-cole/brainageR) (16, 17). The brainageR model is trained on voxel-based morphometry maps (VBM) based on T1-weighted MRI scans from 2,001 healthy individuals (male/female = 1,016/985, mean age SD = 36.95 18.12, age range 18 to 90 y) and uses a Gaussian Processes regression with the kernlab package in R. See ref. 16 for details. The results are shown in SI Appendix, Fig. S1.
The differences between the quantiles of the groups of parous and nulliparous women were investigated using the shift function in the Robust Graphical Methods for Group Comparisons package in R (49) (https://github.com/GRousselet/rogme). In brief, the shift function shows the difference between the quantiles of 2 groups as a function of the quantiles of 1 group.
GWAS.
A GWAS was run on the women in the UK Biobank cohort (n = 271,312, excluding the MRI subsample), using PLINK 2.0 (50) and the UKB v3 imputed genetic data, filtering out SNPs with a minor allele frequency below 0.001 or failing the Hardy–Weinberg equilibrium test at . Nonwhite Europeans and individuals with a brain disorder as indicated by ICD10 were excluded from the study. We then ran a linear regression on the continuous measure number of childbirths, covarying for age and the first 10 genetic principal components, as provided by UK Biobank under field 22009. PRSice v1.25 (51) was used to calculate polygenic scores for number of births across -value thresholds from 0.001 to 0.5, with intervals of 0.001, using PRSice default settings. This includes the removal of the major histocompatibility complex (MHC) (chromosome 6, 26 to 33 Mb) and thinning of SNPs based on linkage disequilibrium and p value. A PCA was run on the polygenic scores across all -value levels (52), and the first component explaining 92.24% of the total variance was used in the subsequent analyses. The PCA component correlated with the PGS at threshold .
Supplementary Material
Acknowledgments
This research has been conducted using the UK Biobank under Application 27412 and received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (Grant 802998), the Research Council of Norway (286838, 273345, 249795, 276082, 223273), the South-East Norway Regional Health Authority (2015073, 2019107), and the Medical Research Council (MR/K006673/1).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910666116/-/DCSupplemental.
References
- 1.Hillerer K. M, Jacobs V. R., Fischer T., Aigner L., The maternal brain: An organ with peripartal plasticity. Neural Plast. 2014, 1–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rolls A., Schori H., London A., Schwartz M., Decrease in hippocampal neurogenesis during pregnancy: A link to immunity. Mol. Psychiatry 13, 468–469 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Eid R. S., et al. , Early and late effects of maternal experience on hippocampal neurogenesis, microglia, and the circulating cytokine milieu. Neurobiol. Aging 78, 1–17 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Kinsley C. H., et al. , Motherhood and the hormones of pregnancy modify concentrations of hippocampal neuronal dendritic spines. Horm. Behav. 49, 131–142 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Oatridge A., et al. , Change in brain size during and after pregnancy: Study in healthy women and women with preeclampsia. Am. J. Neuroradiol. 23, 19–26 (2002). [PMC free article] [PubMed] [Google Scholar]
- 6.Kim P., et al. , The plasticity of human maternal brain: Longitudinal changes in brain anatomy during the early postpartum period. Behav. Neurosci. 124, 695–700 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte-Guterman P., Leuner B., Galea L. A. M., The long and short term effects of motherhood on the brain. Front. Neuroendocrinol. 53, 100740 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Hoekzema E., et al. , Pregnancy leads to long-lasting changes in human brain structure. Nat. Neurosci. 20, 287–296 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Galea L. A. M., Qiu W., Duarte-Guterman P., Beyond sex differences: Short and long-term implications of motherhood on women’s health. Curr. Opin. Physiol. 6, 82–88 (2018). [Google Scholar]
- 10.Orchard E. R., et al. , Cortical changes associated with parenthood are present in late life. bioRxiv:589283 (27 March 2019).
