Significance
Pharmacological receptor theory models are used to predict and interpret how small-molecule ligands impact the function of cellular receptors. Theoretical models have been modified and extended over time to more accurately describe relationships between ligand affinity and receptor functional. However, structural studies are lacking that describe the influence of receptor conformation, or shape, as predicted by these models. Using NMR spectroscopy, we detected how the nuclear receptor transcription factor peroxisome proliferator-activated receptor γ (PPARγ) structurally responds to ligand binding. We observed a correlation between ligand affinity, function, and receptor conformation for chemically similar ligands but not chemical diverse ligands, revealing a potential limitation of theoretical receptor models.
Keywords: nuclear receptor, ligand binding, receptor theory, NMR spectroscopy, X-ray crystallography
Abstract
Ligand–receptor interactions, which are ubiquitous in physiology, are described by theoretical models of receptor pharmacology. Structural evidence for graded efficacy receptor conformations predicted by receptor theory has been limited but is critical to fully validate theoretical models. We applied quantitative structure–function approaches to characterize the effects of structurally similar and structurally diverse agonists on the conformational ensemble of nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ). For all ligands, agonist functional efficacy is correlated to a shift in the conformational ensemble equilibrium from a ground state toward an active state, which is detected by NMR spectroscopy but not observed in crystal structures. For the structurally similar ligands, ligand potency and affinity are also correlated to efficacy and conformation, indicating ligand residence times among related analogs may influence receptor conformation and function. Our results derived from quantitative graded activity–conformation correlations provide experimental evidence and a platform with which to extend and test theoretical models of receptor pharmacology to more accurately describe and predict ligand-dependent receptor activity.
Receptor theory has been used to describe the actions of pharmacological ligands in various forms for nearly a century (1, 2). The idea of a receptor has evolved from a conceptual “black box” to one founded in the principles of biophysics and allostery (3). The 2-state model of receptor activation (4), which extended the Black/Leff operational model of pharmacological agonism (5) with the Monod–Wyman–Changeux model of protein allostery (6) to describe the actions of pharmacological receptor ligands, conceptually represents a minimal theoretical model to describe the action of ligands within the context of a binary ligand–receptor complex. More complex models were subsequently developed that accounted for improved understanding of receptor functions. The extended ternary complex (ETC) model describes how the receptor–ligand complex influences interaction with an effector protein or a signaling pathway (7), whereas the cubic ternary complex (CTC) model extends the ETC model to account for receptor–effector interactions in the absence of ligand (8). These and other theoretical receptor models could be further improved with a more comprehensive experimental understanding of how ligands affect receptor structure and function.
Applying theoretical receptor models to graded activity dose-responsive pharmacological data are common in studies of membrane receptors, including G-protein–coupled receptors (GPCRs), ligand-gated ion channels, and enzyme-linked receptors (2). These receptors bind extracellular ligands and transduce signals across the cell membrane via conformational rearrangement of the intracellular portion of the membrane receptor to affect various downstream signaling pathways, the activities of which can be measured in cellular assays and applied to pharmacological models of receptor function. Conceptually, the principles of receptor theory also apply to nuclear receptors, a superfamily of intracellular transcription factors that recruit chromatin remodeling transcriptional machinery in a ligand-dependent manner to control gene expression (9). Nuclear receptor agonists, which bind to an internal hydrophobic orthosteric pocket within nuclear receptor ligand-binding domain (LBD), activate transcription by stabilizing structural elements that comprise the activation function-2 (AF-2) coregulator interaction surface, located adjacent to the ligand-binding pocket, including helix 3, helix 4/5, and a critical regulatory switch element, helix 12. Agonists stabilize an active AF-2 surface conformation, which increases the binding affinity for and recruitment of transcriptional coactivator proteins that in turn promote chromatin remodeling and increased transcription.
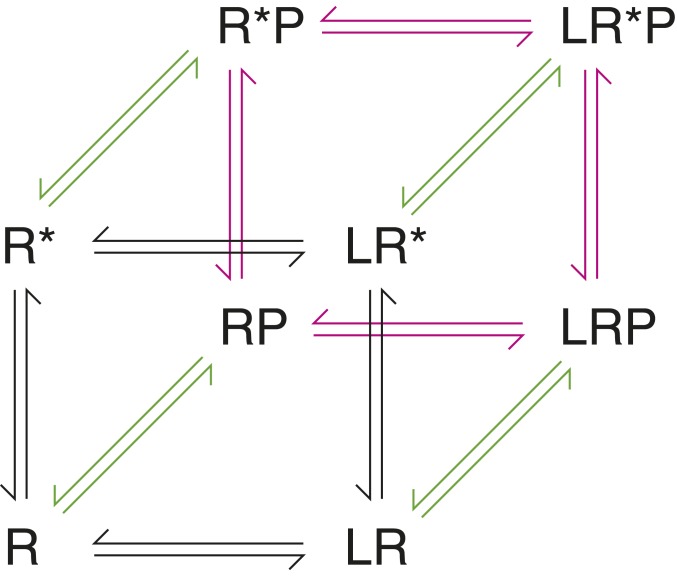
Although receptor theory as practiced in the membrane receptor fields is not used in the nuclear receptor field, in principle the functional endpoints derived from nuclear receptor functional assays can be applied to the same pharmacological models of receptor function. For example, the CTC model (Fig. 1) describes how a receptor exists as a conformational ensemble that fluctuates between resting (R) or active (R*) states capable of binding to ligand (L) and an effector protein or peptide (P). Ligand affinity is described by a combination of L + R ↔ LR and L + R* ↔ LR*. The ligand-bound receptor state (LR/LR*) influences effector binding (LRP/LR*P) by changing the receptor–effector binding affinity constant. Ligand potency is related to ligand affinity but describes the concentration of ligand required to elicit the functional response (efficacy). Assuming that the ligand-free receptor can interact with the effector, which is the case for nuclear receptor LBDs interacting with coactivator peptides, ligand potency is described by RP ↔ LRP and R*P ↔ LR*P.
Fig. 1.
The cubic ternary complex (CTC) model. This theoretical model schematic describes the relationship the receptor resting (R) and active (R*) state when complexed to ligand (L), an effector protein or peptide (P), or both. In the absence of an effector, the apo-state is represented by R and R*, and the ligand-bound state by LR and LR*, also known as the 2-state model of receptor activation.
It has been challenging to structurally observe ligand-bound graded receptor activity conformations predicted by receptor theory. Receptor crystal structures derived from X-ray diffraction data can bias conformations such that the ground state (R or LR) or fully active state (R* or LR*) is observed but the spectrum of graded or partial activity states between R/LR and LR/LR* is not. However, solution NMR spectroscopy studies are capable of detecting graded activity conformational states (10–12). Furthermore, to our knowledge, a direct quantitative assessment has yet to be reported on the relationship between graded ligand potency with functional efficacy and receptor conformation as predicted by the theoretical models of receptor activation. That is, does ligand potency correlate with functional efficacy—where ligands with high potency display high functional efficacy, and ligands with low potency display low functional efficacy—within a structurally related series of ligands or among a structurally diverse set of ligands?
