Significance
Gastrointestinal symptoms are common in patients with autism spectrum disorder (ASD), yet are poorly understood. To further dissect this phenomenon, we investigated a mouse model mimicking the haploinsufficiency seen in patients with FOXP1 syndrome. We found a disturbed structure and function of the gastrointestinal system, atrophy and malfunction of the tunica muscularis, esophageal achalasia, and increased total transit based on altered colon motility. Furthermore, Foxp1 target genes identified in the brain were dysregulated in the Foxp1+/− esophagus. Our findings support the idea that genes relevant in brain function might also cause gastrointestinal disturbances in ASD patients and that these primary defects deserve appropriate treatment.
Keywords: Foxp1, ASD, gastrointestinal tract, achalasia
Abstract
Gastrointestinal dysfunctions in individuals with autism spectrum disorder are poorly understood, although they are common among this group of patients. FOXP1 haploinsufficiency is characterized by autistic behavior, language impairment, and intellectual disability, but feeding difficulties and gastrointestinal problems have also been reported. Whether these are primary impairments, the result of altered eating behavior, or side effects of psychotropic medication remains unclear. To address this question, we investigated Foxp1+/− mice reflecting FOXP1 haploinsufficiency. These animals show decreased body weight and altered feeding behavior with reduced food and water intake. A pronounced muscular atrophy was detected in the esophagus and colon, caused by reduced muscle cell proliferation. Nitric oxide-induced relaxation of the lower esophageal sphincter was impaired and achalasia was confirmed in vivo by manometry. Foxp1 targets (Nexn, Rbms3, and Wls) identified in the brain were dysregulated in the adult Foxp1+/− esophagus. Total gastrointestinal transit was significantly prolonged due to impaired colonic contractility. Our results have uncovered a previously unknown dysfunction (achalasia and impaired gut motility) that explains the gastrointestinal disturbances in patients with FOXP1 syndrome, with potential wider relevance for autism.
Functional disorders of the gastrointestinal tract are common in neurological disorders and have been barely investigated to date. This is particularly true for autism spectrum disorder (ASD), in which core features attributed to CNS dysfunction are defined based on the altered behavior of an individual; co-occurring gastrointestinal disturbances are frequently overlooked. Clinically, ASD is characterized by impaired social interaction and communication, limited interests and activities, and repetitive behaviors and body movements. ASD is diagnosed in early childhood on the basis of behavior, which has directed its focus to the brain. Although the core features of ASD are behavioral, gastrointestinal (GI) disturbances are also common in children with ASD. However, these symptoms are often undiagnosed or are considered comorbid (1).
More than 90% of the 62 genes strongly associated with autism in the SFARI database (https://gene.sfari.org/) are expressed in both brain and GI tissues according to the genotype-tissue GTEx database (https://gtexportal.org/). Therefore, dysregulation of these genes likely affects both the brain and the gut. Forkhead-box protein P1 (FOXP1) is a verified autism gene that is associated with a genetically defined, relatively common ASD subtype. De novo disruptions of the transcription factor FOXP1 cause FOXP1 syndrome, characterized by intellectual disability, language impairment, and social deficits (2, 3). Brain dysfunction was suspected in patients with FOXP1 deficiency based on behavior analysis. Furthermore, MRI scans revealed prominent lateral ventricles in some affected individuals. Feeding difficulties, oromotor dysfunction, esophageal dysmotility, gastroesophageal reflux, and constipation were also reported in patients (3, 4), but these symptoms have not been investigated so far.
We previously defined the neurodevelopmental role of FOXP1 using knockout (KO) mice with conditional deletion of Foxp1 in the central and peripheral nervous system (Nestin-Cre [Foxp1−/−]). Mutant mice displayed a pronounced reduction of the developing striatum and more subtle alterations in the hippocampus, including reduced excitability of pyramidal neurons in the CA1 region and an imbalance between excitatory and inhibitory input. Foxp1 ablation was also associated with various cognitive and social deficits, including hyperactivity, increased repetitive behavior, anxiety, and reduced social interests (5). Interestingly, Nestin-Cre (Foxp1−/−) animals fail to thrive and die between 3 and 5 wk of age unless soft, high-calorie food is provided. This indicates a serious impairment of feeding ability or gastrointestinal function.
Other mouse models have also been used to investigate how Foxp1 contributes to neuronal development. For example, Foxp1 haploinsufficiency in conventional Foxp1+/− mice affects the excitability of striatal medium spiny neurons and correlates with defects in neonatal ultrasonic vocalization (6). Furthermore, Foxp1 is important for normal radial migration and morphogenesis of cortical and hippocampal neurons (7, 8) and neocortical networks regulated by Foxp1 have been uncovered (9).
The goal of this study was to understand the GI-related symptoms of the FOXP1 ASD syndrome. We used conventional heterozygous Foxp1 KO (Foxp1+/−) mice in our study (10), as these animals reflect the FOXP1 haploinsufficiency in patients and therefore represent a good model of the human FOXP1 syndrome. Using this model, we show here that mice lacking 1 allele of Foxp1 display esophageal achalasia and impaired peristalsis in the colon.
Results
Foxp1 Protein Expression in the GI Tract.
To explore whether Foxp1 may play a role in GI function, we analyzed its expression in different tissues of the GI tract (esophagus, stomach, duodenum, ileum, and colon). Immunofluorescence staining on wild-type (WT) tissue revealed that Foxp1 is expressed in all layers of the murine GI tract (SI Appendix, Fig. S1), including the myenteric plexus, which is part of the enteric nervous system and regulates gut peristalsis and transit.
Muscular Hypertrophy in the Esophagus of Nestin-Cre (Foxp1−/−) Mice.
To get an idea of whether the loss of Foxp1 in the nervous system is sufficient to induce a GI phenotype, we examined Nestin-Cre (Foxp1−/−) mice whose severe thriving difficulties with strongly reduced body weight suggested GI impairment. Nestin-Cre (Foxp1−/−) animals also exhibit a pronounced reduction in size compared with WT mice (SI Appendix, Fig. S2 A and B). Cross-sections of the esophagus, stomach, duodenum, ileum, and colon were evaluated for morphological alterations at postnatal day (P)12.5. The tunica muscularis of the esophagus was significantly thicker in Nestin-Cre (Foxp1−/−) animals (SI Appendix, Fig. S2 C and D), with both muscle layers being equally affected (SI Appendix, Fig. S2E) whereas no obvious alterations were detected in other GI segments (SI Appendix, Fig. S3). The thickening of the esophageal muscle layer in Nestin-Cre (Foxp1−/−) mice complies with the current knowledge that patients with achalasia (a motility disorder of the lower esophageal sphincter) typically present with a muscular hypertrophy of the esophagus (11). However, the results from Nestin-Cre (Foxp1−/−) mice cannot be directly correlated with patients with FOXP1 syndrome, who have a body-wide loss of 1 FOXP1 copy. For this reason, we used patient-relevant Foxp1+/− mice in the further course of our study.
