Significance
Antibodies are important research tools because they can be developed to bind to as well as directly inhibit almost any protein. Unfortunately, antibodies cannot cross the plasma membrane and are therefore limited to perturbing membrane or secreted protein activity. We found that by appending anionic polypeptides (ApPs) to immunoglobulin G (IgG) antibodies, they could be complexed with cationic lipids, originally designed for nucleic acid delivery, through electrostatic interactions to enable efficient cytosolic antibody delivery. By fusing ApPs to photoreactive antibody-binding domains, we can rapidly functionalize almost any off-the-shelf IgG without genetic reengineering of the cargo IgG. Our cytosolically delivered antibodies are capable of inhibiting intracellular proteins, establishing a new approach for studying intracellular protein function with inhibitory antibodies.
Keywords: antibody, protein delivery, cytosolic, intracellular, penetrating
Abstract
Antibodies can be developed to directly inhibit almost any protein, but their inability to enter the cytosol limits inhibitory antibodies to membrane-associated or extracellular targets. Developing a cytosolic antibody delivery system would offer unique opportunities to directly inhibit and study intracellular protein function. Here we demonstrate that IgG antibodies that are conjugated with anionic polypeptides (ApPs) can be complexed with cationic lipids originally designed for nucleic acid delivery through electrostatic interactions, enabling close to 90% cytosolic delivery efficiency with only 500 nM IgG. The ApP is fused to a small photoreactive antibody-binding domain (pAbBD) that can be site-specifically photocrosslinked to nearly all off-the-shelf IgGs, enabling easy exchange of cargo IgGs. We show that cytosolically delivered IgGs can inhibit the drug efflux pump multidrug resistance-associated protein 1 (MRP1) and the transcription factor NFκB. This work establishes an approach for using existing antibody collections to modulate intracellular protein function.
Antibodies have become important research tools because they can be developed to bind nearly any exposed protein epitope with high affinity and specificity through either traditional immunization or in vitro display approaches (1). By binding to an appropriate epitope, antibodies can also directly inhibit their antigen’s biological activity by either sterically blocking the antigen from binding to interaction partners or locking the antigen in an inactive conformation (1, 2). Indeed, microinjected antibodies have not only shown that antibody-dependent inhibition of intracellular proteins is possible, but have also been used to uncover the biological roles of oncogenes (3), stress response proteins (4), and regulatory proteins (5, 6). Although physical delivery techniques such as microinjection (3–7) or electroporation (7, 8) are very effective at cytosolic antibody delivery, they are low throughput and can result in significant toxicity, significantly limiting the utility of cytosolic antibodies as research tools.
Unsurprisingly, many alternative approaches have been explored for cytosolic antibody delivery (9, 10). For example, antibody fragments or antibody-like binding proteins can be engineered for cytosolic stability and expressed intracellularly as intrabodies (9, 10). Antibodies or antibody fragments can also be fused to or incubated with cell-penetrating peptides (CPPs) to induce their endocytic uptake into cells followed by endosomal escape into the cytosol (11, 12). Finally, antibodies with an innate ability to enter the cytosol have recently been developed in which the delivery moiety lies within the light chain variable region (13).
Carrier-mediated approaches for cytosolic protein delivery, in which cargo proteins are encapsulated by delivery lipids or polymers, have also been explored and capitalize on advances in nonviral nucleic acid delivery formulations that have already been proven effective (14, 15). Although carrier-mediated cytosolic antibody delivery has been reported multiple times (9, 10), those claims must be evaluated cautiously. A stringent assessment of commercially available carrier-mediated antibody delivery platforms revealed that none were capable of cytosolic delivery to >6% of cells (8). However, recent progress in delivering the Cas9 protein, which is similar in size to IgG antibodies, with lipid nanoparticles for genome editing (16) suggests that carrier-mediated approaches are still viable strategies for cytosolic antibody delivery.