Here, we present a quantitative receptor activity–conformation analysis using 2 distinct sets of agonists of the nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ). We characterized a series of 10 structurally related synthetic PPARγ agonists spanning ∼10,000-fold in affinity using biochemical, biophysical, and cellular assays and structural analysis using X-ray crystallography and NMR spectroscopy. Ligand potency and functional efficacy in this series are correlated to the degree to which the ligands shift the conformational ensemble of PPARγ toward an active state, which we detected by NMR but not in crystal structures, in a manner consistent with relationships predicted by theoretical models of receptor activation. However, using a structurally diverse set of endogenous and synthetic PPARγ ligands, we found that functional efficacy and receptor conformation are correlated independent of ligand potency. Collectively, our studies indicate that data from quantitative structure–function approaches can predict ligand functional efficacy and assess theoretical models of receptor function.
Results
Graded Potency and Functional Efficacy within a Structurally Related Series of PPARγ Agonists.
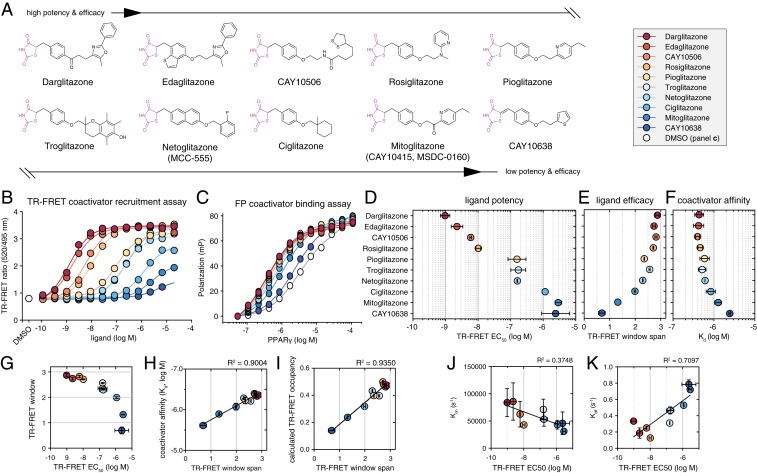
We assembled a series of 10 thiazolidinedione (TZD) PPARγ agonists (Fig. 2A) that include several US Food and Drug Administration-approved antidiabetic drugs (13). All ligands within series contain the conserved TZD head group connected by a linker to a central aromatic moiety and a variable tail group. The linker in all but one of the ligands is a flexible, saturated methylene group that links the TZD head group to a central aromatic moiety; CAY10638 contains an unsaturated linker, which restricts the mobility of the TZD head group. The central aromatic moieties mostly comprise phenyl moieties with the exception of naphthalene and benzothiophene moieties in netoglitazone and edaglitazone, respectively. In contrast to these relatively conservative changes near the TZD head group, the series encompasses a variety of tail moieties extended from the central aromatic core.
Fig. 2.
Quantitative potency and efficacy characterization of a series of thiazolidinedione (TZD) PPARγ agonists. (A) Chemical structures of the TZD ligands. The conserved TZD head group colored purple. (B) TR-FRET assay to determine ligand potency (EC50 values) and efficacy (TR-FRET window span) for recruitment of TRAP220 coactivator peptide to PPARγ LBD fit to a 3-parameter sigmoidal dose–response equation; data represent the mean ± SEM of experimental replicates (ligands, n = 3; DMSO, n = 36). (C) FP assay to determine TRAP220 coactivator peptide affinity to PPARγ LBD when bound to the ligands fit to a one-site–total binding equation; data represent the mean ± SEM of experimental replicates (n = 3). (D) Ligand potencies from the fitted TR-FRET data; data represent the mean ± SD of 2 independent experiments. (E) Ligand efficacies from the fitted TR-FRET data of the experimental replicate shown in B; data represent the TR-FRET ratio window span ± SD from the fit. (F) TRAP220 peptide affinities from the fitted FP data; data represent the mean ± SD of 2 independent experiments. The affinity for apo-PPARγ LBD is indicated on the x axis by a gray arrow. (G) Plot of ligand efficacy from E vs. ligand potency from D. (H) Plot of TRAP220 peptide affinity from F vs. ligand efficacy from E fit with a linear regression equation. (I) Plot of calculated TRAP220 peptide occupancy in the TR-FRET assay (no error bars shown) vs. ligand efficacy fit with a linear regression equation. (J and K) Correlation plots of TRAP220 peptide (J) Kon and (K) Koff values determined in the presence of the TZD ligand series using biolayer interferometry (BLI); BLI data represent the mean ± SD of experimental replicates (n = 2). Errors smaller than the displayed data point size representing the mean value that are too small to display are not shown.
Agonists increase PPARγ-mediated transcription by enhancing binding of transcriptional coactivator proteins, such as TRAP220, also known as MED1 or DRIP205 (14). We assessed the activities of the TZD series in 2 quantitative biochemical assays. We used a time-resolved fluorescence resonance energy transfer (TR-FRET) biochemical assay that measures the ligand-dependent change in the interaction between the PPARγ LBD and a peptide derived from the TRAP220 coactivator (Fig. 2B) containing an “LXXLL” nuclear receptor interaction motif (15). In the TR-FRET coactivator recruitment assay, differences in the overall TR-FRET ratio assay window span, which we refer to as TR-FRET functional efficacy, relates to the relative degree of TRAP220 peptide recruitment and are indicative of ligand-dependent differences in the binding affinity of the TRAP220 coactivator peptide to the PPARγ LBD, which we probed directly using a fluorescence polarization (FP) assay (Fig. 2C).
In the TR-FRET coactivator recruitment assay, the TZD series spans nearly 10,000-fold in potency (EC50) (Fig. 2D) and showed decreasing coactivator peptide recruitment efficacy (Fig. 2E) as ligand potency decreases (Fig. 2G). Coactivator binding affinity was also decreased when PPARγ was bound to less potent ligands (Fig. 2F). Consistent with the correlation between TR-FRET recruitment efficacy and coactivator affinity, there is a correlation between TR-FRET efficacy with coactivator affinity (Fig. 2H) and the coactivator-bound population of the PPARγ LBD in the TR-FRET assay (Fig. 2I). The coactivator peptide concentration used in the TR-FRET assay has a slight effect on ligand potency (higher peptide concentrations result in slightly more potent responses) and a more notable effect on TR-FRET efficacy (higher peptide concentrations result in higher efficacy) (SI Appendix, Fig. S1). These correlated patterns of decreased ligand potency and TR-FRET efficacy are consistent with the allosteric principles of receptor activation theoretical models, which describe the actions of full and partial, or less efficacious agonists where decreasing or graded potency within a ligand series is associated with graded functional efficacy (16, 17). That is, the more potent ligands show higher functional efficacy (e.g., higher TR-FRET assay window span), and the less potent ligands show lower functional efficacy (e.g., lower TR-FRET assay window span). These trends also manifest in differences in the kinetic rate constants for binding TRAP220 coactivator peptide, where Kon and to a larger degree Koff are correlated to ligand potency and functional efficacy from the TR-FRET assay (Fig. 2 J and K and SI Appendix, Fig. S2).
Correlation of TZD Affinity, Receptor Stability, and Cellular Transcription.
