Significance
The transcription factor SKN-1, the Caenorhabditis elegans ortholog of mammalian NRF2, mediates cytoprotective responses to diverse stresses to restore cellular homeostasis. We have discovered that pathogen exposure drives the rapid loss of somatic lipids in a SKN-1–dependent manner. The activation of innate immunity genes by SKN-1 facilitates resistance to pathogen-derived toxins, but this occurs at the expense of organismal lipid homeostasis, which impairs organismal health later in life. Importantly, this program is malleable and the loss of lifelong health can be restored, albeit at the cost of acute pathogen resistance. These data define a new physiological basis for the decline in health associated with pathogen infection.
Keywords: SKN-1, pathogen, H3K4me3, C. elegans, lipid metabolism
Abstract
Early host responses toward pathogens are essential for defense against infection. In Caenorhabditis elegans, the transcription factor, SKN-1, regulates cellular defenses during xenobiotic intoxication and bacterial infection. However, constitutive activation of SKN-1 results in pleiotropic outcomes, including a redistribution of somatic lipids to the germline, which impairs health and shortens lifespan. Here, we show that exposing C. elegans to Pseudomonas aeruginosa similarly drives the rapid depletion of somatic, but not germline, lipid stores. Modulating the epigenetic landscape refines SKN-1 activity away from innate immunity targets, which alleviates negative metabolic outcomes. Similarly, exposure to oxidative stress redirects SKN-1 activity away from pathogen response genes while restoring somatic lipid distribution. In addition, activating p38/MAPK signaling in the absence of pathogens, is sufficient to drive SKN-1–dependent loss of somatic fat. These data define a SKN-1– and p38-dependent axis for coordinating pathogen responses, lipid homeostasis, and survival and identify transcriptional redirection, rather than inactivation, as a mechanism for counteracting the pleiotropic consequences of aberrant transcriptional activity.
The transcription factor SKN-1, the Caenorhabditis elegans ortholog of mammalian NRF2, mediates cytoprotective responses to diverse stresses to restore cellular homeostasis (1). For example, SKN-1 induces the transcription of phase II detoxification genes during oxidative stress, promotes adaptation to proteotoxic and metabolic stress, and drives the induction of innate immune effector genes during pathogen exposure (2, 3).
Although SKN-1 activation in response to stress facilitates survival, when left unchecked, aberrant activation of SKN-1 and its mammalian ortholog NRF2 can have negative pathological outcomes in worms (4, 5) and humans (6, 7). Specifically, constitutive activation of SKN-1 shortens lifespan (5, 8) and causes a reorganization of fat from the soma to the germline, termed age-dependent somatic depletion of fat (Asdf) (4). Thus, although the activation of cytoprotective transcription factors is obligatory for maintaining homeostasis when organisms encounter stressful environments, the inability to turn off or control these transcriptional responses can be detrimental (5, 6, 8).
Here, we show that redirecting activated SKN-1 by modulating the epigenetic landscape abolishes negative metabolic outcomes associated with its aberrant activation, but at the cost of losing increased innate immune function. Specifically, abolishing H3K4me3 epigenetic marks directed SKN-1 activity away from innate immunity targets, thus, reestablishing pathogen sensitivity while also restoring health-promoting, age-dependent outcomes, including homeostatic distribution of lipids, restoration of stress resistance, and increased lifespan.
Results
SKN-1 Activation Drives the Postdevelopmental Expression of Innate Immunity Pathway Genes.
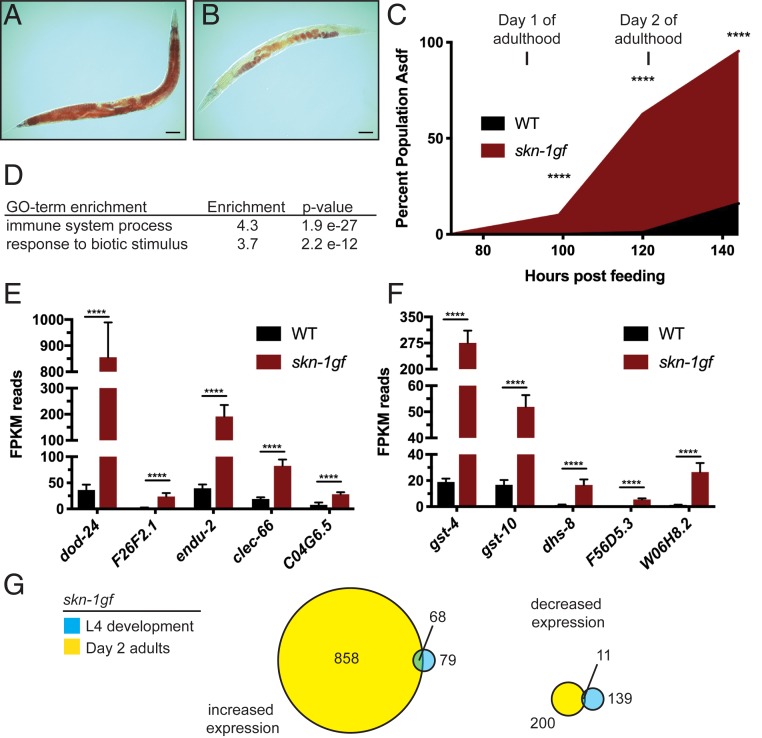
Our previous studies established that activation of SKN-1 results in the age-dependent loss of somatic lipids (4). We demonstrated this finding by activating SKN-1 using environmental factors, loss of a negative regulator, or in a skn-1(lax188) gain-of-function (gf) mutant (skn-1gf). To define the extent of transcriptional dysregulation in skn-1gf mutants at the time of somatic lipid depletion, we performed RNA sequencing (RNA-seq) on day 2 adult worms, when these animals begin to display the Asdf phenotype (4) (Fig. 1 A–C and SI Appendix, Fig. S1). The skn-1gf mutants display dysregulation of 1,986 genes (1,376 up-regulated and 610 down-regulated) as compared to wild type (WT) (Dataset S1).
Fig. 1.
SKN-1 activation causes redistribution of somatic lipids and activation of immune defense genes. (A) Normal lipid distribution in wild-type animals (representative image from n = 873) as compared to (B) somatic depletion of fat (Asdf) in animals with activated SKN-1 (representative image from n = 759) that occurs with age (C). (D) GO-term enrichment analysis of differentially expressed genes from RNA-seq of day 2 adult skn-1gf mutant animals as compared to age-matched WT controls. Analysis of the mRNA reads of the indicated genes related to (E) innate immunity and pathogen resistance and (F) oxidative stress and xenobiotic responses; see Dataset S1 for all RNA-seq measurements. (G) Venn diagram of gene expression changes with at least 2-fold change in skn-1gf(lax188) mutants during development (L4 stage) and day 2 of adulthood. See also Datasets S1 and S4 for all RNA-seq measurements. (Scale bar, 50 μm.) ****P < 0.0001. FPKM, fragments per kilobase of transcript per million mapped reads.