Foxp1 mRNA and Protein Expression Is Reduced in the GI Tract of Foxp1+/− Mice.
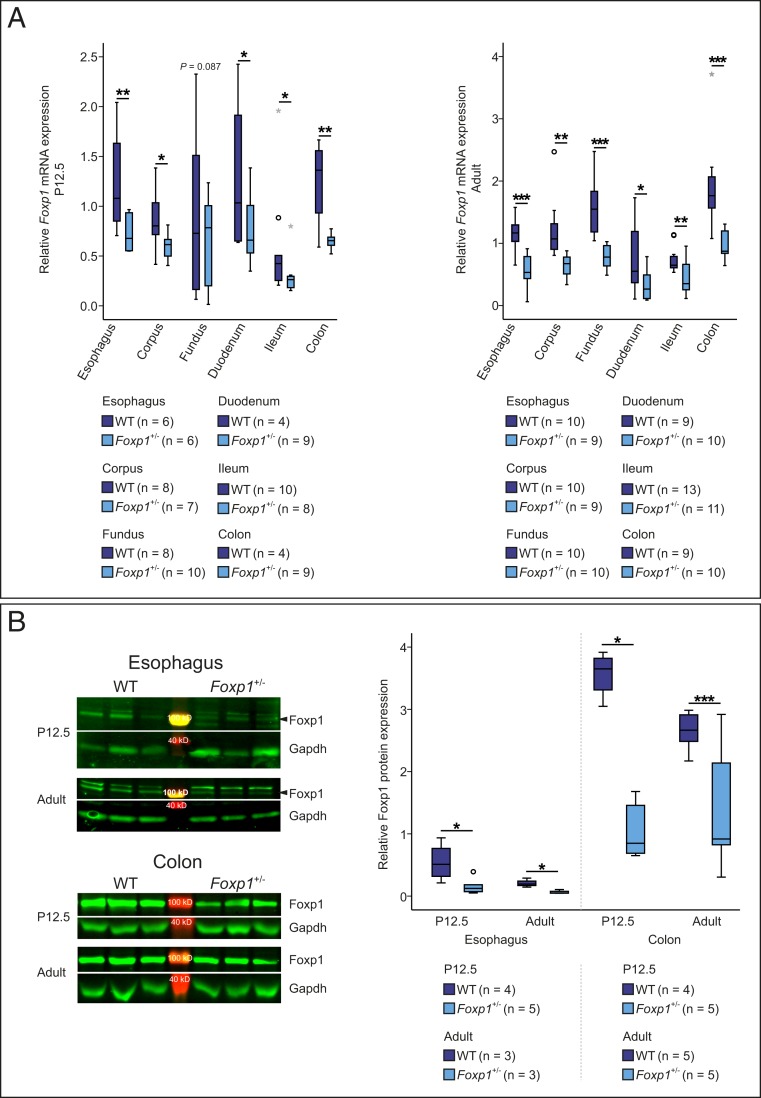
Foxp1+/− and WT mice do not exhibit obvious differences in girth or body length (SI Appendix, Fig. S4). To confirm that Foxp1 expression is reduced in Foxp1+/− animals, we quantified Foxp1 mRNA in the esophagus, stomach, duodenum, ileum, and colon of P12.5 and adult WT and Foxp1+/− mice using real-time PCR. At both developmental stages, Foxp1 mRNA was reduced by 25 to 55% (Fig. 1A). We furthermore confirmed a 20 to 70% down-regulation of Foxp1 protein in the esophagus and colon of Foxp1+/− mice using Western blot analysis (Fig. 1B).
Fig. 1.
Foxp1 mRNA and protein levels are reduced in the GI tract of Foxp1+/− mice. (A) Relative expression of Foxp1 mRNA was compared in the esophagus, corpus, fundus, duodenum, ileum, and colon of WT and Foxp1+/− animals by quantitative real-time PCR. (B) Foxp1 protein expression was quantified in the esophagus and colon of WT and Foxp1+/− mice at P12.5 and at adult stage by Western blot analysis. A comparable number of male and female animals of both genotypes were used in the experiments. Arrowheads mark the specific Foxp1 band. Asterisks indicate significant difference (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; if appropriate, analysis of covariance [ANCOVA] was used; otherwise, unpaired 2-tailed Student’s t test was used).
Numeric Atrophy of the Tunica Muscularis in the Esophagus and Colon Is Caused by Reduced Proliferation.