We determined ligand binding affinities for the TZD series using a TR-FRET assay that measures the displacement of a fluorescent tracer ligand (Fig. 3A). The TZD series spans an affinity (Ki) range of 5 orders of magnitude (Fig. 3B) and is correlated to biochemical potency in the TR-FRET assay (Fig. 3E). Using differential scanning calorimetry, we determined how the TZD series affects the thermal stability of the PPARγ LBD under our experimental conditions (Fig. 3C) and found a linear correlation between ligand binding affinity and receptor stability (Fig. 3F). We also assessed the TZD series in cell-based transcriptional luciferase reporter assay that directly reports on the transcriptional activity of the PPARγ LBD (Fig. 3D) and is highly sensitive to graded PPARγ agonism (18). The efficacy trends in the cellular transcription profile of the TZD series is similar to the efficacy from the quantitative biochemical TR-FRET coactivator recruitment profiles. Due to cellular toxicity of the ligands at higher concentrations, we were unable to determine cellular potency values for some TZDs due to nonsaturating cellular response profiles. However, there is a correlation between the maximal luciferase value derived from a fitted of the transcriptional reporter assay and TR-FRET ligand potency (Fig. 3G) and ligand affinity (Fig. 3H). Thus, ligand affinity, receptor stability, and transcription are correlated to quantitative functional potency and efficacy within the TZD series.
Fig. 3.
Affinity of the TZD series and effects on receptor stability and transcription. (A) TR-FRET assay to determine ligand affinity (Ki values) using a fluorescent tracer ligand fit to the Cheng–Prusoff inhibitor constant equation; data represent the mean ± SEM of experimental replicates (n = 3). (B) Ligand affinities (Ki values) from the fitted TR-FRET data; data represent the mean ± SD of 2 independent experiments. (C) PPARγ LBD thermal transition midpoint receptor stability temperatures from fitted differential scanning calorimetry (DSC) data (n = 1). (D) Cell-based luciferase assay reporting on transcription of the PPARγ LBD fit to a sigmoidal dose–response equation; error bars, data represent the mean ± SEM of experimental replicates (n = 4). (E) Plot of ligand affinity from B vs. ligand potency from the TR-FRET coactivator recruitment assay from Fig. 2D fit with a linear regression equation. (F) Plot of receptor stability from C vs. ligand affinity from B fit with a linear regression equation. (G and H) Plot of transcriptional window of efficacy via maximum luciferase activity from the highest ligand concentration and associated errors from D vs. (G) ligand potency in the TR-FRET coactivator recruitment assay from Fig. 2D and (H) ligand affinity in the TR-FRET ligand displacement assay from B fit with a linear regression equation.
Crystal Structures Provide Some Insight into TZD Affinity but Not Efficacy.
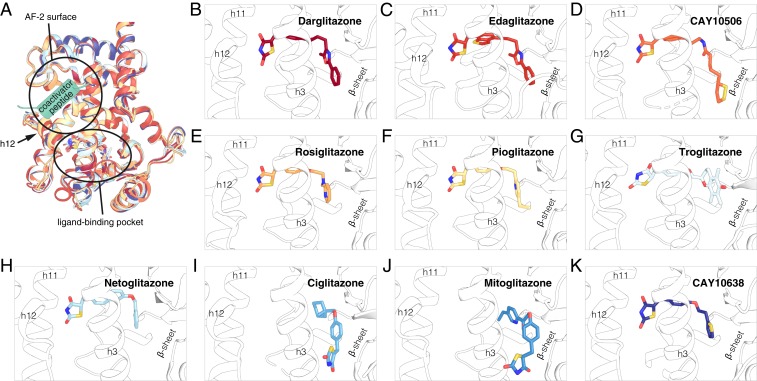
To gain insight into the structural basis for the varied TZD affinity, potency, and functional efficacy, we compared crystal structures of PPARγ LBD bound to each of the TZDs. We solved crystal structures of 6 complexes (SI Appendix, Table S1) of PPARγ LBD bound to darglitazone (Protein Data Bank [PDB]: 6DGL; 1.95-Å resolution) (19), CAY10506 (PDB: 6DGQ; 2.45-Å resolution) (20), troglitazone (PDB: 6DGO; 3.10-Å resolution) (21), ciglitazone (PDB: 6O68; 2.78-Å resolution) (22), mitoglitazone (PDB: 6O67; 2.52-Å resolution) (23), and CAY10638 (PDB: 6DGR; 2.15-Å resolution) (24). We compared our structures to other previously solved PPARγ LBD crystal structures bound to edaglitazone (PDB: 5UGM; 2.1-Å resolution) (25), rosiglitazone (PDB: 4EMA; 2.55-Å resolution) (26), pioglitazone (PDB: 5Y2O; 1.80-Å resolution) (27), and netoglitazone/MCC-155 (PDB: 3B0Q; 2.10-Å resolution). This enabled a complete X-ray crystallographic analysis of the TZD series (Fig. 4A). In all of the structures, PPARγ LBD crystallized with 2 molecules configured as a homodimer in the asymmetric unit, although in solution PPARγ LBD is monomeric (28). Similar to other PPARγ LBD crystal structures solved in the absence of coregulator peptide, in chain A the critical switch element for activation of PPARγ transcription, helix 12, adopts an active conformation. The conformation of helix 12 in chain B is atypical and influenced by a crystallization artifact; it binds to the AF-2 surface of an adjacent molecule (chain A′), which likely stabilizes the active conformation of helix 12 in chain A.
Fig. 4.
X-ray crystallography analysis of the TZD series. (A) Structural overlay of the 10 TZD-bound PPARγ LBD structures (chain A). (B–K) Ligand binding poses of the TZD series; (B) darglitazone (PDB 6DGL), (C) edaglitazone (PDB 5UGM), (D) CAY10506 (PDB 6DGQ), (E) rosiglitazone (PDB 4EMA), (F) pioglitazone (PDB 5Y2O), (G) troglitazone (PDB 6DGO), (H) netoglitazone (PDB 3B0Q), (I) ciglitazone (PDB 6O68), (J) mitoglitazone (PDB 6O67), and (K) CAY10638 (PDB 6DGR). The cartoon structures in A are colored to their corresponding ligand colors in the binding pose views in B–K.
All ligands in the TZD series show clear density in the chain A molecule (Fig. 4 B–K and SI Appendix, Figs. S3 and S4). For 8 of the 10 ligands (all except ciglitazone and mitoglitazone), the TZD head group associates near helix 12, a region of the ligand-binding pocket called the helix 12 subpocket herein, and forms hydrogen bond contacts with the side chains of up to 4 nearby residues Ser289, His323, His449, and Tyr473. Among these ligands, there is a trend in the ligand binding poses whereby the higher affinity, most potent ligands display weaker polar interactions with the side chains of Cys285 and/or Gln286 and contain longer tail groups that wrap around helix 3 and form water-mediated polar interactions with residues in the β-sheet. In contrast to these 8 ligands that adopt “canonical” binding modes, ciglitazone and mitoglitazone, 2 of the least potent ligands in the series, show alternate binding modes in the chain A active conformation where their TZD head groups associate near the flexible, solvent accessible Ω-loop and their relatively shorter extended tail groups insert into the orthosteric pocket alongside the β-sheet surface. Ligand binding to this alternate site has been observed in several other structural studies (29–34). The alternate binding modes of ciglitazone and mitoglitazone may originate from their lower affinity, and in the case of ciglitazone, a lack of atoms in its shorter tail group capable of forming water-mediated polar interactions with residues in the β-sheet (35). Although the significance of these alternate TZD binding modes is not yet clear, it is possible they represent an initial encounter complex binding mode before transitioning to the orthosteric ligand-binding pocket.