Interestingly, genes induced in skn-1gf mutants were strongly enriched for immune and pathogen response by Gene Ontology (GO) analysis (Fig. 1 D and E and Dataset S1), in addition to oxidative stress response and xenobiotic detoxification (Fig. 1 D–F and Dataset S1). Of note, the major SKN-1 reponsive genes were represented in this dataset (7–11) (Dataset S1). The transcription profile of day 2 adults was distinct from previous transcriptional analyses of skn-1gf mutants during development (8), which do not display GO-term enrichment for innate immunity and pathogen-related genes (Fig. 1G and Dataset S1). This finding suggests that the age-related negative outcomes of skn-1gf mutants, like the loss of somatic fat at day 2 of adulthood, may stem from an increase in expression of innate immune response genes.
H3K4 Methylation by WDR-5 Is Required for SKN-1–Dependent Loss of Somatic Lipids.
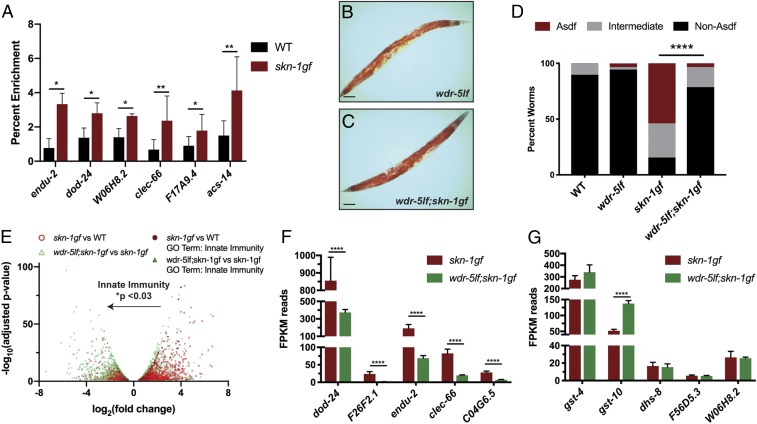
We performed chromatin immunoprecipitation (ChIP) followed by qPCR of SKN-1 and SKN-1gf protein at day 2 of adulthood and observed enrichment of SKN-1gf relative to wild-type SKN-1 at the promoters of several genes, which we identified by RNA-seq (Fig. 1 and Dataset S1), selecting representative genes from the innate immunity, oxidative stress, and metabolism GO classes, including: endu-2, dod-24, W06H8.2, clec-66, F17A4.9, and acs-14 (Fig. 2A and Dataset S1). We hypothesized that manipulating epigenetic modifications to restrict the transcriptional activity of the skn-1gf mutants could mitigate the pleiotropic effects of SKN-1 constitutive activation. Methylation of Histone H3 at lysine 4 (H3K4me) is an established effector of transcriptional activity and several conserved protein complexes regulate H3K4 di- and trimethylation states (12). With this in mind, we screened a panel of epigenetic chromatin modifiers by RNA interference (RNAi). We found that reducing the expression of wdr-5 or rbbp-5—2 SET1/MLL-like proteins that influence the H3K4me3 state—restored somatic fat in skn-1gf mutant animals (SI Appendix, Fig. S2). This finding suggests that H3K4me3 mediates physiological outcomes in the skn-1gf mutant.
Fig. 2.
Loss of Histone H3 trimethylation restricts SKN-1gf transcriptional activity and suppresses the loss of somatic lipids. (A) ChIP-qPCR reveals SKN-1gf enrichment at the promoter of target genes relative to wild-type SKN-1. (B and C) ORO staining of lipid stores in C. elegans. (B) wdr-5lf(ok1417) mutants display normal distribution of lipids across tissues (representative image from n = 415). (C) The loss of somatic lipid stores observed in SKN-1gf mutant animals is restored in wdr-5lf(ok1417);skn-1gf(lax188) double mutants (representative image from n = 892). (D) Quantification of lipid distribution across tissues. All experiments were performed in a minimum of 3 biological replicates and lipid distribution was assessed in at least 300 animals. (See also SI Appendix, Fig. S3 for all measurements.) (E) Volcano plot of differentially expressed genes in skn-1gf compared to WT (red), genes altered by loss of wdr-5lf (green), and innate immunity genes (black); changes in immune genes analyzed by unpaired, nonparametric, t test (Mann–Whitney). (F and G) Analysis of the mRNA reads of the indicated genes related to (F) innate immunity and pathogen resistance and (G) oxidative stress and xenobiotic responses. See Datasets S2 and S4 for RNA-seq measurements. (Scale bar, 50 μm.) *P < 0.05; **P < 0.01; ****P < 0.0001. FPKM, fragments per kilobase of transcript per million mapped reads.
We confirmed this genetic relationship in wdr-5lf(ok1417);skn-1gf double mutants at day 2 of adulthood, noting that the wdr-5lf mutant had wild-type lipid distribution between the soma and germline and that skn-1gf does not impact H3K4me3 levels in the wdr-5lf background (Fig. 2 B–D and SI Appendix, Fig. S3 A and B). In support of the molecular connection between SKN-1gf activity and H3K4me3 epigenetic marks, the loss of wdr-5 attenuated the shortened lifespan (8) phenotype of skn-1gf mutants (SI Appendix, Fig. S3C). As previously documented, the skn-1gf(lax188) allele enhances resistance to hydrogen peroxide exposure when reproduction begins (SI Appendix, Fig. S3D), but this resistance is lost when reproduction ceases, as somatic lipids are depleted (SI Appendix, Fig. S3E) (4). Loss of wdr-5 restored resistance to acute oxidative stress in the skn-1gf mutant background at this later stage of life. Recent studies have linked oxidative stress (13) and lipid mobilization (14) to cold stress tolerance. Similar to the oxidative stress responses, skn-1gf mutant animals at day 2 of adulthood were more sensitive to exposure at 2 °C than wild-type animals and this sensitivity was suppressed in the absence of wdr-5 (SI Appendix, Fig. S3 F and G); thus, cold stress resistance is associated with the abundance of somatic lipids. It should be noted that both wdr-5lf and wdr-5lf;skn-1gf worms develop at a slightly slower rate as compared to wild-type and skn-1gf animals (SI Appendix, Fig. S3H). However, even if examined 24 h later, at day 3 of adulthood, wdr-5lf;skn-1gf worms continue to display a suppression of Asdf (SI Appendix, Fig. S3I). Taken together, these data reveal the importance of H3K4me3 marks for SKN-1–dependent metabolic and stress responses.
Loss of H3K4 Trimethylation Impacts SKN-1 Activation of Innate Immunity Genes.
Next, we performed RNA-seq analysis on wdr-5lf;skn-1gf double mutants to measure the impact of loss of H3K4me3 on SKN-1 transcriptional activity (Fig. 2E and Dataset S2). The genes with the most significant reduction in expression, when compared to skn-1gf mutants, were the pathogen resistance GO-term class of genes (Fig. 2F, SI Appendix, Fig. S4A, and Dataset S2). Surprisingly, the expression of oxidative and redox homeostasis genes remained largely unchanged (Fig. 2G, SI Appendix, Fig. S4B, and Dataset S2). These data revealed that H3K4me3 can influence the transcriptional focus of constitutively activated SKN-1 between classes of target genes and that redirecting the transcriptional focus away from pathogen response genes, but not oxidative stress genes, can alleviate the negative metabolic outcomes of deregulated SKN-1 transcriptional activity (SI Appendix, Fig. S4C).