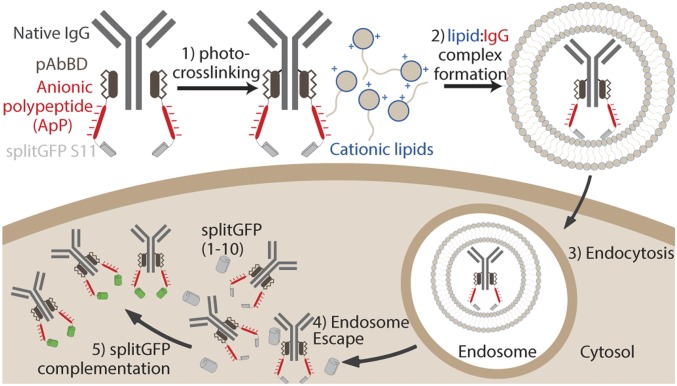
Inspired by strategies for complexing proteins with cationic lipids (16), polymers (17–19), and nanoparticles (20), we hypothesized that IgGs functionalized with anionic polypeptides (ApPs) could mimic the polyanionic nature of nucleic acids and be complexed with cationic lipids designed for nucleic acid delivery. Rather than engineering IgGs directly, we fused ApPs to a photoreactive antibody-binding domain (pAbBD) that could be photocrosslinked to each heavy chain of an IgG to create highly negatively charged IgG-(pAbBD-ApP)2 conjugates without perturbing binding affinity (Fig. 1) (21). Because functionality is built into the pAbBD rather than the IgG, cargo IgGs can be easily exchanged without genetic reengineering, allowing most off-the-shelf IgGs to be easily functionalized.
Fig. 1.
Schematic of antibody delivery approach. (1) pAbBD is purified as a fusion to an ApP as well as the splitGFP S11 reporter peptide and then photocrosslinked to a cargo IgG. (2) IgG-(pAbBD-ApP-S11)2 conjugates are complexed with cationic delivery lipids. (3) Lipid-IgG complexes are taken up by cells via endocytosis. (4) Delivery lipids promote endosome escape and cytosolic IgG-(pAbBD-ApP-S11)2 delivery. (5) Cytosolic IgG-(pAbBD-ApP-S11)2 delivery can be detected by splitGFP complementation and turn-on fluorescence.
Here we report that our cytosolic antibody delivery approach enables close to 90% delivery efficiency at a concentration of only 500 nM IgG. Our modular antibody functionalization strategy is compatible with IgGs from many different species and isotypes and our complexation approach is compatible with a diverse set of cationic lipids. Finally, we demonstrate that cytosolically delivered IgGs are functional and can inhibit not only the drug efflux pump MRP1 to sensitize cancer cells to chemotherapeutic drugs, but also the transcription factor NFκB. These findings establish that our method enables easy and efficient cytosolic delivery of almost any off-the-shelf IgG antibody and can extend the potential applications of existing IgG antibody collections.
Results
Stringent Detection of Cytosolic Protein Delivery.
When developing intracellular protein delivery technologies, it is imperative to use an assay that only detects cargo proteins that have been successfully delivered to the cytosol. Endosome escape is the major bottleneck for cytosolic delivery, resulting in large false-positive rates for assays that measure total cellular uptake of cargo proteins (22, 23). Accordingly, we used a stringent self-assembling splitGFP (24) reporter system in which one half of the splitGFP, the S11 peptide, is fused to pAbBD-ApP, whereas the other half, splitGFP(1–10), is expressed in reporter cells (25–28). Only once IgG-(pAbBD-ApP-S11)2 is successfully delivered into the cytosol does splitGFP complementation and turn-on fluorescence occur (Fig. 1). Furthermore, fluorescence intensity is directly correlated with the amount of protein that is cytosolically delivered.