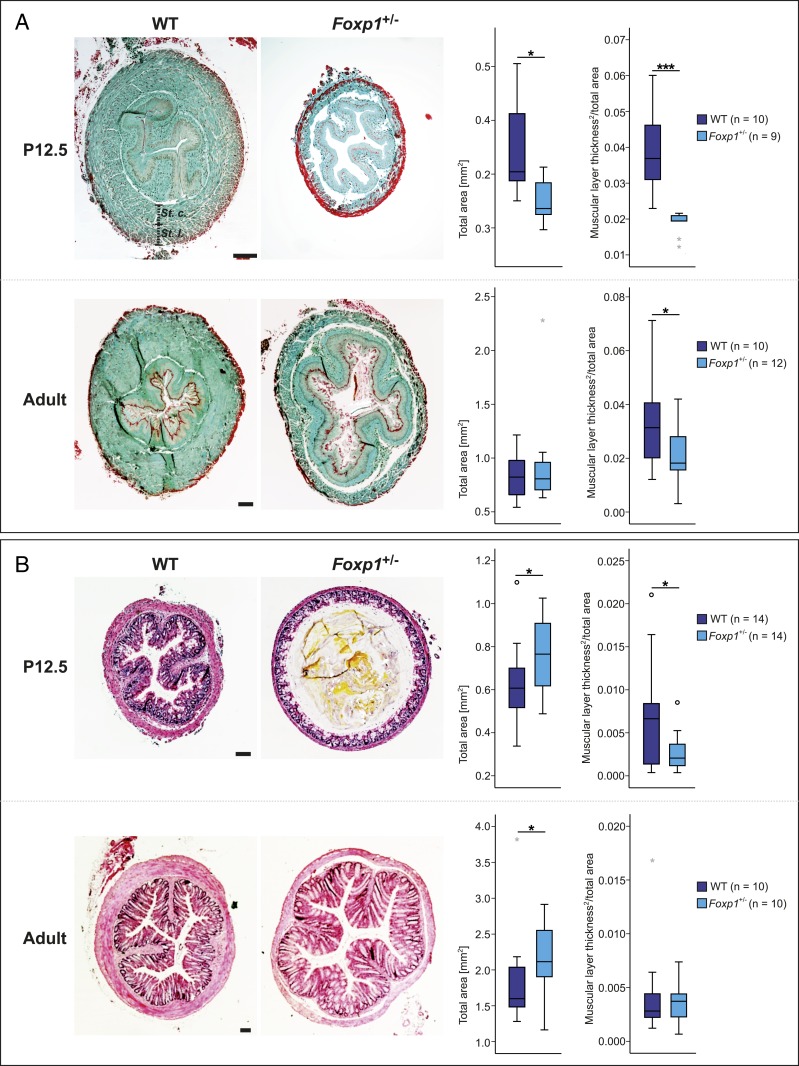
To investigate the consequences of reduced Foxp1 expression in the GI tract of Foxp1+/− mice, we examined P12.5 and adult cross-sections. No morphological alterations were detectable in the stomach, duodenum, and ileum, but the tunica muscularis was significantly thinner in the esophagus and colon (Fig. 2). At P12.5, the tunica muscularis was reduced by 52% in the esophagus and 61% in the colon. In adult Foxp1+/− mice, muscle thickness was reduced in the esophagus (40%) but not in the colon (Fig. 2). We also compared the ratio of longitudinal to circular muscle thickness in the tunica muscularis of WT and Foxp1+/− esophagus at P12.5. Both muscular layers were equally affected (SI Appendix, Fig. S5).
Fig. 2.
Tunica muscularis of the esophagus and the colon is altered in Foxp1+/− mice. (A) The esophagus is significantly smaller in Foxp1+/− animals compared with WT littermates. Sections from Foxp1+/− organs harvested at P12.5 and adult stage are 25 and 11% smaller compared with WT sections, respectively. The thickness of the tunica muscularis is significantly reduced in Foxp1+/− mice at both stages, by 52% at P12.5 and 40% at adult stage. (B) The colon of Foxp1+/− animals reveals a strong reduction in muscle layer thickness by 61% together with a dilated lumen at P12.5. In the adult colon, the total size of Foxp1+/− sections is significantly increased by 14% compared with WT sections, whereas morphological differences regarding the thickness of the muscular layer were not observed. A comparable number of male and female animals of both genotypes were used in the experiments. Esophageal sections were stained with Masson–Goldner trichrome, and colon sections were stained with hematoxylin and eosin. Asterisks indicate significant difference (*P ≤ 0.05, ***P ≤ 0.001; ANCOVA). (Scale bars, 100 µm.)
To find out whether the reduction of muscle mass in the esophagus is caused by fewer cells (numeric atrophy) or lower cell volume (volumetric atrophy), we analyzed cell number and cell density in the tunica muscularis at P12.5 (SI Appendix, Fig. S6). Cell numbers were lower in Foxp1+/− mice (SI Appendix, Fig. S6A) but cell density was not affected (SI Appendix, Fig. S6B). To identify the pathological mechanism of muscular atrophy, we analyzed the rate of apoptosis and proliferation in the tunica muscularis. TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) staining did not reveal differences in the number of apoptotic cells between Foxp1+/− mice and WT littermates but fewer cells stained positive for the proliferation marker Ki67 (SI Appendix, Fig. S6 C and D). These data strongly suggest that muscular atrophy in Foxp1+/− mice is caused by reduced cell proliferation.
Weight Loss Increases with Age in Foxp1+/− Animals Because of Reduced Food and Water Intake.
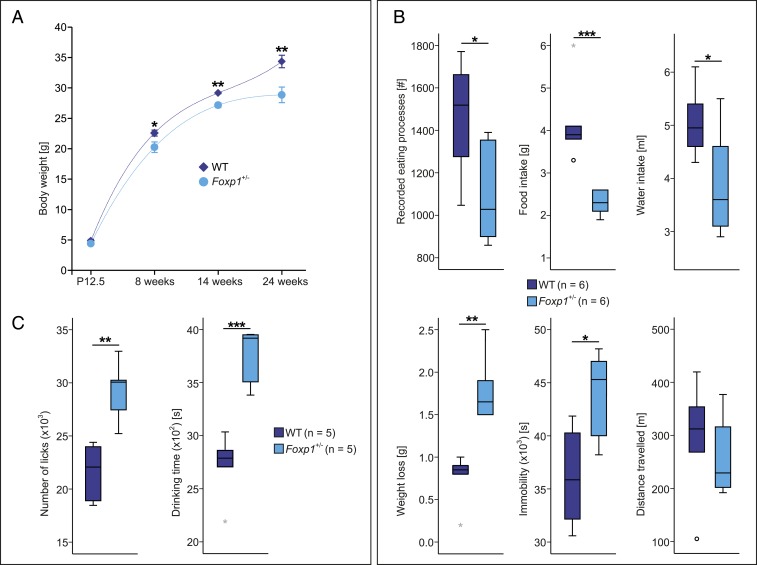
The altered development of esophageal and colonic tissue in Foxp1+/− mice suggested difficulties in thriving. We compared the body weight of male Foxp1+/− mice with corresponding WT littermates at different developmental stages (P12.5 and 8, 14, and 24 wk) (Fig. 3). No reduction in body weight was detected in Foxp1+/− mice at P12.5, but from 8 wk onward, the body weight of Foxp1+/− mice was significantly lower than WT littermates (10% at 8 wk, 7% at 14 wk, and 16% at 24 wk). In addition, we also explored the body weight of female Foxp1+/− animals at the age of 8 wk and observed a comparable reduction (6% compared with WT females) (SI Appendix, Fig. S4B).
Fig. 3.