Redirection of Activated SKN-1 from Innate Immunity Genes Abates Metabolic Dysfunction.
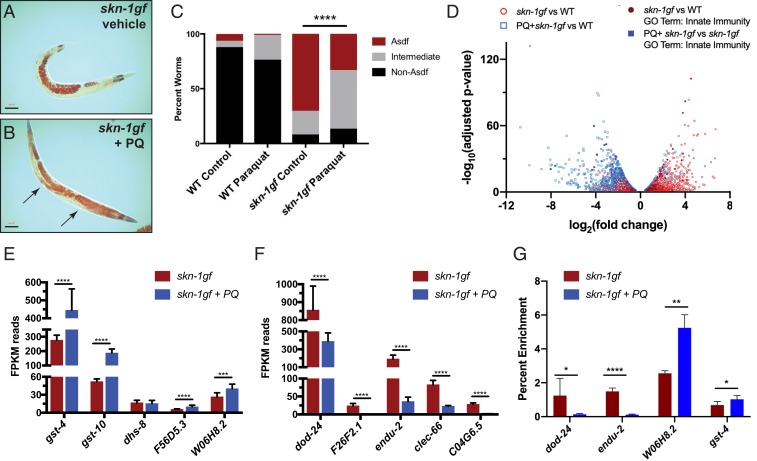
Our finding that oxidative stress gene targets were generally unaltered in the wdr-5lf;skn-1gf double mutants was unexpected, given the established role of H3K4me3 as a transcriptionally activating epigenetic mark and the impact its loss has on skn-1gf stress responses. Our previous study has shown that acute exposure to hydrogen peroxide can induce a loss of fat phenotype in wild-type animals (4). We therefore examined how continuous expression of oxidative stress response genes would affect fat levels in an age-dependent manner. We used a subliminal amount of the superoxide-generating oxidant paraquat (PQ) (75 μM), in order to chronically induce an oxidative stress response, and found no apparent change in somatic or germline lipid pools in wild-type animals (SI Appendix, Fig. S5A). Unexpectedly, this treatment suppressed the Asdf phenotype of skn-1gf animals, restoring their somatic lipid stores (Fig. 3 A–C and SI Appendix, Fig. S5B). We next compared the transcriptional profiles of skn-1gf mutant animals treated with paraquat to vehicle treatment (Fig. 3D and Dataset S3). Exposure to paraquat maintained, and for some genes enhanced, the expression of SKN-1–dependent antioxidant pathway genes (Fig. 3E, SI Appendix, Fig. S6A, and Dataset S3) but, remarkably, paraquat treatment reduced the expression of pathogen response genes that are activated in skn-1gf (Fig. 3E, SI Appendix, Fig. S6B, and Dataset S3). Consistent with this finding, chromatin immunoprecipitation revealed that the association of SKN-1gf with the promoter regions of innate immunity genes was markedly reduced (Fig. 3G), utilizing dod-24 and endu-2 as representative reporters of innate immunity and SKN-1 responsive genes (Fig. 1E) (8, 10, 15, 16). Importantly, association of SKN-1gf with the promoter of the oxidative stress genes gst-4 and W06H8.2, previously identified as SKN-1 responsive to oxidative stress (17), was enhanced with PQ treatment (Fig. 3G). SKN-1 regulates transcriptional targets that perform highly diverse functions (8, 10, 18–21), but these data reveal that even when activated, the protective responses regulated by SKN-1 can be refined to suit the current need to restore homeostatic balance (SI Appendix, Fig. S6C). Moreover, our discovery reveals the important finding that the focus of activated transcription factors, as probed using SKN-1gf mutants, can be interrupted and redirected to improve health.
Fig. 3.
Oxidative stress redirects SKN-1gf transcriptional activity while restoring somatic lipid distribution. (A–C) The absence of somatic lipids in SKN-1gf mutants treated with vehicle (control) (A) is restored in animals exposed to 75 µM PQ (B) (representative image from n = 237); arrows indicate somatic lipids. (C) Quantification of lipid distribution across tissues. All experiments were performed in a minimum of biological triplicates and lipid distribution was assessed in at least 300 animals. (See also SI Appendix, Fig. S5 and Dataset S3 for all measurements.) (D) Volcano plot of differentially expressed genes in skn-1gf compared to WT (red), genes altered by PQ exposure (blue), and innate immunity genes (black); changes in immune genes analyzed by unpaired, nonparametric, t test (Mann–Whitney). (E and F) Analysis of the mRNA reads of the indicated genes related to (E) oxidative stress and xenobiotic responses and (F) innate immunity and pathogen resistance. See also Datasets S3 and S4 for RNA-seq measurements. (G) ChIP-qPCR reveals PQ treatment redirection of SKN-1gf activity is associated with a loss of recruitment at innate immunity gene promoters. (Scale bar, 50 μm.) *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. FPKM, fragments per kilobase of transcript per million mapped reads.
Pathogen Exposure Drives the Rapid Loss of Somatic Lipids.