We prepared pAbBD-ApP-S11 with 10 to 30 residues-long polyaspartate or polyglutamate ApPs and confirmed that they could photocrosslink to rituximab (Ritux) (SI Appendix, Fig. S1 A and B). We chose rituximab because its antigen, CD20, is not expressed in our reporter cells, removing a potential confounding factor for delivery. To validate that our protein cargos are compatible with the splitGFP reporter system, we confirmed that splitGFP complementation occurred when either pAbBD-S11 or Ritux-(pAbBD-S11)2 were incubated with purified splitGFP(1–10) or physically delivered into HEK293T splitGFP(1–10) reporter cells by electroporation (SI Appendix, Fig. S1 C–F). Due to the time required for chromophore maturation (SI Appendix, Fig. S1 C and D), reporter cells were assessed for splitGFP fluorescence 6 h following electroporation with either flow cytometry or live-cell fluorescence microscopy. As expected, fluorescence microscopy revealed diffuse splitGFP fluorescence with both pAbBD-S11 and Ritux-(pAbBD-S11)2, but fluorescence was depleted from the nucleus only with Ritux-(pAbBD-S11)2 (SI Appendix, Fig. S1F). Because pAbBD-S11 is only ∼11 kDa, it is capable of passively translocating across the nuclear pore complex, whereas Ritux-(pAbBD-S11)2 is ∼170 kDa and is very inefficient at passive nuclear translocation (29).
Cytosolic pAbBD Delivery.
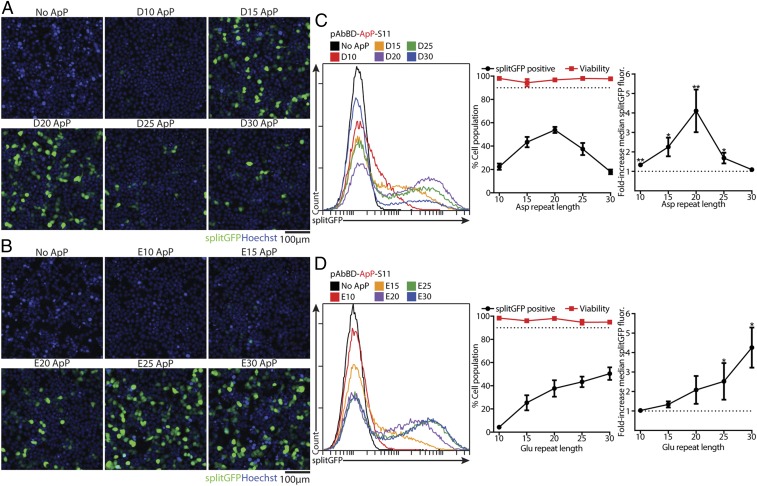
Initially, we wondered whether pAbBD-ApP-S11 alone, without IgG, could be cytosolically delivered into HEK293T splitGFP(1–10) cells once complexed with a commercially available cationic transfection lipid, Lipofectamine (Lipo) 2000. We tested simple polyaspartate or polyglutamate ApPs that were 10 to 30 residues long. Live-cell fluorescence microscopy showed robust splitGFP fluorescence, indicating substantial cytosolic delivery, once polyaspartate ApPs were at least 15 residues long (D15), with a peak at 20 aspartate residues (D20) (Fig. 2A). With polyglutamate ApPs, splitGFP fluorescence increased with length up to 30 glutamate residues (E30) (Fig. 2B). Even though the base pAbBD-S11 has a net charge of −7, no cytosolic delivery could be detected without ApPs (Fig. 2 A and B).
Fig. 2.
Optimizing ApPs for cytosolic pAbBD delivery. A total of 500 nM pAbBD-S11 (negative control) or pAbBD-ApP-S11 with either polyaspartate or polyglutamate ApPs 10 15, 20, 25, or 30 residues long were complexed with 2 μL Lipo 2000 and added to HEK293T splitGFP(1–10) cells for 6 h. (A and B) Representative live-cell fluorescence microscopy images following delivery with polyaspartate (A) and polyglutamate (B) ApPs shows diffuse splitGFP fluorescence indicating significant cytosolic delivery. (C and D) Flow cytometry of splitGFP fluorescence following delivery with polyaspartate (C) and polyglutamate (D) ApPs. (Left) Representative flow cytometry histograms. (Center) Percent of cells splitGFP-positive. (Right) Fold increase in median splitGFP fluorescence over negative control. The dotted line indicates either 90% of the cell population (Center) or no increase in fluorescence (Right). Viability was determined with the LDH assay. Data are mean ± SEM, n = 4; *P < 0.05, **P < 0.01 (1-sided 1-sample t test of log ratios).