Whereas the structural contributions to the graded ligand binding affinity within the TZD series may be apparent from the crystallized ligand binding modes (Fig. 4 B–K), the structures do not explain the graded functional efficacy of the TZD series. In principle, the differences in graded efficacy should manifest in conformational differences in the structural elements that comprise the AF-2 coregulator interaction surface. However, there are no obvious structural changes in the AF-2 surface among the crystal structures for this TZD series (Fig. 4A). This is likely due to the aforementioned helix 12/AF-2 surface crystal contacts, although another contribution could be that the crystallized conformations do not fully represent the conformational ensemble of PPARγ in solution. Namely, the crystallized conformations we captured may represent a low energy minima of a broader energy landscape that are not sensitive to differences caused by ligand binding in X-ray data collected under cryogenic temperatures (36–39).
NMR Reveals Graded TZD Potency and Efficacy Are Correlated to Receptor Active State.
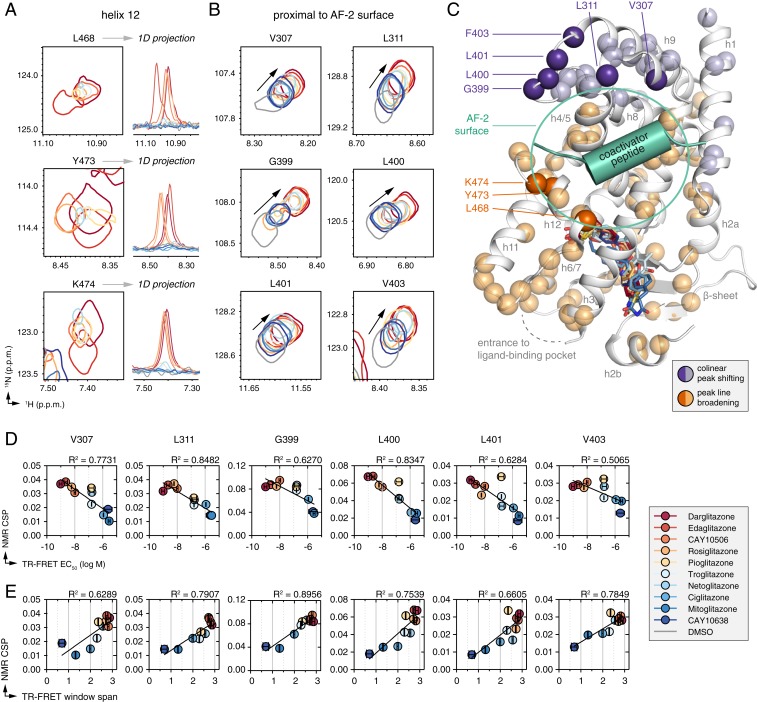
Solution NMR spectroscopy is a powerful approach to study dynamic allosteric properties in proteins (40). We therefore envisioned that NMR may detect a shift in the PPARγ LBD conformational ensemble toward the fully active state (R/LR → graded activity conformations → LR*) for the TZD series in a manner consistent with the theoretical models of receptor activation, which relate the rate of transformation of the receptor conformational ensemble from a ground state to the ligand-bound active state (17, 41). That is, ligands with higher potency and efficacy would shift the conformational ensemble of PPARγ more toward an “active” state than ligands with less potency and efficacy.
Previous NMR studies have shown that the ligand-binding pocket and helix 12 of apo-PPARγ LBD is dynamic and switches between 2 or more conformations on the microsecond-to-millisecond timescale, also known as the intermediate-exchange NMR timescale, resulting in the appearance of approximately one-half of the expected NMR peaks (18, 42). Exchange between these 2 conformations—thought to be one that is similar to a transcriptionally active conformation where the AF-2 surface can bind coactivator and another that is similar to a transcriptionally repressive conformation where the AF-2 can bind corepressor—is the likely origin of the NMR peak broadening in the apo-state. NMR peaks missing in the apo-form, which include residues within ligand-binding pocket and the AF-2 coregulator interaction surface (helix 3, helix 4/5, and helix 12), are stabilized upon binding potent full agonists that robustly activate PPARγ but remain absent, or persist, upon binding potent non-TZD partial agonists that weakly activate PPARγ (18, 42–44). Agonist binding selects for a transcriptionally active conformation, so there could be at least 2 potential origins of the NMR peak broadening when bound to partial agonists: broadening due to protein dynamics in the ligand-bound state, and “competition” between the less potent agonists binding to the active conformation and the repressive conformation. In either case, microsecond-to-millisecond timescale dynamics that persist in the AF-2 surface in the apo-form and ligand-bound forms cause less favorable interaction with coactivators (45).
We performed differential NMR analysis comparing 2D [1H,15N]-transverse relaxation optimized spectroscopy (TROSY)–heteronuclear single quantum coherence (HSQC) NMR spectra of 15N-labeled PPARγ LBD bound to each ligand within the TZD series to the apo-form under the same ligand vehicle DMSO condition. Ligands within the series contain different extended side-chain moieties, including various aromatic groups that interact with the β-sheet region and wrap around helix 3 resulting in ring current effects that impart unpredictable nonlinear NMR peak shifting for residues within this region of the pocket. NMR peak shifting is also observed for residues near the ligand entry/exit site, likely due in part to slight differences in the relative binding mode of the ligands. Despite these general binding mode-induced NMR peak shifting, 2 general NMR observable conformational trends are apparent for the TZD series.
We found that the less potent, and therefore less efficacious, ligands within the series that caused partial or graded functional efficacy resulted in NMR peak line broadening (Fig. 5A and SI Appendix, Fig. S5), indicative of microsecond-to-millisecond timescale dynamics, that persist from the apo-form for residues within the ligand entry/exit site, ligand-binding pocket, and structural elements comprising the AF-2 coregulator interaction surface (Fig. 5C). The structural regions with persistent microsecond-to-millisecond timescale dynamics when bound to the less potent TZDs comprise similar structural regions with persistent microsecond-to-millisecond timescale dynamics when bound to potent non-TZD partial agonists. In the case of the less potent TZD ligands, the persistent microsecond-to-millisecond timescale dynamics likely originates from relatively fast ligand off-exchange compared to the more potent TZDs (46). However, for the potent non-TZD partial agonists, the persistent microsecond-to-millisecond timescale dynamics likely originates from a different mechanism not related to ligand off-exchange; these ligands were developed to retain high affinity but lack a TZD-comparable head group capable of forming hydrogen bond contacts to residues within the pocket and on helix 12 that stabilize the AF-2 surface. Thus, when bound to PPARγ at high affinity, the non-TZD partial agonists enable the dysfunctional microsecond-to-millisecond timescale dynamics present in the apo-form to persist.
Fig. 5.