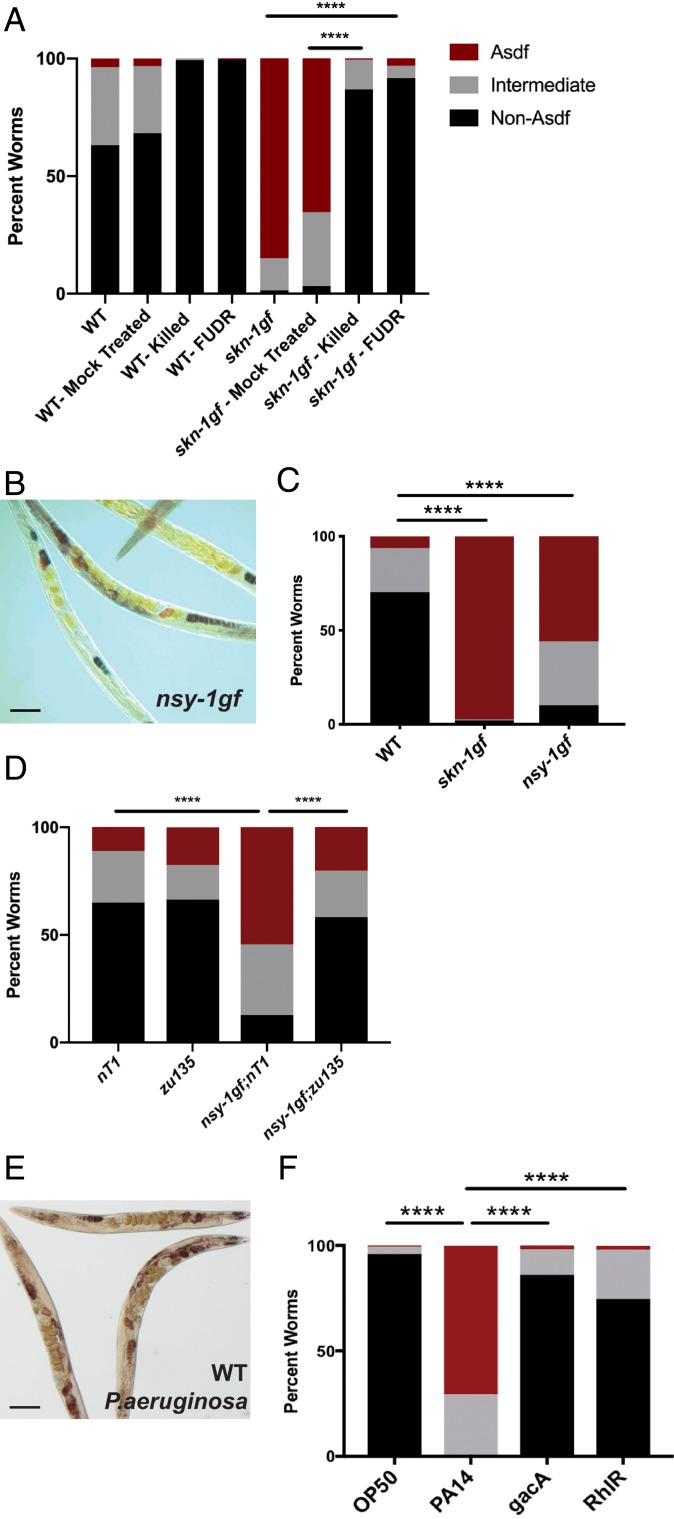
Because C. elegans feed on bacteria, they represent an established model to study host–microbe interactions (22). Although live bacteria are routinely used as a food source, C. elegans can be maintained on dead bacteria and the loss of colonization and proliferation of bacterial cells in the intestine can increase lifespan (23), supporting the notion that, compared to other microbial diets, the standard OP50 Escherichia coli diet is modestly pathogenic (23, 24). With this in mind, we examined lipid distribution in skn-1gf animals raised on dead bacteria and observed suppression of Asdf (Fig. 4A). We also examined animals moved to plates with 5-fluoro-2′-deoxyuridine (FUdR)-treated bacteria, which inhibits bacterial proliferation, and observed a similar suppression of Asdf (Fig. 4A). Taken together, these data suggest that the mobilization of lipids from the soma to the germline at the end of reproduction in our skn-1gf animals requires an interaction with live and proliferating bacteria.
Fig. 4.
Exposure to pathogens drives a rapid and SKN-1–dependent loss of somatic lipids. (A) Somatic lipid depletion requires live bacteria. (A) Bactericidal treatment of OP50 bacteria with UV+antibiotics (“killed”) or FUdR reduces Asdf in skn-1gf animals relative to mock-treated bacteria. (B and C) Constitutive activation of the p38 MAPK pathway in nsy-1gf(ums8) mutant animals displays age-dependent somatic depletion of fat (Asdf) at 144 h postfeeding; representative image (B) and quantification of population (C). (D) Lipid reallocation in nsy-1gf(ums8) mutant animals requires skn-1. (E and F) Exposure of 72-h postfed animals to P. aeruginosa results in the rapid loss of somatic lipid stores; representative image (E) and quantification of population (F). All experiments were performed in a minimum of biological triplicate and lipid distribution was assessed in at least 200 animals. (Scale bar, 50 μm.) ****P < 0.0001.
Based on this finding, we explored whether immune activation itself was sufficient to drive lipid redistribution. In C. elegans, the p38 MAPK PMK-1 pathway is a canonical regulator of innate immune defenses (25–27). Following pathogen exposure, NSY-1/MAPKKK (the C. elegans ASK1 homolog) phosphorylates a p38 MAPK cascade with one of the final targets being SKN-1 (2, 3, 28). We first examined the lipid distribution of animals harboring a nsy-1gf allele, which drives constitutive immune activation (29). Interestingly, nsy-1gf animals mobilize lipids from the soma to the germline in an age-dependent manner, just like the Asdf phenotype observed in skn-1gf mutants (Fig. 4 B and C and SI Appendix, Fig. S7A). Importantly, this loss of fat phenotype requires SKN-1 activity as both skn-1(zu135) loss-of-function (lf) allele and skn-1 RNAi suppressed lipid redistribution in the nsy-1gf mutant background, thus confirming the placement of SKN-1 downstream of NSY-1/MAPKKK signaling for changes in metabolic homeostasis (Fig. 4D and SI Appendix, Fig. S7B). Taken together with the transcriptome profiling data of the skn-1gf mutants, these data indicate that activation of SKN-1 by the NSY-1/PMK-1 pathways and the subsequent transcription of innate immunity target genes, drives the loss of somatic lipids.
To determine whether physiological activation of innate immune defenses in the context of pathogen infection is sufficient to drive changes in metabolic homeostasis, we exposed wild-type and skn-1gf mutant animals to the opportunistic human pathogen Pseudomonas aeruginosa, which also infects and kills nematodes (22, 30–32). Postdevelopmental exposure to P. aeruginosa resulted in the rapid depletion of somatic lipids (Fig. 4 E and F and SI Appendix, Fig. S8A). Importantly, exposure to the virulence-attenuated pseudomonal mutant gacA or rhlR (31, 33, 34), which do not robustly activate C. elegans immune defenses (35), did not cause loss of somatic fat (Fig. 4F and SI Appendix, Fig. S8 B and C). We previously demonstrated that the loss of somatic lipids in skn-1gf mutants was mediated by lipid transport from the soma to the germline by vitellogenins (4). Similarly, vit-5 RNAi-treated animals displayed reduced somatic fat loss when exposed to P. aeruginosa, indicating a shared mechanism of lipid loss between skn-1gf mutants and pathogen exposure (SI Appendix, Fig. S8D). In addition, loss of somatic lipids during infection with P. aeruginosa was accelerated in skn-1gf mutants (SI Appendix, Fig. S8E) and delayed in skn-1 RNAi-treated animals (SI Appendix, Fig. S8 F and G). It is noteworthy that the genes with the largest changes in expression in wild-type animals exposed to pathogens are also activated in skn-1gf mutant animals on nonpathogenic bacteria (e.g., fmo-2, clec-71, cpt-4, sodh-1, clec-60, cyp-34A4, F53A9.8, C34C6.7, T07G12.5, M60.2, ugt-18, and Y65B4BR0.4; for full list see Dataset S1) (16, 25, 36). Taken together, these findings demonstrate that the immune response to pathogenic bacteria involves SKN-1–dependent redistribution of somatic lipids and provides evidence of a sophisticated relationship between these previously unconnected essential aspects of organismal homeostasis.
Somatic Lipid Abundance Is Associated with Pathogen Sensitivity.