Using flow cytometry, we quantified splitGFP fluorescence as either the percentage of splitGFP-positive cells, which reflects delivery efficiency, or the fold increase in splitGFP fluorescence, which reflects the amount of protein delivered (Fig. 2 C and D and SI Appendix, Fig. S2). Flow cytometry confirmed the trends seen between ApP length and delivery efficiency identified via microscopy. Increasing the ratio of cationic lipid to protein generally improved delivery efficiency, but at the cost of increased toxicity (SI Appendix, Fig. S2). We were able to identify regimes, though, where viability remained greater than 90% with excellent delivery efficiency (Fig. 2 C and D and SI Appendix, Fig. S2). The best polyglutamate and polyaspartate ApPs were D20 and E30 with delivery efficiencies of 53.9 ± 2.6% and 50.5 ± 5.4%, respectively, when 500 nM pAbBD-ApP-S11 was complexed with 2 µL Lipo 2000 (Fig. 2 C and D).
Together, these results demonstrate that a small protein, pAbBD-S11 (∼11 kDa), can be efficiently delivered into the cytosol simply by fusing it to polyaspartate or polyglutamate ApPs via complexation with cationic lipids. We believe that this strategy can be easily adopted for cytosolic delivery of small antibody-like binding proteins such as affibodies (∼6 kDa), monobodies (∼10 kDa), or nanobodies (∼15 kDa), but ApP length may need to be reoptimized for maximal delivery.
Cytosolic IgG Delivery.
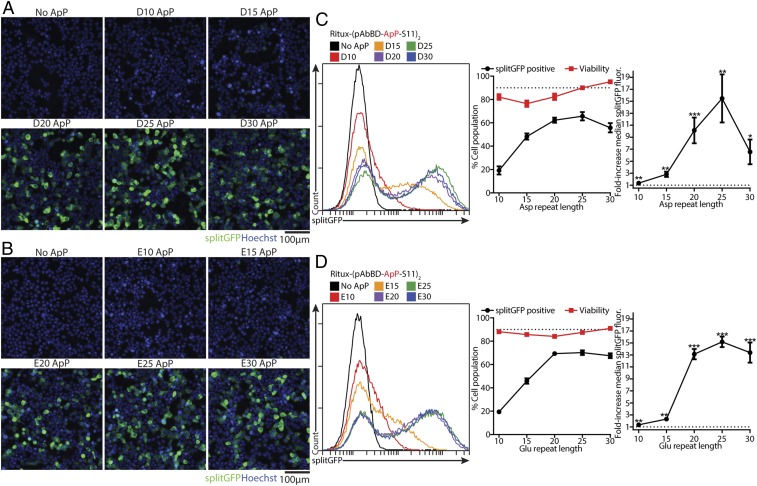
Next, we tested whether Ritux-(pAbBD-ApP-S11)2 could also be cytosolically delivered when complexed with Lipo 2000. As expected, no splitGFP fluorescence could be detected by microscopy with Ritux-(pAbBD-S11)2, which has a base net charge of +4 (Fig. 3 A and B). Once both polyaspartate and polyglutamate ApPs reached at least 20 residues long, however, microscopy revealed diffuse splitGFP fluorescence with nuclear depletion (Fig. 3 A and B). Because nuclear depletion indicates that the S11 reporter peptide remained linked to Ritux-(pAbBD-ApP-S11)2 following endosomal escape, we are confident that the splitGFP fluorescence is reflective of significant cytosolic delivery of Ritux-(pAbBD-ApP-S11)2.
Fig. 3.