Reduced food and water intake of Foxp1+/− mice and its impact on body weight. (A) Body weight in WT and Foxp1+/− mice compared at P12.5 and 8, 14, and 24 wk of age. At P12.5, the weight of Foxp1+/− pups does not differ compared with WT animals. At adult stage, Foxp1+/− mice display significant differences with 10, 7, and 16% less body weight at 8, 14, and 24 wk, respectively, compared with WT animals. Data shown are means ± SEM of n = 5 to 18 per genotype and age. (B) WT and Foxp1+/− mice at the age of 8 wk were monitored for 24 h using LABORAS. The number of recorded eating processes is reduced by 25% in Foxp1+/− animals. Food and water intake is 45 and 23% lower in Foxp1+/− compared with WT animals, respectively. Mice commonly lose weight during the time course of the experiment. In Foxp1+/− animals, weight loss is much stronger compared with WT mice (282% higher), although Foxp1+/− mice display 22% more time periods of immobility than WT animals. In addition, WT and Foxp1+/− mice travel comparable distances during monitoring. (C) IntelliCage measurements were performed for 5 consecutive days to analyze the drinking pattern in WT and Foxp1+/− animals. Foxp1+/− mice exhibit an increased number of recorded licks by 35% and the drinking time is significantly increased by 38% compared with WT animals. For all experiments only male animals were used. Asterisks indicate significant difference (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; ANCOVA was used in A; unpaired 2-tailed Student’s t test was used in B and C).
To investigate whether lower body weight in adult Foxp1+/− animals is caused by reduced food and water intake, we monitored the animals using LABORAS and IntelliCage. LABORAS is a noninvasive device that automatically monitors eating and drinking behavior through animal movements. IntelliCage further validated changes in drinking behavior and assessed home cage behavior and cognitive performance of 16 mice in a social environment. LABORAS data showed that the number of feeds was lower in Foxp1+/− mice than WT littermates (Fig. 3B). Food and water uptake were significantly lower in Foxp1+/− animals and weight loss was higher compared with WT mice (Fig. 3B).
Foxp1+/− animals are hyperactive in the open field (6); therefore, the body weight in Foxp1+/− animals might be lower because these animals are more active and burn more calories. However, Foxp1+/− mice moved significantly less in a familiar environment than they did in the open field and the distance traveled by Foxp1+/− mice over 24 h did not differ from that of WT animals (Fig. 3B). IntelliCage monitoring for 5 consecutive days also confirmed that Foxp1+/− mice have altered drinking behavior. Both the number of licks and drinking time were significantly increased in Foxp1+/− animals, suggesting difficulties in swallowing food and water (Fig. 3C).
Relaxation Induced by Nitric Oxide Is Altered in the Lower Esophageal Sphincter of Foxp1+/− Mice.
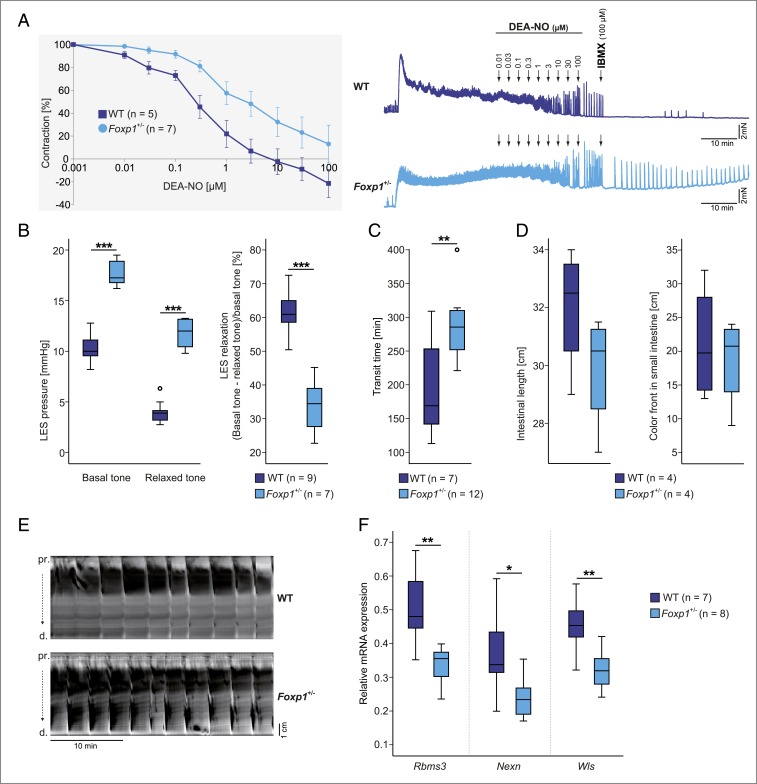
To analyze nitric oxide-induced relaxation within the GI tract of Foxp1+/− mice, we performed organ bath experiments using esophageal sphincter, fundus, and pylorus tissue. In these experiments, tissue samples from WT and Foxp1+/− animals were precontracted with 1 μM carbachol and then relaxed with increasing concentrations of the nitric oxide donor 2-(N,N-diethylamino)-diazenolate-2-oxide.diethylammonium salt (DEA-NO). Nitric oxide-induced relaxation did not differ between WT and Foxp1+/− mice in the fundus and pylorus (SI Appendix, Fig. S7). However, nitric oxide-induced relaxation was significantly reduced in the esophageal sphincter of Foxp1+/− animals compared with WT animals (Fig. 4A).
Fig. 4.
Altered contractility of the esophagus and the colon and dysregulated Foxp1 target genes in the esophagus of Foxp1+/− mice. (A) Quantitative analysis of organ bath experiments from WT and Foxp1+/− mice (Left) and representative original traces of organ bath experiments (Right) showing DEA-NO–induced relaxation in the lower esophageal sphincter (LES) from WT and Foxp1+/− animals after precontraction with 1 μM carbachol. The normally occurring dose-dependent relaxation in response to the NO donor DEA-NO is significantly decreased in the Foxp1+/− esophagus compared with WT tissue. Data shown are means ± SEM. IBMX, 3-isobutyl-1-methylxanthine. (B) Esophageal manometry of basal and swallow-induced LES tone in male WT and Foxp1+/− animals. Both basal and relaxed tones are significantly increased in the Foxp1+/− LES. The relaxation ability of the LES is reduced to 45% in Foxp1+/− mice. (C) Total transit time is increased by 44% in the Foxp1+/− GI tract. (D) The intestinal length of WT and Foxp1+/− animals is comparable and gastric emptying is not altered in Foxp1+/− mice compared with WT animals. (E) Representative original spatiotemporal map of the WT and Foxp1+/− colon. Foxp1+/− colons display an atypical contraction pattern with strongly contracted segments within both proximal (pr) and distal (d) parts. (F) Expression of Rbms3, Nexn, and Wls in esophageal tissue from adult Foxp1+/− and WT animals. mRNA levels were compared between Foxp1+/− and WT tissue via quantitative real-time PCR. Rbms3, Nexn, and Wls are significantly down-regulated in Foxp1+/− tissue. Asterisks indicate significant difference (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; unpaired 2-tailed Student’s t test was used in A–D; ANCOVA was used in F).