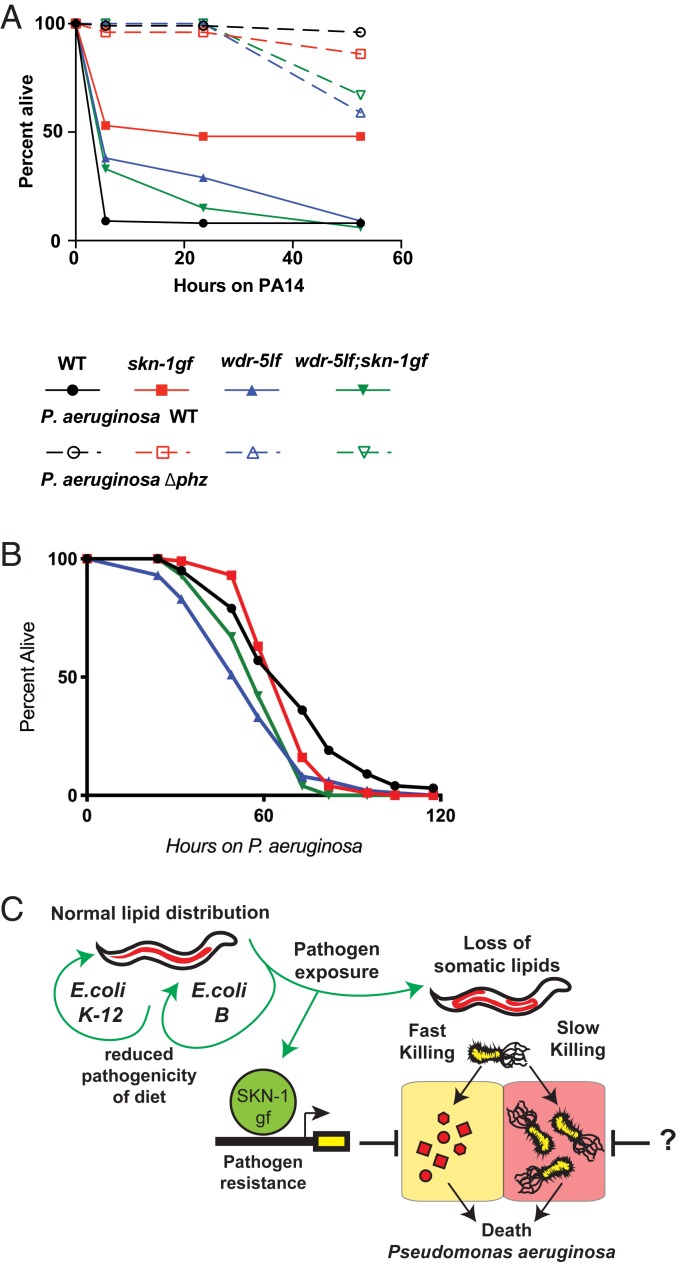
Our data suggest that skn-1gf animals would potentially display enhanced resistance to pathogen exposure. The effects of pathogens on host physiology are a result of the presence of the microbe itself and the toxins it secretes (37). Exposure of wild-type animals to P. aeruginosa can result in “slow-killing” of C. elegans, which reflects the colonization of C. elegans intestine and subsequent lethal infection. Altering the media on which the P. aeruginosa are grown prior to C. elegans exposure results in hyperproduction of phenazine toxins, which causes rapid intoxication of nematodes, an assay which is also called “fast-killing” (30, 32). Although each of the modes of killing have been well studied, several aspects of pathogenesis remain unanswered, but it is clear that it is multifactorial. We challenged wild-type and skn-1gf animals to the fast-kill and slow-kill models of pathogen-induced death. Interestingly, skn-1gf mutants were resistant to P. aeruginosa fast killing, in a manner dependent on the production of phenazine toxins (Fig. 5A and SI Appendix, Fig. S9A), but surprisingly the effect in the slow-killing scenario was unremarkable (Fig. 5B).
Fig. 5.
Redirection of SKN-1gf negates pathogen resistance. (A–B) Pathogen sensitivity assays. Wild-type (black), skn-1gf(lax188) mutants (red), wdr-5lf(ok1417) mutants (blue), or wdr-5lf(ok1417);skn-1gf(lax188) double mutants (green) were exposed to P. aeruginosa (A) “fast-kill” or (B) “slow kill” pathogen stress plates. Activated SKN-1 supports resistance to pathogenic P. aeruginosa specifically on fast-kill plates, which is abolished in the absence of H3K4me3. (C) Model for the impact of SKN-1gf on physiological responses to P. aeruginosa secreted factors “fast killing.” “?”, the role of SKN-1gf on colonization “slow killing” is unclear and requires further study. All experiments were performed in a minimum of biological triplicate.
The pathogen resistance phenotype of skn-1gf animals was suppressed in the absence of wdr-5, which is consistent with our findings that loss of H3K4me3 epigenetic marks on chromatin diminishes the transcriptional output of SKN-1–associated pathogen defense genes. Collectively, these findings support a biological framework for host–pathogen responses where the presence of a pathogen drives SKN-1 activation of pathogen defense genes (Fig. 5C). This program enables resistance to pathogen-derived toxins, but occurs at the expense of organismal lipid homeostasis, which impairs organismal health later in life. Importantly, this program is malleable and the loss of lifelong health can be restored, albeit at the cost of changes in pathogen susceptibility, by dampening SKN-1 activity at these specific gene targets.
Activated SKN-1 Drives Preferential Utilization of Diet-Derived and Short Unsaturated Lipids.
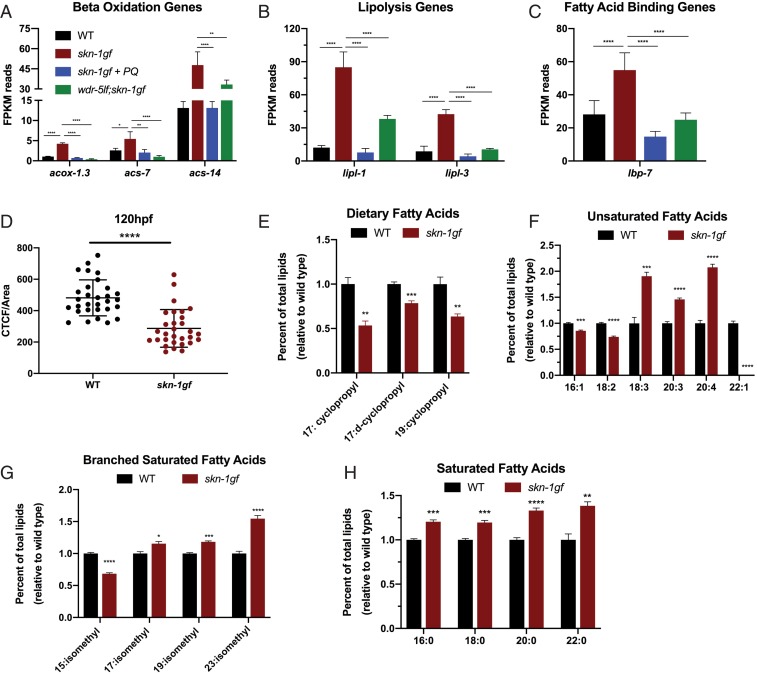
Several events that immediately precede somatic lipid depletion in animals with SKN-1 activation have been described (4); however, a full account of the metabolic state once somatic lipids have been depleted is lacking. We applied a tandem transcriptomic and lipidomic assessment of animals at the peak of somatic lipid depletion (day 2 of adulthood). We examined our RNA-seq data (Datasets S1–S4) for changes in the expression of lipid metabolism genes in animals with activated SKN-1 that was dampened in the absence of H3K4me3 or when SKN-1 was redirected by PQ treatment. Several metabolism genes fit these criteria (Fig. 6 and SI Appendix, Fig. S10), including mitochondrial and peroxisomal B-oxidation genes acox-1.3, acs-7, and acs-14 (Fig. 6A); lipolysis genes lipl-1 and lipl-3 (Fig. 6B); and lipid binding genes like lbp-7 (Fig. 6C). We also measured a reduction in several lipid biosynthesis genes, including cpt-6 and fat-5, whose expression is restored when SKN-1 is redirected (SI Appendix, Fig. S10 A and B). These data reveal that SKN-1 activation induces a metabolic state where lipid utilization pathways are activated while de novo biosynthesis is suppressed, both of which could contribute to the observed depletion of somatic lipids.