NMR-detected changes in the PPARγ LBD conformational ensemble correlate with graded ligand potency and efficacy. (A and B) Snapshots of [1H,15N]-TROSY-HSQC NMR spectra of 15N-labeled PPARγ LBD that show NMR changes as a function of graded potency and efficacy, including residues (A) in helix 12 that show NMR peak line broadening, along with 1D projections; or (B) proximal to the AF-2 coactivator interaction surface that show colinear shifting. (C) Analysis of all well-dispersed NMR peaks with reasonable peak separation that could be faithfully analyzed shows widespread correlations throughout the PPARγ LBD that group into 2 surfaces sensitive either to NMR peak line broadening or colinear shifting. The dark orange and dark purple spheres correspond to residues shown in A and B; the light orange and light purple spheres correspond to other residues that also show line broadening (SI Appendix, Fig. S5) or colinear shifting (SI Appendix, Fig. S6) in the TZD series. (D and E) Plot of NMR chemical shift perturbation for each liganded state shown in B relative to the apo-protein state vs. (D) ligand potency from Fig. 2D and (E) ligand efficacy in the TR-FRET coactivator recruitment assay from Fig. 2E fit with a linear regression equation.
In contrast to these NMR line broadening changes, we observed colinear NMR chemical shift perturbations for residues distal from the ligand-binding pocket but proximal to the AF-2 surface (Fig. 5B and SI Appendix, Fig. S6), which includes residues that form a surface connected by helix 1, helix 4/5, helix 8, helix 8–9 loop, and helix 9 (Fig. 5C). In most cases, one NMR peak is visible for each ligand-bound state; however, in select cases for a few ligands with intermediate or weak potency, a combination shifting, line broadening, and 2 receptor populations are observed. The coincident shifting and broadening suggest conversion on the fast (peak shifting) to intermediate (peak broadening) NMR timescale. The 2 receptor populations observed for residues in the helix 8–9 loop (G399 and L400) indicate there is exchange between 2 ligand-bound receptor populations (SI Appendix, Fig. S7). One potential contribution to this phenomenon is the racemic nature of the TZD head group, which isomerize between (R)- and (S)- conformers, each of which have different affinities for binding PPARγ. A previous study used chromatographic methods to separate rosiglitazone enantiomers for use in a ligand displacement assay (47). (S)-Rosiglitazone displayed higher affinity than (R)-rosiglitazone (30 nM vs. 2 μM, respectively), although after separation the ligands spontaneously racemize with a half-life of ∼3 h, which precludes a detailed NMR analysis of the individual isomers. Using molecular docking approaches, the (S)-pioglitazone is also predicted to display higher affinity than (R)-pioglitazone (35). Thus, if the overall racemic affinity ratio, (S)/(R)- for intermediate-affinity TZDs becomes closer to 1, the difference in affinity between isomers becomes less significant allowing both to bind. In this scenario, the different TZD head group isomers will interact differently with the receptor, which could in principle cause the peak doubling observed that correspond to 2 receptor populations. A second potential contribution is that possible isomerization of a proline residue (P398) nearby G399 and L400 may occur in a ligand-specific manner and contribute to the 2 populations. Notwithstanding these limitations, we found that the degree to which the NMR peaks shift colinearly from a ground state conformation (apo-form) toward an “active” conformation (i.e., toward the NMR peaks corresponding to the most efficacious TZDs) is correlated to ligand potency (Fig. 5D) and efficacy (Fig. 5E) in the TR-FRET coactivator recruitment assay.
There are a few limitations to our quantitative NMR chemical shift analysis. For the 2 residues that display 2 receptor populations in certain ligand-bound states (G399 and L400), we used most abundant receptor population (i.e., NMR chemical shift values of the peak with the highest intensity) in the analysis, rationalizing that this conformation may dominate in functional assays. In these cases, the less abundant population also falls along the colinear shifting trajectory, indicating this conformation may be more (e.g., CAY10638) or less (e.g., rosiglitazone, troglitazone, pioglitazone, netoglitazone) functionally active than the most abundant conformation (SI Appendix, Fig. S7). Another limitation is that for the bound occupancy of the lower affinity ligands will be lower (∼97 to 98%) than the higher affinity ligands (>99%) under the conditions we performed NMR, which could in principle affect the NMR peak positions and cause extra NMR signal broadening induced by the exchange between free and bound states. However, no significant spectral changes are observed for CAY10638 between 1 and 4 molar equivalents (SI Appendix, Fig. S8), suggesting that any effects caused by a 1 to 3% ligand-free population are likely minor. This analysis also assumes that the exchange rate (Kex) between the 2 states is far larger than the chemical shift difference in all cases, and that the chemical shift values of the 2 exchanging states are not largely affected by the bound ligands.
One interesting observation is that the NMR chemical shift values are proportional to the log EC50 values of the ligands as opposed to the (nonlog) EC50 values. A similar relationship was noted in a study of the ligand dissociation mechanism of dihydrofolate reductase where a series of inhibitors displayed a log-linear relationship between an NMR measured rate parameter with Ki and Koff (46). In our study, the log-linear relationship could indicate there is a conformational exchange process associated with reaching the ligand-bound active state that is not directly measured in our experiments, such as a competition between ligand binding to a transcriptionally active state with a transcriptionally repressive state.
Ligand Efficacy Is Correlated to Receptor Active State within a Larger Set of Structurally Diverse Ligands.
We wondered whether there may be a correlation between ligand efficacy and receptor conformation within a different set of structurally diverse ligands. We assessed 17 natural/endogenous and synthetic PPARγ ligands not previously reported to span a range of graded transcriptional activation efficacy (SI Appendix, Fig. S9). Unlike the TZD series, which share a common head group that interacts with the helix 12 subpocket, this diverse ligand set contains different types of head groups and chemotypes, some capable of hydrogen bonding to the helix 12 subpocket and others lacking such moieties that function primarily as partial agonists. Given the diversity of this ligand set, some of which were optimized for high-affinity binding with low transcriptional efficacy by incorporating different types of head groups, we initiated these studies understanding that ligand potency will not be correlated to efficacy as we observed in the more structurally conserved TZD series.
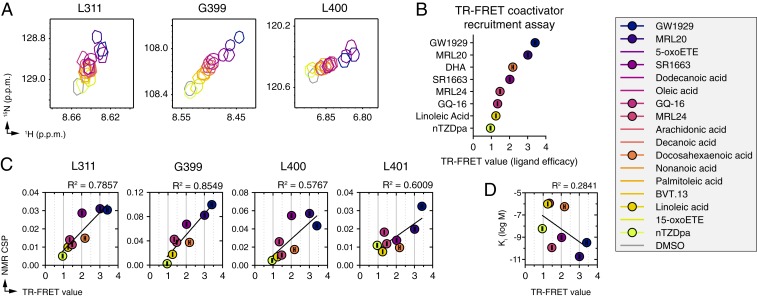
We compared 2D [1H,15N]-TROSY-HSQC NMR spectra of 15N-labeled PPARγ LBD bound to each ligand within the structurally diverse set. Due to the diversity of chemotypes present in this ligand set, there are larger differences in the NMR spectra compared to the TZD series. However, several residues with well-resolved NMR peaks that showed colinear shifting in the TZD series also showed colinear shifting in this structurally diverse ligand set (Fig. 6A), indicating a correlation between ligand efficacy and receptor conformation. Compared to the TZD series NMR data, the majority of natural/endogenous PPARγ ligands show shifted NMR peak positions indicating they are partial agonists, which is consistent with the idea that PPARγ displays basal transcriptional activity and can be further activated by synthetic agonists (25). To assess this more directly, we performed our TR-FRET coactivator recruitment assay using a single ligand concentration (5 μM) to assess ligand efficacy (Fig. 6B). In contrast to the NMR experiments where the receptor concentration is relatively high (200 μM) and ligands added stoichiometrically result in ∼100% bound occupancy, in the TR-FRET assay the receptor concentration is low (4 nM) and ligands titrated increase binding of the peptide (efficacy) in proportion to their respective binding affinities. Because high concentrations of excess ligand can result in compound precipitation or aggregation, we limited this analysis to PPARγ ligands with affinities better than ∼1 to 2 μM (SI Appendix, Fig. S10) that would show appreciable complex formation under the TR-FRET assay conditions. For this ligand subset, we observed a correlation between TR-FRET coactivator recruitment efficacy and colinear NMR peak shifting (Fig. 6C). However, for these ligands, there is a poor correlation between TR-FRET ligand affinity and efficacy (Fig. 6D). For example, MRL24 is a partial agonist with a low TR-FRET efficacy value similar to GQ-16; however, MRL20 is much more potent than GQ-16 and is also more potent than the superagonist GW1929, which has the highest TR-FRET efficacy value. Thus, for the structurally diverse ligand set, these data show that the receptor conformation shifts from a ground state conformation (apo-form) toward an active conformation in a manner that is correlated to ligand efficacy but independent of ligand affinity.