In Vivo Manometry of Basal and Relaxed Tone in the Lower Esophageal Sphincter after Swallowing Reveals Achalasia in Foxp1+/− Animals.
To verify our organ bath results in vivo, we compared the lower esophageal sphincter tone of WT and Foxp1+/− mice using esophageal manometry in anesthetized mice. Basal and relaxed tone of the lower esophageal sphincter was significantly increased in Foxp1+/− animals, which explains the impaired relaxation of the lower esophageal sphincter (Fig. 4B). This finding confirms our in vitro observations and is consistent with the idea that Foxp1+/− mice suffer from achalasia.
Total Gut Transit Is Significantly Prolonged in Foxp1+/− Mice.
To analyze whether reduced Foxp1 expression affects gut motility, we measured the total transit time in WT and Foxp1+/− animals. Transit time was significantly increased by 44% in the Foxp1+/− GI tract, indicating impaired motility and peristalsis (Fig. 4C). As differences in intestinal length and gastric retention can influence the transit time, we examined intestinal length and gastric emptying in Foxp1+/− mice. Compared with WT animals, no difference in intestinal length or abnormalities in gastric food transport was detected (Fig. 4D). Consequently, neither a longer intestinal tract nor defective gastric peristalsis seems to be responsible for the prolonged transit time in Foxp1+/− mice. Spatiotemporal maps depicting the contraction patterns of the colon, however, displayed differences in contractility in Foxp1+/− animals. WT colons exhibited regular long-distance contractions from the proximal to the distal parts, but Foxp1+/− organs showed partial retrograde contractions, together with strongly contracted segments within the proximal and distal parts of the colon (Fig. 4E). This is shown in Movies S1 and S2, which demonstrate peristalsis with pendular (swaying) movements in the Foxp1+/− colon, a sign of severely impaired colon motility.
Foxp1+/− Animals Display Significant Down-Regulation of Nexn, Rbms3, and Wls in the Esophagus.
To understand how Foxp1 haploinsufficiency may impact on the GI tract, we investigated Foxp1 target genes that were identified in striatal tissue of Nestin-Cre (Foxp1−/−) mice. Thirty-seven genes were significantly dysregulated in the striatum at both embryonic day (E)18.5 (ref. 12; Dataset S1) and P1.5 (Dataset S1) (5), when striatal degeneration is first detected (SI Appendix, Fig. S8). To find out whether Foxp1 targets in striatal tissue are also dysregulated in the esophagus of haploinsufficient Foxp1+/− mice, we consulted RNA (https://www.gtexportal.org) and protein (https://www.proteinatlas.org) expression databases. Five of the dysregulated genes in the Nestin-Cre (Foxp1−/−) striatum (Epha7, Nexn, Rbms3, Vcl, and Wls) are also highly expressed in esophageal tissue (SI Appendix, Fig. S9). Therefore, we tested the expression of these genes in the esophagus of WT and Foxp1+/− mice by quantitative real-time PCR at P12.5 and adult stage. Three genes, Nexn, Rbms3, and Wls, were significantly down-regulated by 36, 34, and 29% in the esophagus of adult Foxp1+/− animals, respectively (Fig. 4F).
Discussion
Only recently, genetic defects affecting brain function in ASD have been explored regarding their impact on the gastrointestinal tract and enteric nervous system (13). This is based on the observation that individuals with ASD are more likely to develop GI disturbances than healthy individuals (14). Moreover, autism severity apparently associates with an increased probability of having GI problems (15). The pathophysiology of GI dysfunction in ASD is still poorly understood because of its heterogeneous profile. Unfortunately, GI distress is difficult to assess in children with ASD due to their difficulties in communicating and their altered perception of pain (14, 16). Therefore, it remains unclear whether GI symptoms in ASD individuals are caused by primary GI impairment or are a secondary effect of altered behavior or psychotropic medication.
Using Foxp1+/− mice as a model of FOXP1 haploinsufficiency, we demonstrated that loss of 1 Foxp1 copy leads to a pronounced atrophy of the tunica muscularis in the esophagus and colon, accompanied by a motility dysfunction. We revealed that reduced muscle cell proliferation is the likely cause of muscular atrophy in these animals, whereas achalasia patients often present with increased muscular thickness. Nestin-Cre (Foxp1−/−) mice with deletion of Foxp1 exclusively in the nervous system also display muscular changes in the esophagus, with an increased thickness of the muscular layer. But despite the fact that Nestin-Cre (Foxp1−/−) and Foxp1+/− mice exhibit contrary muscle malformation, neuronal dysfunctions may contribute to the phenotype in both mouse models. Reduced muscle thickness was also reported in the esophagus of Foxp1+/−/Foxp2−/− mice (17), although the authors could not explain the underlying mechanism. Foxp1 and Foxp2 interact during development, but it was not clear to what extent the heterozygous reduction of Foxp1 contributed to the reduced muscle thickness in this study. Our data now suggest that heterozygous Foxp1 deficiency is sufficient to cause esophageal aberration.
Foxp1+/− mice showed an increased number of licks and a longer drinking time, which can probably be explained by swallowing problems. Investigation of esophageal function using organ bath experiments and manometry in living animals confirmed this hypothesis. Foxp1+/− mice had significantly higher basal and relaxed tone in the lower esophageal sphincter, which are characteristics of achalasia. The causes of achalasia are unknown, but it can either occur in isolation or as part of a syndrome. Several mechanisms have been suggested, including autoimmune dysfunction, neurodegenerative or infectious contributions, and genetics (18, 19). Recently, impaired relaxation of the esophagus was attributed to interstitial cells of Cajal (ICCs), the pacemaker cells of peristalsis, which control the contraction of smooth muscle cells in the GI tract and mediate enteric motor responses (20). Although we did not detect any obvious differences in the number of ICCs in the esophagus of WT and Foxp1+/− animals (immunofluorescence staining of cKit) and the NO-sensitive guanylyl cyclase which modulates the lower esophageal sphincter tone (SI Appendix, Fig. S10), we cannot totally exclude the possibility that ICC function is impaired or that structural or functional changes in the enteric nervous system exist.