Fig. 6.
SKN-1 activation drives lipid utilization during Asdf. (A–C) Analysis of the mRNA reads of the indicated genes related to lipid utilization: (A) lipid beta-oxidation and (B) lipolysis; and (C) lipid binding, which collectively reveal a scenario where activated SKN-1 drives lipid utilization while suppressing de novo synthesis (see also SI Appendix, Fig. S10). (D) Fixed Nile Red staining reveals skn-1gf(lax188) mutants have a ∼60% reduction in total fat (n = 30). (E–H) GCMS analysis of total fatty acids in the triglyceride fraction of wild-type (black) and skn-1gf(lax188) mutant (red) animals fed an OP50 diet at day 2 of adulthood when Asdf is most prominent, 2-tailed t test. See also Datasets S1–S4 for all RNA-seq measurements. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. FPKM, fragments per kilobase of transcript per million mapped reads.
Our previous studies of L4-stage skn-1gf mutant animals identified the differential abundance of multiple lipid species as compared to age-matched wild-type animals before Asdf occurs (4). To connect the transcriptomic analysis with the lipid signature of animals with a depletion of somatic lipids, we used quantitative Nile Red staining of lipids (38) and gas chromatography coupled to mass spectrometry (GCMS) (39). At day 2 of adulthood, skn-1gf animals have 60% less total fat compared to wild-type animals (Fig. 6D and SI Appendix, Fig. S10C) and we identified several classes of lipids with differential abundance in skn-1gf mutants as compared to wild-type animals (Fig. 6 E–H). Specifically, bacterial diet-derived cyclopropane fatty acids (Fig. 6E) and synthesized 16:1 and 18:2 species (Fig. 6F) were all reduced. There was also a modest increase in monomethyl branched-chain fatty acids (Fig. 6G) and a more significant increase in synthesized mono- and polyunsaturated fat species of longer lengths, including eicosatetraenoic acid 20:4(n-3) lipid species (Fig. 6 F and H). The relative abundance of these lipids in skn-1gf mutants is an important biomarker as it defines the lipid species that are a hallmark of Asdf and also which lipid species are lost in response to SKN-1 activation.
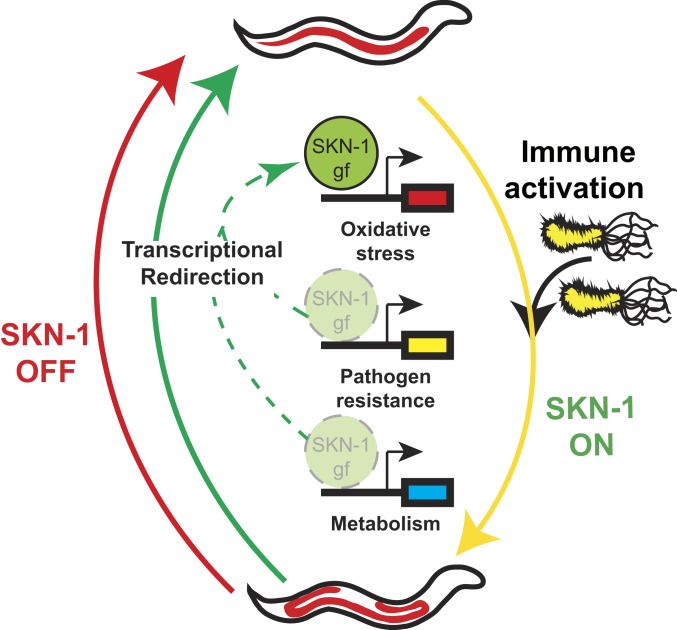
Taken together, our data identify redirection of an activated cytoprotective transcription factor, like SKN-1, as a powerful mechanism to abate pleiotropic consequences of unchecked transcriptional activity, including age-dependent loss of lipid homeostasis that normally accompanies pathogen infection and chemosensitivity, respectively (Fig. 7).
Fig. 7.
Transcriptional redirection of SKN-1 activity mitigates pleiotropic outcomes. In the presence of pathogens, SKN-1 is activated (yellow arrow) and induces the expression of several classes of target genes (e.g., oxidative stress and pathogen resistance). The induction of pathogen resistance genes drives the depletion of somatic lipids with age that leads to poor health outcomes. Transcriptional redirection (green arrows) does not completely turn off all SKN-1 activity (red arrow), but rather focuses the gene targets that are activated. Redirection of activated SKN-1 away from pathogen resistance genes and toward oxidative stress genes abates the loss of metabolic homeostasis.
Discussion
We report a mechanism by which the pleiotropic outcomes stemming from the activation of SKN-1 can be ameliorated by redirecting transcriptional activity. Modulation of transcriptional outputs, rather than inactivation of the transcription factor, provides a model for the treatment of pathologies stemming from transcriptional dysregulation.
Importantly, we report the rapid loss of somatic lipid stores as one of the earliest documented pathological symptoms of pathogen exposure, which is dependent on SKN-1 activity. Although early responses to pathogens are essential for optimal health, the long-term effects of these responses are uncharacterized. Our data reveal that SKN-1 activation depletes somatic lipids while providing pathogen resistance. However, aberrant SKN-1 activation ultimately impedes oxidative stress resistance and shortens lifespan. Dampening the innate immunity axis of SKN-1 activation increases lifespan, restores oxidative stress resistance, and reestablishes the healthy distribution of lipids, but compromises the ability to survive pathogen challenge.
We previously found that depletion of the monounsaturated fatty acid oleate drives the redistribution of somatic lipids in skn-1 gain-of-function mutants, or Asdf phenotype (4). Of note, oleate is also necessary for the pathogen-mediated induction of host defense genes and resistance to diverse bacterial pathogens (40). Taken together with data from this study that immune activation during pathogen infection drives the Asdf phenotype, these studies emphasize that oleate sufficiency is a key marker of health, depletion of which triggers protective reallocation of somatic fat and suppression of immune defenses, thereby allowing energy reserves to be devoted to the preservation of evolutionary fitness. Oleate is also required for proper immune defenses in plants (41–43), which suggests that cytoprotective redistribution of host lipids and energy stores is evolutionarily ancient.