- 11.Pawluski J. L., Lambert K. G., Kinsley C. H., Neuroplasticity in the maternal hippocampus: Relation to cognition and effects of repeated stress. Horm. Behav. 77, 86–97 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Barban N., et al. , Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat. Genet. 48, 1462–1472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong A., et al. , Selection against variants in the genome associated with educational attainment. Proc. Natl. Acad. Sci. U.S.A. 114, E727–E732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith S. M., Vidaurre D., Alfaro-Almagro F., Nichols T. E., Miller K. L., Estimation of brain age delta from brain imaging. Neuroimage 200, 528– 539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le T. T., et al. , A nonlinear simulation framework supports adjusting for age when analyzing BrainAGE. Front. Aging Neurosci. 10, 317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole J. H., et al. , Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage 163, 115–124 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Cole J. H., et al. , Brain age predicts mortality. Mol. Psychiatry 23, 1385–1392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization , The ICD-10 Classification of Mental and Behavioural Disorders: Clinical Descriptions and Diagnostic Guidelines (World Health Organization, Geneva, Switzerland, 1992). [Google Scholar]
- 19.World Health Organization , The ICD-10 Classification of Mental and Behavioural Disorders: Diagnostic Criteria for Research (World Health Organization, Geneva, Switzerland, vol. 2, 1993). [Google Scholar]
- 20.Barha C. K., Lieblich S. E., Chow C., Galea L. A. M., Multiparity-induced enhancement of hippocampal neurogenesis and spatial memory depends on ovarian hormone status in middle age. Neurobiol. Aging 36, 2391–2405 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Champagne F. A., Curley J. P., Plasticity of the maternal brain across the lifespan. New Dir. Child Adolesc. Dev. 2016, 9–21 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Galea L. A. M., Leuner B., Slattery D. A., Hippocampal plasticity during the peripartum period: Influence of sex steroids, stress and ageing. J. Neuroendocrinol. 26, 641–648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim P., Strathearn L., Swain J. E., The maternal brain and its plasticity in humans. Horm. Behav. 77, 113–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinsley C. H., Lambert K. G., Reproduction-induced neuroplasticity: Natural behavioural and neuronal alterations associated with the production and care of offspring. J. Neuroendocrinol. 20, 515–525 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Kinsley C. H., Franssen R. A., Meyer E. A., “Reproductive experience may positively adjust the trajectory of senescence” in Behavioral Neurobiology of Aging, Pardon M.-C., Bondi M. W., Eds. (Springer, 2011), pp. 317–345. [DOI] [PubMed] [Google Scholar]
- 26.Beeri M. S., et al. , Number of children is associated with neuropathology of Alzheimer’s disease in women. Neurobiol. Aging 30, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colucci M., et al. , The number of pregnancies is a risk factor for Alzheimer’s disease. Eur. J. Neurol. 13, 1374–1377 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Jang H., et al. , Differential effects of completed and incomplete pregnancies on the risk of Alzheimer disease. Neurology 91, e643–e651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Y., et al. , Parity and all-cause mortality in women and men: A dose-response meta-analysis of cohort studies. Sci. Rep. 6, 19351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Natri H., Garcia A. R., Buetow K. H., Trumble B. C., Wilson M. A., The pregnancy pickle: Evolved immune compensation due to pregnancy underlies sex differences in human diseases. Trends Genet. 35, 478–488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simerly R. B., Wired for reproduction: Organization and development of sexually dimorphic circuits in the mammalian forebrain. Annu. Rev. Neurosci. 25, 507–536 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Fox M., Berzuini C., Knapp L. A., Cumulative estrogen exposure, number of menstrual cycles, and Alzheimer’s risk in a cohort of British women. Psychoneuroendocrinology 38, 2973–2982 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Prince M. J., et al. , Reproductive period, endogenous estrogen exposure and dementia incidence among women in Latin America and China; A 10/66 population-based cohort study. PLoS One 13, e0192889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barha C. K., Galea L. A. M., The maternal ‘baby brain’ revisited. Nat. Neurosci. 20, 134–135 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Boddy A. M., Fortunato A., Wilson Sayres M., Aktipis A., Fetal microchimerism and maternal health: A review and evolutionary analysis of cooperation and conflict beyond the womb. BioEssays 37, 1106–1118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng X. X., et al. , Pregnancy-associated progenitor cells differentiate and mature into neurons in the maternal brain. Stem Cells Dev. 19, 1819–1830 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Mor G., Cardenas I., Abrahams V., Guller S., Inflammation and pregnancy: The role of the immune system at the implantation site. Ann. N. Y. Acad. Sci. 1221, 80–87 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luppi P., How immune mechanisms are affected by pregnancy. Vaccine 21, 3352–3357 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Fox M., Berzuini C., Knapp L. A., Glynn L. M., Women’s pregnancy life history and Alzheimer’s risk: Can immunoregulation explain the link? Am. J. Alzheimers Dis. Other Demen. 33, 516–526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyss-Coray T., Rogers J., Inflammation in Alzheimer disease - a brief review of the basic science and clinical literature. Cold Spring Harbor Perspect. Med. 2, a006346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui J., et al. , Amyloid precursor protein mutation disrupts reproductive experience-enhanced normal cognitive development in a mouse model of Alzheimer’s disease. Mol. Neurobiol. 49, 103–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corbo R. M., et al. , Combined effect of apolipoprotein E genotype and past fertility on age at onset of Alzheimer’s disease in women. Dement. Geriatr. Cognit. Disord. 24, 82–85 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Turkheimer E., “Genome wide association studies of behavior are social science” in Philosophy of Behavioral Biology, Plaisance K. S., Reydon T. A. C., Eds. (Springer, 2012), pp. 43–64. [Google Scholar]
- 44.Fischl B., et al. , Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Kaufmann T., et al. , Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat. Neurosci. 22, 1617–1623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glasser M. F., et al. , A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosen A. F. G., et al. , Quantitative assessment of structural image quality. Neuroimage 169, 407–418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke K., Gaser C., Ten years of BrainAGE as a neuroimaging biomarker of brain aging: What insights have we gained? Front. Neurol. 10, 789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rousselet G. A., Pernet C. R., Wilcox R. R., Beyond differences in means: Robust graphical methods to compare two groups in neuroscience. Eur. J. Neurosci. 46, 1738–1748 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Chang C. C., et al. , Second-generation plink: Rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Euesden J., Lewis C. M., O’Reilly P. F.. PRSice: Polygenic risk score software. Bioinformatics 31, 1466–1468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alnæs D., et al. , Brain heterogeneity in schizophrenia and its association with polygenic risk. JAMA Psychiatry 76, 739–748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.