Adult Foxp1+/− mice exhibit a lower body weight than WT animals, most likely caused by reduced food and water intake. Foxp1+/− mice are hyperactive under stress (6), and we confirmed increased traveling in the open-field test. However, this hyperactivity is unlikely to contribute to weight loss because Foxp1+/− mice moved significantly less than WT animals when monitored in a familiar environment for several days. Weight loss in Foxp1+/− mice increased with age and body weight was only significantly lower in adult mice; this is probably explained by changes in diet during development. Until P21, juvenile animals are still suckling milk, which is easier to swallow than solid food. Therefore, swallowing problems due to achalasia may be exacerbated at late juvenile stages, causing significant weight loss in adulthood.
Achalasia has been reported in several mouse models, including Gucy1b1, Rassf1a, nNOS, Kit, and Spry2 knockout mice (20–24). Mutation analysis confirmed that homozygous loss of NOS1 causes early-onset achalasia (25) and, interestingly, these patients were also diagnosed with autism (26). Homozygous or compound heterozygous mutations in GMPPA, AAAS, and GUCY1A genes were reported to cause achalasia in Triple-A syndrome [Mendelian Inheritance in Man (MIM) 231550], AAMR syndrome (MIM 615510), and Moyamoya disease-6 syndrome (MIM 615750) (26–29), respectively. Foxp1 is the first gene reported to cause achalasia in the heterozygous state.
Interestingly, genome-wide association studies have revealed a susceptibility locus 75 kb downstream of FOXP1 that modifies the association of gastroesophageal reflux with Barrett’s esophagus and esophageal adenocarcinoma (30, 31). Barrett’s esophagus is characterized by metaplastic changes to the cell lining of the lower esophagus. These changes are caused by chronic acid exposure from reflux esophagitis. Gastroesophageal reflux was also reported in patients with FOXP1 syndrome (32). In contrast to achalasia, gastroesophageal reflux disease develops when a reflux of gastric contents causes symptoms and complications. The lower esophageal sphincter is hypertensive and does not relax after swallowing in achalasia. But in gastroesophageal reflux disease, the lower esophageal sphincter is hypotensive and frequently relaxes. It remains controversial whether these 2 conditions coexist or whether one disease transforms into the other (33, 34). Heartburn and regurgitation, the main symptoms of gastroesophageal reflux disease, commonly occur during the early stages of achalasia and are consequently poor indicators of esophageal motility disorder (35). Therefore, children with FOXP1 syndrome who present with gastroesophageal reflux may be displaying early signs of achalasia.
In addition to esophageal dysfunction, GI transit was significantly prolonged in Foxp1+/− mice. However, GI length or gastric emptying—which may strongly affect transit time—was not altered. Pronounced atrophy of the tunica muscularis in the colon suggested motility defects. Indeed, propulsive peristalsis was severely disrupted and pendular swaying movements were observed, which prolonged the transport of chyme. These findings may also explain why constipation is frequently reported in patients with FOXP1 syndrome. Other ASD genes have been previously implicated in slow-transit constipation, including CHD8, TCF4, SLC6A4, and SHANK3 (13, 36–39). These observations together with our findings in Foxp1 mouse models suggest that genetic defects affecting the central nervous system in ASD might also affect the enteric nervous system and GI tract, which would explain the high prevalence of GI symptoms in individuals with ASD (40).
Expression of Foxp1 in the GI tract, enteric nervous system, and brain throughout development suggests that this transcription factor plays a fundamental role in their development, probably by orchestrating the regulation of target genes that are common or distinct to different tissues. To better understand Foxp1-mediated gene regulation in different tissues, we analyzed 5 genes that were dysregulated in the striatum of our Nestin-Cre (Foxp1−/−) mice and discovered a significant down-regulation of 3 of these genes (Nexn, Rbms3, and Wls) in the esophagus of adult Foxp1+/− mice compared with WT mice.
Nexilin (encoded by Nexn) is an actin filament-binding protein present at cell-matrix adherens junctions, and is expressed in the Z discs of heart and skeletal muscle (41, 42). Loss of nexilin destabilizes Z discs, causes heart failure, and reduces actin polymerization and cell migration (43). As Nexn expression is reduced in Foxp1+/− mice, we examined its protein expression in the esophagus of WT animals and the structure of sarcomeres and F-actin filaments in esophageal tissue sections of Foxp1+/− and WT mice (SI Appendix, Figs. S11 and S12). We observed no obvious alterations in Z-disc structure or disruptions in the actin filaments of smooth muscle cells in the mid and distal esophagus of 8-wk-old Foxp1+/− and WT animals by confocal microscopy (SI Appendix, Fig. S12). It is possible, however, that mild structural abnormalities may be detected only by higher-resolution methods, such as transmission electron microscopy. Furthermore, actin filaments may only be structurally altered over time by mechanical stress; therefore, any changes might only be detected in older animals.
The second dysregulated gene, Rbms3, encodes an RNA-binding protein and acts as a tumor suppressor in esophageal squamous cell carcinoma and other cancers (44, 45). Missense mutations in RBMS3 have been detected in mucosal biopsies of patients with Barrett’s esophagus and have been associated with esophageal adenocarcinoma (46). Protein expression was detected in the epithelium mucosae of the esophagus (SI Appendix, Fig. S13). Therefore, Rbms3 down-regulation in the adult Foxp1+/− esophagus may promote dysplasia of the squamous epithelium. This may be exacerbated by gastroesophageal reflux. The third dysregulated gene, Wls (Wntless), encodes a Wnt receptor and plays a role in the morphogenesis of pulmonary vasculature and is also expressed in smooth and skeletal muscle of the esophagus (SI Appendix, Fig. S14). Wnt/β-catenin signaling regulates the proliferation of smooth muscle cells and is required for smooth muscle maintenance (47). Decreased Wls expression might therefore be responsible for the reduced proliferation of smooth muscle cells that we found in the Foxp1+/− esophagus at P12.5.