Optimizing ApPs for cytosolic IgG delivery. A total of 500 nM Ritux-(pAbBD-S11)2 (negative control) or Ritux-(pAbBD-ApP-S11)2 with either polyaspartate or polyglutamate ApPs 10, 15, 20, 25, or 30 residues long were complexed with 2 μL Lipo 2000 and added to HEK293T splitGFP(1–10) cells for 6 h. (A and B) Representative live-cell fluorescence microscopy images following delivery with polyaspartate (A) and polyglutamate (B) ApPs shows diffuse splitGFP fluorescence with nuclear depletion indicating significant cytosolic delivery. (C and D) Flow cytometry of splitGFP fluorescence following delivery with polyaspartate (C) and polyglutamate (D) ApPs. (Left) Representative flow cytometry histograms. (Center) Percent of cells splitGFP-positive. (Right) Fold increase in median splitGFP fluorescence over negative control. The dotted line indicates either 90% of the cell population (Center) or no increase in fluorescence (Right). Viability was determined with the LDH assay. Data are mean ± SEM, n = 4; *P < 0.05, **P < 0.01, ***P < 0.001 (1-sided 1-sample t test of log ratios).
Flow cytometry of splitGFP fluorescence corresponded well to microscopy, but additionally revealed that delivery peaked with 25 aspartates (D25), but plateaued with 20 glutamates (E20) (Fig. 3 C and D and SI Appendix, Fig. S3). Lipo RNAiMax, a cationic lipid designed for siRNA delivery, was more effective than Lipo 2000 at delivering Ritux-(pAbBD-ApP-S11)2 with short ApPs, but worse with longer ApPs (SI Appendix, Fig. S4). Similarly to pAbBD-ApP-S11 delivery, increasing the ratio of either lipids to Ritux-(pAbBD-ApP-S11)2 generally improved delivery, but also increased toxicity (SI Appendix, Figs. S3 and S4). After significant optimization, we achieved a maximal delivery efficiency of 65.7 ± 3.6% in HEK293T splitGFP(1–10) cells with >90% viability when 500 nM Ritux-(pAbBD-D25-S11)2 was complexed with 2 µL Lipo 2000 (Fig. 3C). Interestingly, delivery efficiencies for Ritux-(pAbBD-ApP-S11)2 were higher than that of pAbBD-ApP-S11 alone, likely due to having 2 ApPs linked to each rituximab molecule.
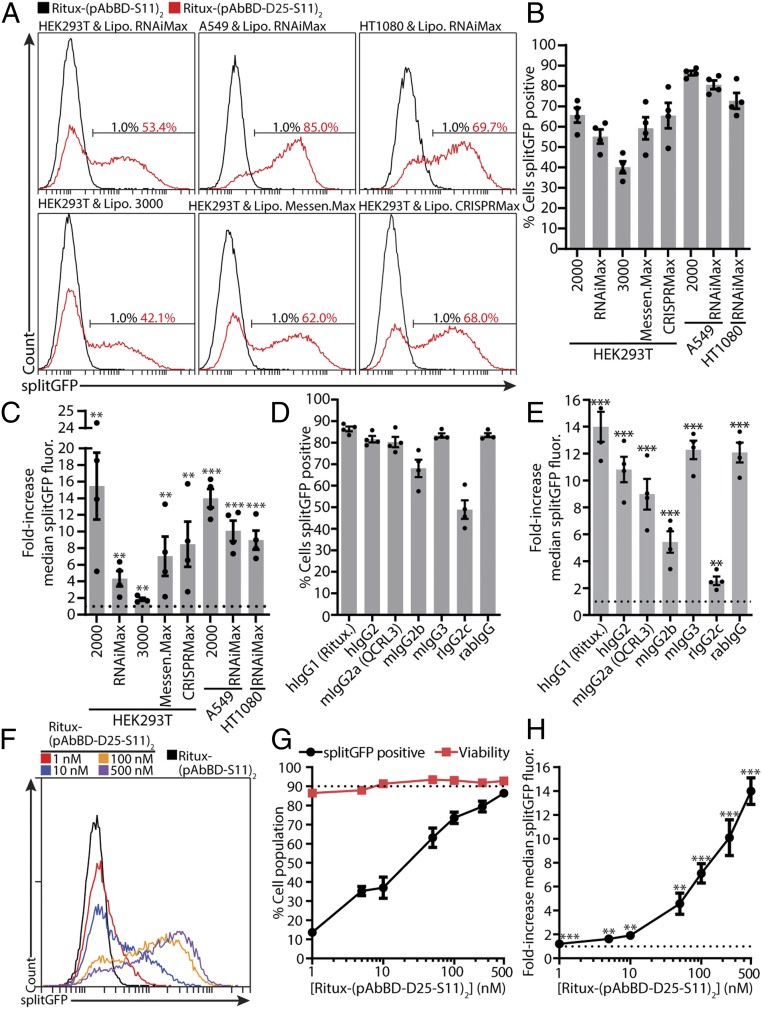