Fig. 6.
NMR-detected correlations PPARγ LBD active state and efficacy of a structurally diverse ligand set. (A) Snapshots of [1H,15N]-TROSY-HSQC NMR spectra of 15N-labeled PPARγ LBD that show colinear NMR peak shifting for residues proximal to the AF-2 coactivator interaction surface. (B) TR-FRET assay to determine ligand efficacy for recruitment of TRAP220 coactivator peptide to PPARγ LBD at a single concentration of ligand (5 μM) fit to a sigmoidal dose–response equation; data represent the mean ± SD of experimental replicates (n = 4). In the legend, ligands with a filled circle correspond to ligands with affinities (Ki values) better than ∼1 to 2 μM (from SI Appendix, Fig. S10) that were included in the TR-FRET assay. (C) Plot of NMR chemical shift perturbation for each liganded state relative to the apo-protein state (n = 1) vs. ligand efficacy in the TR-FRET coactivator recruitment assay from B fit with a linear regression equation. (D) Binding affinity of ligands included in the TR-FRET and NMR cross-correlation analysis (SI Appendix, Fig. S10; n = 1) show poor correlation to TR-FRET efficacy. Ligands ordered and colored in the legend by the approximate degree to which the ligands shift the NMR peak in A relative to DMSO.
Discussion
The objective of receptor theory is to understand and predict the relationship between ligand potency and functional efficacy. Theoretical receptor models, which are used to extract quantitative information from functional activity studies on GPCRs, ion channels, and enzyme-linked receptors, have evolved over the years to account for new findings in membrane receptor function: the realization that GPCRs can affect multiple signaling pathways and therefore have multiple distinct active states, the discovery of orthosteric vs. allosteric ligand binding sites, among others. Computer simulations are heavily used in the development and assessment of theoretical receptor models by varying mathematical parameters to determine relationships between ligand potency, functional efficacy, and other model system parameters. However, the manner in which the ligand affects receptor conformation in these theoretical studies is a conceptual “black box”—there is no knowledge or description of how ligand binding pose or chemical modifications influence ligand binding affinity, receptor conformation, and receptor functional activity. To gain insight into this relationship, here we studied the influence of chemical modifications within a structurally related ligand series (TZDs) and a structural distinct ligand set using quantitative experimental approaches that enable assessment of ligand potency, graded functional efficacy or activity, and receptor conformation.
To our knowledge, theoretical receptor models have not been utilized or considered in studies of nuclear receptor transcription factors, but our work here shows they are relevant. We used several quantitative functional assays that report on various ligand-responsive biophysical and functional endpoints related to nuclear receptor function including direct physical readouts on the PPARγ LBD (ligand displacement and thermal stability assays) as well as coactivator affinity for and recruitment to the PPARγ LBD (TR-FRET and FP assays), the latter of which has a direct relation to cellular transcription (luciferase assay). As predicted by receptor theory, we observed correlations between the various functional activity readouts for PPARγ agonists within the structurally related TZD series. To relate the graded activities of the TZD series to receptor conformation, we performed structural analysis of ligand-bound PPARγ LBD complexes using X-ray crystallography and NMR spectroscopy. We did not observe any apparent graded activity conformations in crystal structures of PPARγ LBD bound to the TZD series. However, NMR detected a graded shift in the conformational ensemble of the PPARγ LBD toward an active conformation correlated to the degree of ligand potency and efficacy, and therefore also correlated to the other functional activity readouts (ligand affinity, receptor stability, coactivator affinity). These NMR-observed structural findings are consistent with underpinnings of allostery in receptor theory models of receptor activation. We were unable to directly measure the binding kinetics of the TZD series due to apparent fast binding kinetics in surface plasmon resonance experiments, suggesting a 2-state binding mechanism. However, assuming that all of the TZDs display similar association rate constants, the notion that all of the functional efficacy and conformational data correlate to ligand affinity indicates that ligand residence times (i.e., ligand dissociation kinetic rate constants) may be involved in the functional activities of the TZD series.
In contrast to our results with the structurally related TZD agonist series, we found that the predicted relationship between ligand potency and efficacy breaks down for structurally diverse endogenous and synthetic PPARγ agonists. On the structural level, this can be rationalized because different ligand scaffolds or small chemical modifications result in different chemical bonding patterns between ligand and receptor that can influence affinity distinct from functional efficacy. The TZD series have a conserved head group capable of hydrogen bonding to residues within the helix 12 subpocket, whereas the varied length of tail groups that contribute to ligand affinity are all structurally distal from the AF-2 surface. In contrast, the structurally distinct ligands possess different types of head groups, some capable of robustly stabilizing the AF-2 surface and others that are not. Furthermore, within this diverse ligand set, there are many other chemical variations in the tail groups that distinctly contribute to affinity. Although these diverse ligands contradicted the potency–efficacy correlation predicted by receptor theory models, our NMR analysis revealed a correlation between efficacy and graded activity conformation state for all ligands, independent of chemical structure.
Related to our work here on nuclear receptors, NMR studies have been used to define how ligand binding influences the conformational ensemble of GPCRs in ways not detectable in ligand-bound crystal structures. 19F NMR studies of the GPCRs β2-adrenergic receptor (β2AR) and adenosine A2 receptor (A2AR) revealed that pharmacologically distinct classes of ligands or nanobodies differentially stabilize the receptor conformational ensemble into G protein inactive and active states (48–54). We similarly used 19F NMR to show that pharmacologically distinct PPARγ ligands differentially affect the conformation of the AF-2 helix 12, and that the conformations we observed are predictive of ligand efficacy (55). Furthermore, other work has revealed the existence of ligand-bound GPCR conformational states that exist in the continuum between the inactive/ground state and fully active conformations previously captured in crystal structures (56–65). Among these, most related to our study here is an NMR analysis of backbone amide groups of valine residues in β1-adrenergic receptor (β1AR) bound to structurally diverse agonists and antagonists (62), which observed a heterogenous conformational response to the ligands. Within the structurally diverse β1AR ligand set, there did not seem to be any relationship between ligand affinity and functional efficacy. The conformation of several residues within the extracellular ligand binding pocket were differentially correlated to ligand affinity and ligand chemical composition, whereas the conformation of a different group of residues on the intracellular side of transmembrane helix 5 (TM5) correlated to G-protein ligand efficacy independent of ligand affinity. This is similar to our finding here that structurally distinct PPARγ agonists stabilize an active conformation to a degree correlated to their functional efficacy independent of ligand potency.