Our previous work showed that diet can influence loss of somatic lipids in the context of our skn-1gf worms, as the bacteria E. coli OP50/B leads to fat loss but not E. coli HT115/K-12 (4). This was not the case for exposure to P. aeruginosa, as virulence-attenuated strains of the pathogen did not lead to depletion of somatic lipids. Thus, some aspect associated with P. aeruginosa virulence drives somatic lipid depletion, which is partially dependent on SKN-1, as RNAi against skn-1 attenuates the lipid depletion. Our observation that animals with reduced SKN-1 activity eventually lose somatic fat following exposure to P. aeruginosa indicates the presence of additional factors that mediate pathogen-dependent fat loss. Additionally, we observed that wild-type animals exposed to P. aeruginosa displayed a rapid loss of somatic lipids—about 4 to 6 h for fat loss to occur. The speed of this response is intriguing as the “slow-killing” model requires bacteria to colonize the intestine and during the first 24 h of exposure there are neither disease symptoms nor appreciable mortality. Moreover, SKN-1gf mutant animals, which are already primed for an innate immune response, deplete somatic lipids even more rapidly than wild-type animals and loss of SKN-1 attenuates the somatic lipid depletion in animals with constitutive activation of p38 signaling. As these host–pathogen interactions are mediated, at least in part, by intestinal epithelial cells (16, 27, 29, 44), the loss of lipids in this tissue is one of the first detectable physiological responses to pathogen exposure.
H3K4 methylation marks are epigenetic signatures of active gene expression, but they are not essential for all transcriptional activity (45) and their deposition at loci after transcription to maintain an active transcriptional state has been documented (46). Our transcriptomic analysis revealed that loss of H3K4me3 resulted in reduced expression of certain genes but not others. For instance, in the skn-1gf animals, the loss of H3K4me3 marks suppressed the expression of innate immune targets, but not oxidative stress response genes. The observation that loss of H3K4me3 marks has differential effects on the expression of specific stress response genes could indicate a role for these marks for the recruitment of additional transcriptional regulators beyond SKN-1. Additionally, H3K4me3 can also facilitate transcriptional reinitiation, which can impact total transcriptional output of specific loci (47). It should be of note that our RNA-seq was performed on whole worms, which precludes tissue-specific resolution of H3K4me3-sensitive targets. Nevertheless, our study identifies that the maintenance of H3K4me3 epigenetic marks is critical for the regulation of lipid homeostasis and pathogen resistance. Our previous study indicates a relationship between the Asdf phenotype and a reallocation of lipids to the germline to increase reproductive output. Loss of WDR-5 activity can impact reproduction, but the loss of wdr-5 exerts effects on progeny output at temperatures higher than those used in this study (48).
In contrast to acute hydrogen peroxide exposure, chronic exposure to a low dose of paraquat (a superoxide-producing agent) suppressed Asdf. Our discovery that paraquat exposure refines the transcriptional focus of activated SKN-1 suggests that transcriptional redirection is a powerful approach to curb the negative pleiotropies of unregulated transcriptional activity. It remains possible that low-dose exposure to paraquat reduces the pathogenicity of the OP50 E. coli diet. Exposure to much higher concentrations of paraquat can result in reduced E. coli B strain growth; however, treatment with 75 μM only slightly impaired OP50 viability (SI Appendix, Fig. S5 C and D). Intriguingly, E. coli K-12 strains that do not induce Asdf (4) are resistant to the effects of paraquat (49), which supports further examination of dietary effects on SKN-1–dependent pathogen responses. Nevertheless, our results reveal that the induced expression of oxidative stress response genes does not drive the metabolic pleiotropies observed in skn-1gf animals. The shared suppression of pathogen response genes suggests the activation of immune responses drives the Asdf phenotype in skn-1gf mutants (Dataset S4). Moreover, the similarities in phenotypes between wild-type worms exposed to pathogens and our skn-1gf worms support our transcriptional data that it is the expression of innate immunity genes in our mutant worms that is driving its associated pleiotropies. Importantly, our data also reveal that once activated, SKN-1 remains responsive and can be redirected away from pathogen response genes to alleviate negative health outcomes. Determining whether the ability to redirect other transcription factors, like NRF2, is a generalizable approach to treat diseases with aberrant transcription is an exciting avenue for future investigation.
Materials and Methods
C. elegans and Bacterial Strains Used and Culturing Methods.
Worms were grown following standard culture protocols at 20 °C, unless otherwise noted. The following strains were used: WT, N2 Bristol strain; SPC227, skn-1(lax188); RB1304, wdr-5lf(ok1417); SPC415, wdr-5lf(ok1417);skn-1gf(lax188); RPW43, nsy-1(ums8); LG340, skn-1(zu135); and SPC425, nsy-1gf(ums8);skn-1(zu135).
Oil Red O Staining.
Oil Red O (ORO) staining of lipids was performed as previously described (4, 38, 50, 51). In brief, synchronized worms were collected using 1 mL of 1× PBS + 0.01% Triton X-100 (PBST) into a microcentrifuge tube and allowed to gravity settle. Supernatant was aspirated and washed 3 additional times with 1 mL PBST. After the final wash, supernatant was removed until 100 μL was left and then 600 μL of 60% isopropanol was added, and samples were rocked for 3 min at room temperature. Samples were spun down at 25 × g and supernatant was removed until 100 μL was left. Worms were then stained for 2 h with 600 μL of ORO working solution (0.5 g of ORO in 100 mL of 100% isopropanol) while rotating at room temperature and destained in 600 μL of PBST for 30 min. All staining was done on synchronous populations of day 2 adult animals (120 h postfeeding starting from a synchronous L1 population), unless otherwise stated.
Asdf Quantification.
ORO-stained worms were placed on glass slides and a coverslip was placed over the sample. Worms were scored, and images were taken using a Zeiss microscope at 10× magnification. Fat levels of worms were placed into 3 categories: non-Asdf, intermediate, and Asdf. Non-Asdf worms display no loss of fat and are stained a dark red throughout most of the body (somatic and germ cells). Intermediate worms display significant fat loss from the somatic tissues, with portions of the intestine being clear, but ORO-stained fat deposits are still visible (somatic < germ cells). Asdf worms have had most, if not all, observable somatic fat deposits depleted (germ cells only).
Hydrogen Peroxide Stress Survival Assay.
Acute exposure to H2O2 was performed as previously described (4, 52). In brief, age-synchronized worms were exposed to 10 mM H2O2 in M9T at room temperature while rotating for 20 min. Samples were washed in 1 mL of M9T 3 times and then dropped directly on the OP50 bacterial lawn on a nematode growth media (NGM) plate. Scoring of dead or alive worms was done 24 h post H2O2 exposure. Worms were considered dead either when they were unresponsive to gentle prodding from a platinum-tipped wire or when they showed signs of internal larval hatching.
Pathogen Exposure Experiment.