Regulatory networks among transcription factors are highly conserved (48). Also, patterns of motility in the esophagus and colon are remarkably similar between humans and mice (21, 49). Therefore, our findings that Foxp1+/− mice suffer from achalasia and impaired colon motility may be extrapolated to humans and might explain the feeding difficulties, gastroesophageal reflux, and constipation seen in individuals with FOXP1 syndrome. Our data show that these clinical GI symptoms are not just comorbidities or consequences of medication but are part of the phenotype of FOXP1 syndrome. Our results, which reveal a FOXP1-dependent pathomechanism for GI phenotypes in ASD, should be considered in order to prevent serious sequelae such as Barrett’s esophagus and esophageal adenocarcinoma. In other words, regular checkups with gastroenterologists and symptomatic treatment for the gastrointestinal disturbances are indicated. On a more global level, our data support the idea that GI dysfunction should be seriously taken into account in ASD patients and appropriate treatments should be administered. This may also have a positive impact on the overall well-being of these patients, as GI problems often exacerbate existing behavioral abnormalities because of pain, stress, or discomfort (50).
Materials and Methods
Animals.
Mice were kept in a specific pathogen-free biomedical animal facility under a 12-h light–dark cycle and given ad libitum access to water and food. All procedures were conducted in strict compliance with the NIH Guidelines for the Care and Use of Laboratory Animals (51) and approved by the National Institute of Mental Health Animal Care and Use Committee. Animal studies were approved by the Regierungspräsidium Karlsruhe (approval nos. 35-9185.81/G-105/16 and 35-9185.81/G-86/14). The day of birth was considered as postnatal day 0.5.
Generation of Nestin-Cre (Foxp1−/−) Mice.
Homozygous floxed Foxp1 mice (52) were crossed with Nestin-Cre transgenic mice [B6.Cg-Tg(Nes-cre)1Kln/JIn] (53) heterozygous for the floxed Foxp1 allele (see also SI Appendix, Materials and Methods).
Generation of Foxp1+/− Animals.
WT female mice were crossed with male mice heterozygous for the Foxp1 KO allele (Foxp1+/−) (10).
Measurement of Food and Water Intake.
Food and water intake of our animals was analyzed over a time period of 24 h using the LABORAS home cage monitoring system (Metris), an automated animal behavior recognition system. Drinking behavior was further validated by using the IntelliCage system (TSE Systems).
mRNA Expression and Protein Analysis.
cDNA synthesis, quantitative real-time PCR, and protein isolation were performed using standard protocols as described in SI Appendix, Materials and Methods. Western blot analysis was executed using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Histological Staining.
Staining by hematoxylin and eosin stain as well as Masson–Goldner trichrome (Carl Roth) was performed on 3-µm paraffin sections according to the manufacturer’s instructions.
Analysis of Muscle Thickness in the Esophagus and Colon.
An exact description of the histological analysis for both mouse models used in this study is provided in SI Appendix, Fig. S15.
Immunofluorescence Analysis.
Immunostaining and TUNEL staining were performed as described in SI Appendix, Materials and Methods.
Whole-Colon Preparation, Isometric Force Studies, Total Gut Transit, and Esophageal Manometry.
Colonic motility patterns were recorded and esophageal manometry was performed as described earlier (54, 55). A detailed description of the methods is provided in SI Appendix, Materials and Methods.
Statistics.
Data analysis was performed as described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge the expert advice of Dr. Claudia Pitzer and the support from the Interdisciplinary Neurobehavioral Core Unit, and we thank Dr. Carsten Sticht and Dr. Carolina De la Torre from the University Medical Centre of the Medical Faculty Mannheim (head: Dr. Norbert Gretz) for performing the microarray experiments and related services. We also thank Dr. Rolf Sprengel, Institute for Anatomy and Cell Biology, Heidelberg and the Nikon Imaging Center at the University of Heidelberg for technical support, Dr. Claire Bacon and Bianca Christian for helpful comments, and Dr. Phil Tucker and Dr. Geneviève Konopka for kindly providing the Foxp1+/− mice. This project was funded by the Deutsche Forschungsgemeinschaft (RA 380/15-1 and FR 1725/5-1) and by the Medical Faculty of the University of Heidelberg. G.R. is a member of the CellNetworks Cluster of Excellence (EXC 81), Interdisciplinary Center for Neurosciences, and Center of Rare Disease.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data (E18.5 striatal tissue from WT and Nestin-Cre [Foxp1−/−] animals) reported in this paper are available via the Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138337, and in Dataset S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911429116/-/DCSupplemental.
References
- 1.Holingue C., Newill C., Lee L. C., Pasricha P. J., Daniele Fallin M., Gastrointestinal symptoms in autism spectrum disorder: A review of the literature on ascertainment and prevalence. Autism Res. 11, 24–36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn D., et al. , Identification of FOXP1 deletions in three unrelated patients with mental retardation and significant speech and language deficits. Hum. Mutat. 31, E1851–E1860 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meerschaut I., et al. , FOXP1-related intellectual disability syndrome: A recognisable entity. J. Med. Genet. 54, 613–623 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Siper P. M., et al. , Prospective investigation of FOXP1 syndrome. Mol. Autism 8, 57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacon C., et al. , Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behaviour. Mol. Psychiatry 20, 632–639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araujo D. J., et al. , FoxP1 orchestration of ASD-relevant signaling pathways in the striatum. Genes Dev. 29, 2081–2096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araujo D. J., et al. , Foxp1 in forebrain pyramidal neurons controls gene expression required for spatial learning and synaptic plasticity. J. Neurosci. 37, 10917–10931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X., et al. , Foxp1 regulates cortical radial migration and neuronal morphogenesis in developing cerebral cortex. PLoS One 10, e0127671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Usui N., et al. , Foxp1 regulation of neonatal vocalizations via cortical development. Genes Dev. 31, 2039–2055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B., et al. , Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development 131, 4477–4487 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Dogan I., Puckett J. L., Padda B. S., Mittal R. K., Prevalence of increased esophageal muscle thickness in patients with esophageal symptoms. Am. J. Gastroenterol. 102, 137–145 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Fröhlich H., Rappold G., Gene expression in the striatum of Nestin-Cre (Foxp1-/-) mice in comparison to WT animals at embryonic day (E)18.5. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138337. Deposited 2 October 2019.