Our findings raise a question as to whether theoretical receptor models can be updated to better predict the relationship between ligand potency with functional efficacy and receptor conformation. This would not be a trivial task as it would require knowledge of ligand binding poses and a precise understanding of how small chemical modifications affect ligand affinity and stabilization of functional surfaces. However, it is difficult if not possible to predict how small chemical changes in ligand composition affect potency and efficacy (66, 67). Structural differences in how different ligand-binding proteins bind synthetic ligands could also differentially influence potency–efficacy relationships. In nuclear receptors, residues within the ligand-binding pocket that contact the ligand are connected to nearby structural elements in the AF-2 coregulator binding surface. In contrast, the extracellular ligand-binding pocket of GPCRs is structurally distant from the intracellular effector protein binding surface. It is therefore possible that small chemical differences in nuclear receptor ligands, which could impact structural elements that are closer to functional surfaces, could impact affinity–function relationships differently than membrane receptors. Nonetheless, our studies here on the PPARγ nuclear receptor as well as the aforementioned studies on GPCRs show that NMR analysis can predict ligand efficacy and provide structural insight into the influence of ligands on activity-related receptor conformational ensembles.
Methods
Materials and Reagents.
Ligands and peptides were purchased or synthesized from commercial vendors, or synthesized in-house. See SI Appendix for full details.
Protein Expression and Purification.
Human PPARγ LBD (residues 203 to 477, isoform 1 numbering) was expressed in Escherichia coli BL21(DE3) cells using autoinduction ZY media, or M9 minimal media supplemented with 15N-labeled ammonium chloride, as a tobacco etch virus-cleavable N-terminal His-tagged (6×-His) fusion protein using a pET46 Ek/LIC vector (Novagen) and purified using Ni-NTA affinity chromatography and gel filtration chromatography as previously described (18).
Biochemical Experiments.
TR-FRET, FP, biolayer interferometry, and differential scanning calorimetry experiments were performed as detailed in SI Appendix.
Cellular Transactivation.
The luciferase reporter assay was performed in HEK293T cells (ATCC CRL03216) as detailed in SI Appendix.
Crystallization and Structure Determination.
Crystal structures of PPARγ bound to ciglitazone, troglitazone, mitoglitazone, CAY10506, and CAY10638 were determined from data collected at Advanced Light Source Beamline 5.0.2 at Berkeley Center for Structural Biology (Lawrence Berkeley National Laboratory) or an in-house MicroMax007 HF X-ray generator equipped with the mar345 detector as detailed in SI Appendix.
NMR Spectroscopy.
The 2D [1H,15N]-TROSY HSQC NMR data of 15N-labeled PPARγ LBD were acquired at 298 K on a Bruker 700-MHz NMR instrument equipped with a QCI cryoprobe and analyzed NMR chemical shift assignments previously reported for ligand-bound PPARγ (18) as detailed in SI Appendix.
Data Availability.
Crystal structures generated in the current study have been deposited in the PDB under accession codes 6DGL, 6DGO, 6DGQ, 6DGR, 6O67, and 6O68. NMR chemical shift assignments used in our analysis were obtained from the Biological Magnetic Resonance Data Bank (BMRB), http://www.bmrb.wisc.edu/ (ID codes 17975, 17976, and 17977).
Supplementary Material
Acknowledgments
We thank Sarah Mosure and Paola Munoz-Tello for helpful discussions. This work was supported in part by National Institutes of Health Grants R01DK101871 (to D.J.K.) and F32DK108442 (to R.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The crystal structures generated in the current study have been deposited in the Protein Data Bank (PDB), http://www.wwpdb.org/ (ID codes 6DGL, 6DGO, 6DGQ, 6DGR, 6O67, and 6O68).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909016116/-/DCSupplemental.
References
- 1.Maehle A. H., Prüll C. R., Halliwell R. F., The emergence of the drug receptor theory. Nat. Rev. Drug Discov. 1, 637–641 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Kenakin T., Principles: Receptor theory in pharmacology. Trends Pharmacol. Sci. 25, 186–192 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Changeux J. P., Allostery and the Monod-Wyman-Changeux model after 50 years. Annu. Rev. Biophys. 41, 103–133 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Leff P., The two-state model of receptor activation. Trends Pharmacol. Sci. 16, 89–97 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Black J. W., Leff P., Operational models of pharmacological agonism. Proc. R. Soc. Lond. B Biol. Sci. 220, 141–162 (1983). [DOI] [PubMed] [Google Scholar]
- 6.Monod J., Wyman J., Changeux J. P., On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12, 88–118 (1965). [DOI] [PubMed] [Google Scholar]
- 7.Samama P., Cotecchia S., Costa T., Lefkowitz R. J., A mutation-induced activated state of the beta 2-adrenergic receptor. Extending the ternary complex model. J. Biol. Chem. 268, 4625–4636 (1993). [PubMed] [Google Scholar]
- 8.Weiss J. M., Morgan P. H., Lutz M. W., Kenakin T. P., The cubic ternary complex receptor–occupancy model I. Model description. J. Theor. Biol. 181, 151–167 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Weikum E. R., Liu X., Ortlund E. A., The nuclear receptor superfamily: A structural perspective. Protein Sci. 27, 1876–1892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boehr D. D., Nussinov R., Wright P. E., The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kar G., Keskin O., Gursoy A., Nussinov R., Allostery and population shift in drug discovery. Curr. Opin. Pharmacol. 10, 715–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casiraghi M., et al. , NMR analysis of GPCR conformational landscapes and dynamics. Mol. Cell. Endocrinol. 484, 69–77 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Willson T. M., et al. , The structure-activity relationship between peroxisome proliferator-activated receptor gamma agonism and the antihyperglycemic activity of thiazolidinediones. J. Med. Chem. 39, 665–668 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Ge K., et al. , Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417, 563–567 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Savkur R. S., Burris T. P., The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 63, 207–212 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Black J. W., Leff P., Shankley N. P., Wood J., An operational model of pharmacological agonism: The effect of E/[A] curve shape on agonist dissociation constant estimation. Br. J. Pharmacol. 84, 561–571 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenakin T., A scale of agonism and allosteric modulation for assessment of selectivity, bias, and receptor mutation. Mol. Pharmacol. 92, 414–424 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Hughes T. S., et al. , Ligand and receptor dynamics contribute to the mechanism of graded PPARγ agonism. Structure 20, 139–150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang J., Kojetin D., Crystal structure of human PPARgamma ligand binding domain in complex with darglitazone. Protein Data Bank. 10.2210/pdb6DGL/pdb. Deposited 17 May 2018. [DOI]
- 20.Shang J., Kojetin D., Crystal structure of human PPARgamma ligand binding domain in complex with CAY10506. Protein Data Bank. 10.2210/pdb6DGQ/pdb. Deposited 17 May 2018. [DOI]
- 21.Shang J., Kojetin D., Crystal structure of human PPARgamma ligand binding domain in complex with troglitazone. Protein Data Bank. 10.2210/pdb6DGO/pdb. Deposited 17 May 2018. [DOI]
- 22.Shang J., Kojetin D., Crystal structure of human PPARgamma ligand binding domain in complex with ciglitazone. Protein Data Bank. 10.2210/pdb6O68/pdb. Deposited 5 March 2019. [DOI]
- 23.Shang J., Kojetin D., Crystal structure of human PPARgamma ligand binding domain in complex with mitoglitazone. Protein Data Bank. 10.2210/pdb6O67/pdb. Deposited 5 March 2019. [DOI]
- 24.Shang J., Kojetin D., Crystal structure of human PPARgamma ligand binding domain in complex with CAY10638. Protein Data Bank. 10.2210/pdb6DGR/pdb. Deposited 18 May 2018. [DOI]