An overnight culture of P. aeruginosa strain PA14 (WT), PA14ΔgacA, or PA14ΔrhIR mutant was seeded on 6 cm “slow-killing” media plates, prepared as previously described (31). Synchronized L1 stage C. elegans of the indicated genotypes were grown to day 0 adult stage then transferred to seeded plates and incubated at 20 °C for indicated time points. Plates were incubated at 20 °C to slow the kinetics of the P. aeruginosa-induced Asdf phenotype. Worms were then collected and stained for fat and scored for fat levels as previously described (31, 53). P. aeruginosa “fast kill” pathogenesis assays were conducted as previously described (30, 32). Late L4 animals obtained from timed egg lays were used.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center, funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for providing some strains, and WormBase. We also thank Melanie Trombly for comments on the manuscript. This work was supported by NIH grants R01GM109028 (S.P.C.), R01AG058610 (S.P.C.), R01AI130289 (R.P.-W.), T32AG000037 (J.D.N., D.L.R., and H.M.D.), T32GM118289 (C.D.T.), T32AI007349 (S.M.A.), F31AG051382 (H.M.D.), R01AG058256 (A.A.S.), and P30DK040561 (A.A.S.); and research support from the American Federation for Aging Research (C.-A.Y. and S.P.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Sequencing data are deposited to the Gene Expression Omnibus (accession no. GSE123531).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909666116/-/DCSupplemental.
References
- 1.Sykiotis G. P., Bohmann D., Stress-activated cap’n’collar transcription factors in aging and human disease. Sci. Signal. 3, re3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papp D., Csermely P., Sőti C., A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog. 8, e1002673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoeven Rv., McCallum K. C., Cruz M. R., Garsin D. A., Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 7, e1002453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynn D. A., et al. , Omega-3 and -6 fatty acids allocate somatic and germline lipids to ensure fitness during nutrient and oxidative stress in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 112, 15378–15383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang S., Curran S. P., Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab. 19, 221–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X. J., et al. , Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29, 1235–1243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo J. Y., Spatola B. N., Curran S. P., WDR23 regulates NRF2 independently of KEAP1. PLoS Genet. 13, e1006762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paek J., et al. , Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 16, 526–537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwell T. K., Steinbaugh M. J., Hourihan J. M., Ewald C. Y., Isik M., SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88, 290–301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira R. P., et al. , Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8, 524–541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang S., Lynn D. A., Lo J. Y., Paek J., Curran S. P., SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 5, 5048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black J. C., Van Rechem C., Whetstine J. R., Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 48, 491–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad T. K., Anderson M. D., Martin B. A., Stewart C. R., Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell 6, 65–74 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F., Xiao Y., Ji X. L., Zhang K. Q., Zou C. G., The cAMP-PKA pathway-mediated fat mobilization is required for cold tolerance in C. elegans. Sci. Rep. 7, 638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapira M., et al. , A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 14086–14091 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irazoqui J. E., et al. , Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6, e1000982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Przybysz A. J., Choe K. P., Roberts L. J., Strange K., Increased age reduces DAF-16 and SKN-1 signaling and the hormetic response of Caenorhabditis elegans to the xenobiotic juglone. Mech. Ageing Dev. 130, 357–369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X., et al. , Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 7, e1002119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewald C. Y., et al. , NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. eLife 6, e19493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinbaugh M. J., et al. , Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. eLife 4, e07836 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tullet J. M., et al. , Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025–1038 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pukkila-Worley R., Surveillance immunity: An emerging paradigm of innate defense activation in Caenorhabditis elegans. PLoS Pathog. 12, e1005795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garigan D., et al. , Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics 161, 1101–1112 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portal-Celhay C., Bradley E. R., Blaser M. J., Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12, 49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shivers R. P., Youngman M. J., Kim D. H., Transcriptional responses to pathogens in Caenorhabditis elegans. Curr. Opin. Microbiol. 11, 251–256 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D. H., et al. , A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Pukkila-Worley R., Ausubel F. M., Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr. Opin. Immunol. 24, 3–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue H., et al. , The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 19, 2278–2283 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheesman H. K., et al. , Aberrant activation of p38 MAP kinase-dependent innate immune responses is toxic to Caenorhabditis elegans. G3 (Bethesda) 6, 541–549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cezairliyan B., et al. , Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. 9, e1003101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan M. W., Mahajan-Miklos S., Ausubel F. M., Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 96, 715–720 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahajan-Miklos S., Tan M. W., Rahme L. G., Ausubel F. M., Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96, 47–56 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Feinbaum R. L., et al. , Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 8, e1002813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estes K. A., Dunbar T. L., Powell J. R., Ausubel F. M., Troemel E. R., bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 107, 2153–2158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Troemel E. R., et al. , p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2, e183 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Head B. P., Olaitan A. O., Aballay A., Role of GATA transcription factor ELT-2 and p38 MAPK PMK-1 in recovery from acute P. aeruginosa infection in C. elegans. Virulence 8, 261–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darby C., “Interactions with microbial pathogens” in WormBook, D. H. A. Fitch, Ed. (The C. elegans Research Community, 2005). [DOI] [PMC free article] [PubMed]
- 38.Escorcia W., Ruter D. L., Nhan J., Curran S. P., Quantification of lipid abundance and evaluation of lipid distribution in Caenorhabditis elegans by nile red and oil red O staining. J. Vis. Exp., e57352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pino E. C., Webster C. M., Carr C. E., Soukas A. A., Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J. Vis. Exp., e50180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson S. M., et al. , The fatty acid oleate is required for innate immune activation and pathogen defense in Caenorhabditis elegans. PLoS Pathog. 15, e1007893 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandal M. K., et al. , Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis. Plant Cell 24, 1654–1674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandstrom A., et al. , Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kachroo A., Kachroo P., Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176 (2009). [DOI] [PubMed] [Google Scholar]
- 44.McEwan D. L., Kirienko N. V., Ausubel F. M., Host translational inhibition by Pseudomonas aeruginosa exotoxin A triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11, 364–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavri R., et al. , Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Cruz C., et al. , Tri-methylation of histone H3 lysine 4 facilitates gene expression in ageing cells. eLife 7, e34081 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orphanides G., Reinberg D., RNA polymerase II elongation through chromatin. Nature 407, 471–475 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Wang S., Fisher K., Poulin G. B., Lineage specific trimethylation of H3 on lysine 4 during C. elegans early embryogenesis. Dev. Biol. 355, 227–238 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Kitzler J. W., Minakami H., Fridovich I., Effects of paraquat on Escherichia coli: Differences between B and K-12 strains. J. Bacteriol. 172, 686–690 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pradhan A., Hammerquist A. M., Khanna A., Curran S. P., The C-Box region of MAF1 regulates transcriptional activity and protein stability. J. Mol. Biol. 429, 192–207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khanna A., Johnson D. L., Curran S. P., Physiological roles for mafr-1 in reproduction and lipid homeostasis. Cell Rep. 9, 2180–2191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalton H. M., Curran S. P., Hypodermal responses to protein synthesis inhibition induce systemic developmental arrest and AMPK-dependent survival in Caenorhabditis elegans. PLoS Genet. 14, e1007520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan M. W., Rahme L. G., Sternberg J. A., Tompkins R. G., Ausubel F. M., Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. U.S.A. 96, 2408–2413 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.