- 13.Margolis K. G., et al. , Serotonin transporter variant drives preventable gastrointestinal abnormalities in development and function. J. Clin. Invest. 126, 2221–2235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McElhanon B. O., McCracken C., Karpen S., Sharp W. G., Gastrointestinal symptoms in autism spectrum disorder: A meta-analysis. Pediatrics 133, 872–883 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Wang L. W., Tancredi D. J., Thomas D. W., The prevalence of gastrointestinal problems in children across the United States with autism spectrum disorders from families with multiple affected members. J. Dev. Behav. Pediatr. 32, 351–360 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Buie T., et al. , Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: A consensus report. Pediatrics 125 (suppl. 1), S1–S18 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Shu W., et al. , Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development 134, 1991–2000 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Boeckxstaens G. E., Zaninotto G., Richter J. E., Achalasia. Lancet 383, 83–93 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Gockel I., et al. , Common variants in the HLA-DQ region confer susceptibility to idiopathic achalasia. Nat. Genet. 46, 901–904 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Groneberg D., et al. , Dominant role of interstitial cells of Cajal in nitrergic relaxation of murine lower oesophageal sphincter. J. Physiol. 593, 403–414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivarao D. V., Mashimo H. L., Thatte H. S., Goyal R. K., Lower esophageal sphincter is achalasic in nNOS(−/−) and hypotensive in W/W(v) mutant mice. Gastroenterology 121, 34–42 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Taketomi T., et al. , Loss of mammalian Sprouty2 leads to enteric neuronal hyperplasia and esophageal achalasia. Nat. Neurosci. 8, 855–857 (2005). [DOI] [PubMed] [Google Scholar]
- 23.van der Weyden L., Happerfield L., Arends M. J., Adams D. J., Megaoesophagus in Rassf1a-null mice. Int. J. Exp. Pathol. 90, 101–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward S. M., Morris G., Reese L., Wang X. Y., Sanders K. M., Interstitial cells of Cajal mediate enteric inhibitory neurotransmission in the lower esophageal and pyloric sphincters. Gastroenterology 115, 314–329 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Müller M., et al. , Murine genetic deficiency of neuronal nitric oxide synthase (nNOS(−/−)) and interstitial cells of Cajal (W/W(v)): Implications for achalasia? J. Gastroenterol. Hepatol. 29, 1800–1807 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Shteyer E., et al. , Truncating mutation in the nitric oxide synthase 1 gene is associated with infantile achalasia. Gastroenterology 148, 533–536.e4 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Koehler K., et al. , Mutations in GMPPA cause a glycosylation disorder characterized by intellectual disability and autonomic dysfunction. Am. J. Hum. Genet. 93, 727–734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tullio-Pelet A., et al. , Mutant WD-repeat protein in triple-A syndrome. Nat. Genet. 26, 332–335 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Wallace S., et al. , Disrupted nitric oxide signaling due to GUCY1A3 mutations increases risk for Moyamoya disease, achalasia and hypertension. Clin. Genet. 90, 351–360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai J. Y., et al. , A newly identified susceptibility locus near FOXP1 modifies the association of gastroesophageal reflux with Barrett’s esophagus. Cancer Epidemiol. Biomarkers Prev. 24, 1739–1747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine D. M., et al. , A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett’s esophagus. Nat. Genet. 45, 1487–1493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers A., et al. , FOXP1 haploinsufficiency: Phenotypes beyond behavior and intellectual disability? Am. J. Med. Genet. A 173, 3172–3181 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Jaakkola A., Reinikainen P., Ovaska J., Isolauri J., Barrett’s esophagus after cardiomyotomy for esophageal achalasia. Am. J. Gastroenterol. 89, 165–169 (1994). [PubMed] [Google Scholar]
- 34.Sprung D. J., Gibb S. P., Barrett’s esophagus in a patient with achalasia. Am. J. Gastroenterol. 80, 330–333 (1985). [PubMed] [Google Scholar]
- 35.Patti M. G., et al. , Importance of preoperative and postoperative pH monitoring in patients with esophageal achalasia. J. Gastrointest. Surg. 1, 505–510 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Barnard R. A., Pomaville M. B., O’Roak B. J., Mutations and modeling of the chromatin remodeler CHD8 define an emerging autism etiology. Front. Neurosci. 9, 477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James D. M., et al. , Intestinal dysmotility in a zebrafish (Danio rerio) shank3a;shank3b mutant model of autism. Mol. Autism 10, 3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sweatt J. D., Pitt-Hopkins syndrome: Intellectual disability due to loss of TCF4-regulated gene transcription. Exp. Mol. Med. 45, e21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veenstra-VanderWeele J., et al. , Autism gene variant causes hyperserotonemia, serotonin receptor hypersensitivity, social impairment and repetitive behavior. Proc. Natl. Acad. Sci. U.S.A. 109, 5469–5474 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao M., Gershon M. D., The bowel and beyond: The enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol. Hepatol. 13, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassel D., et al. , Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat. Med. 15, 1281–1288 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Ohtsuka T., et al. , Nexilin: A novel actin filament-binding protein localized at cell-matrix adherens junction. J. Cell Biol. 143, 1227–1238 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu B., et al. , Nexilin/NEXN controls actin polymerization in smooth muscle and is regulated by myocardin family coactivators and YAP. Sci. Rep. 8, 13025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., et al. , Downregulation of RBMS3 is associated with poor prognosis in esophageal squamous cell carcinoma. Cancer Res. 71, 6106–6115 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Zhang T., et al. , Low expression of RBMS3 and SFRP1 are associated with poor prognosis in patients with gastric cancer. Am. J. Cancer Res. 6, 2679–2689 (2016). [PMC free article] [PubMed] [Google Scholar]
- 46.Streppel M. M., et al. , Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett’s esophagus. Oncogene 33, 347–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang M., et al. , Gpr177 regulates pulmonary vasculature development. Development 140, 3589–3594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stergachis A. B., et al. , Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515, 365–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spencer N. J., Motility patterns in mouse colon: Gastrointestinal dysfunction induced by anticancer chemotherapy. Neurogastroenterol. Motil. 28, 1759–1764 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Gorrindo P., et al. , Gastrointestinal dysfunction in autism: Parental report, clinical evaluation, and associated factors. Autism Res. 5, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 52.Feng X., et al. , Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood 115, 510–518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tronche F., et al. , Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Beck K., et al. , Cell-specific effects of nitric oxide on the efficiency and frequency of long distance contractions in murine colon. Neurogastroenterol. Motil. 31, e13589 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Zizer E., et al. , Loss of Lsc/p115 protein leads to neuronal hypoplasia in the esophagus and an achalasia-like phenotype in mice. Gastroenterology 139, 1344–1354 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.