- 25.Shang J., et al. , Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPARγ. eLife 7, e43320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberato M. V., et al. , Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) γ activators and pan-PPAR partial agonists. PLoS One 7, e36297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M. A., Tan L., Yang H., Im Y. G., Im Y. J., Structures of PPARγ complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs. Sci. Rep. 7, 16837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernardes A., et al. , Low-resolution molecular models reveal the oligomeric state of the PPAR and the conformational organization of its domains in solution. PLoS One 7, e31852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes T. S., et al. , An alternate binding site for PPARγ ligands. Nat. Commun. 5, 3571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae H., et al. , Mechanistic elucidation guided by covalent inhibitors for the development of anti-diabetic PPARγ ligands. Chem. Sci. 7, 5523–5529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes T. S., et al. , Probing the complex binding modes of the PPARγ partial agonist 2-chloro-N-(3-chloro-4-((5-chlorobenzo[d]thiazol-2-yl)thio)phenyl)-4-(trifluoromethyl)benzenesulfonamide (T2384) to orthosteric and allosteric sites with NMR spectroscopy. J. Med. Chem. 59, 10335–10341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brust R., et al. , Modification of the orthosteric PPARγ covalent antagonist scaffold yields an improved dual-site allosteric inhibitor. ACS Chem. Biol. 12, 969–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jang J. Y., et al. , Structural basis for differential activities of enantiomeric PPARγ agonists: Binding of S35 to the alternate site. Biochim. Biophys. Acta. Proteins Proteom. 1865, 674–681 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Laghezza A., et al. , Identification of the first PPARα/γ dual agonist able to bind to canonical and alternative sites of PPARγ and to inhibit its cdk5-mediated phosphorylation. J. Med. Chem. 61, 8282–8298 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Mosure S. A., et al. , Structural basis of altered potency and efficacy displayed by a major in vivo metabolite of the anti-diabetic PPARgamma drug pioglitazone. J. Med. Chem. 62, 2008–2023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraser J. S., et al. , Hidden alternative structures of proline isomerase essential for catalysis. Nature 462, 669–673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraser J. S., et al. , Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 108, 16247–16252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyka M. D., et al. , Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 405, 607–618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keedy D. A., et al. , Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography. eLife 4, e07574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grutsch S., Brüschweiler S., Tollinger M.; NMR Methods to Study Dynamic Allostery , NMR methods to study dynamic allostery. PLoS Comput. Biol. 12, e1004620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenakin T., What is pharmacological “affinity”? Relevance to biased agonism and antagonism. Trends Pharmacol. Sci. 35, 434–441 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Johnson B. A., et al. , Ligand-induced stabilization of PPARgamma monitored by NMR spectroscopy: Implications for nuclear receptor activation. J. Mol. Biol. 298, 187–194 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Berger J. P., et al. , Distinct properties and advantages of a novel peroxisome proliferator-activated protein [gamma] selective modulator. Mol. Endocrinol. 17, 662–676 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Marciano D. P., et al. , Pharmacological repression of PPARγ promotes osteogenesis. Nat. Commun. 6, 7443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kojetin D. J., Burris T. P., Small molecule modulation of nuclear receptor conformational dynamics: Implications for function and drug discovery. Mol. Pharmacol. 83, 1–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carroll M. J., et al. , Evidence for dynamics in proteins as a mechanism for ligand dissociation. Nat. Chem. Biol. 8, 246–252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parks D. J., Tomkinson N. C., Villeneuve M. S., Blanchard S. G., Willson T. M., Differential activity of rosiglitazone enantiomers at PPAR gamma. Bioorg. Med. Chem. Lett 8, 3657–3658 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Liu J. J., Horst R., Katritch V., Stevens R. C., Wüthrich K., Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335, 1106–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim T. H., et al. , The role of ligands on the equilibria between functional states of a G protein-coupled receptor. J. Am. Chem. Soc. 135, 9465–9474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manglik A., et al. , Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell 161, 1101–1111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staus D. P., et al. , Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 535, 448–452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye L., Van Eps N., Zimmer M., Ernst O. P., Prosser R. S., Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 533, 265–268 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Prosser R. S., Ye L., Pandey A., Orazietti A., Activation processes in ligand-activated G protein-coupled receptors: A case study of the adenosine A2A receptor. Bioessays 39, 1700072 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Sušac L., Eddy M. T., Didenko T., Stevens R. C., Wüthrich K., A2A adenosine receptor functional states characterized by 19F-NMR. Proc. Natl. Acad. Sci. U.S.A. 115, 12733–12738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chrisman I. M., et al. , Defining a conformational ensemble that directs activation of PPARγ. Nat. Commun. 9, 1794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bokoch M. P., et al. , Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463, 108–112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kofuku Y., et al. , Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun. 3, 1045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nygaard R., et al. , The dynamic process of β2-adrenergic receptor activation. Cell 152, 532–542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kofuku Y., et al. , Functional dynamics of deuterated β2-adrenergic receptor in lipid bilayers revealed by NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 53, 13376–13379 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Okude J., et al. , Identification of a conformational equilibrium that determines the efficacy and functional selectivity of the μ-opioid receptor. Angew. Chem. Int. Ed. Engl. 54, 15771–15776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sounier R., et al. , Propagation of conformational changes during μ-opioid receptor activation. Nature 524, 375–378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isogai S., et al. , Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 530, 237–241 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Clark L. D., et al. , Ligand modulation of sidechain dynamics in a wild-type human GPCR. eLife 6, e28505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Solt A. S., et al. , Insight into partial agonism by observing multiple equilibria for ligand-bound and Gs-mimetic nanobody-bound β1-adrenergic receptor. Nat. Commun. 8, 1795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eddy M. T., et al. , Allosteric coupling of drug binding and intracellular signaling in the A2A adenosine receptor. Cell 172, 68–80.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujioka M., Omori N., Subtleties in GPCR drug discovery: A medicinal chemistry perspective. Drug Discov. Today 17, 1133–1138 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Dosa P. I., Amin E. A., Tactical approaches to interconverting GPCR agonists and antagonists. J. Med. Chem. 59, 810–840 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structures generated in the current study have been deposited in the PDB under accession codes 6DGL, 6DGO, 6DGQ, 6DGR, 6O67, and 6O68. NMR chemical shift assignments used in our analysis were obtained from the Biological Magnetic Resonance Data Bank (BMRB), http://www.bmrb.wisc.edu/ (ID codes 17975, 17976, and 17977).