To demonstrate the generalizability of our IgG delivery approach, we tested several other commercially available cationic lipids and reporter cell lines. All conditions resulted in efficient cytosolic delivery of Ritux-(pAbBD-D25-S11)2 and Ritux-(pAbBD-E25-S11)2, albeit to varying degrees (Fig. 4 A–C and SI Appendix, Figs. S5 A–C, S6, and S7). Delivery efficiency was significantly higher in HT1080 and A549 splitGFP(1–10) cells, reaching up to 72.7 ± 3.9% and 86.4 ± 1.1%, respectively, with 500 nM Ritux-(pAbBD-D25-S11)2 (Fig. 4B and SI Appendix, Fig. S7). Other than rituximab (human IgG, hIgG1), pAbBD-D25-S11 could also photocrosslink and cytosolically deliver hIgG2, mouse IgG2a (mIgG2a), mIgG2b, mIgG3, rat IgG2c (rIgG2c), and rabbit IgG (rabIgG) into A549 splitGFP(1–10) cells (Fig. 4 D and E). Delivery efficiency remained high with all tested IgG species and isotypes, except for rIgG2c, which had a moderate delivery efficiency (Fig. 4D).
Fig. 4.
IgG delivery scope. (A–C) A total of 500 nM Ritux-(pAbBD-S11)2 (negative control) or Ritux-(pAbBD-D25-S11)2 was complexed with 2 μL of the indicated cationic lipid and added to the indicated splitGFP(1–10) reporter cells for 6 h. Representative flow cytometry histograms of splitGFP fluorescence are shown in A. Flow cytometry data were quantified as the percent of cells splitGFP-positive (B) and the fold increase in median splitGFP fluorescence over negative control (C). (D and E) Same as for B and C but 500 nM IgG-(pAbBD-D25-S11)2 of the indicated species and isotype was complexed with 2 μL Lipo 2000 and added to A549 splitGFP(1–10) cells. (F–H) Same as for A–C, but indicated concentrations of Ritux-(pAbBD-D25-S11)2 were complexed with 2 μL Lipo 2000 and added to A549 splitGFP(1–10) cells. The dotted line indicates either no increase in fluorescence (C, E, and H) or 90% of the cell population (G). Viability was determined with the LDH assay. Data are mean ± SEM, n = 4; **P < 0.01, ***P < 0.001 (1-sided 1-sample t test of log ratios).
Finally, we measured how delivery efficiency varied with Ritux-(pAbBD-D25-S11)2 and Ritux-(pAbBD-E25-S11)2 dose (Fig. 4 F–H and SI Appendix, Fig. S5 D–F). At 100 nM Ritux-(pAbBD-D25-S11)2, a 5-fold decrease in concentration, delivery efficiency was only slightly reduced to 73.5 ± 3.0% in A549 splitGFP(1–10) cells with Lipo 2000 (Fig. 4G). Even at 1 nM Ritux-(pAbBD-D25-S11)2 or Ritux-(pAbBD-E25-S11)2, cytosolic delivery was still detectable, albeit low (Fig. 4 F–H and SI Appendix, Fig. S5 D–F).
Collectively, these results demonstrate the versatility of our IgG delivery approach with regards to the following factors: 1) We can deliver off-the-shelf IgGs from a variety of species and isotypes. 2) It is compatible with all tested cationic lipids and cell lines thus far. 3) Delivery efficiency is maintained at low IgG-(pAbBD-ApP-S11)2 concentrations. We observe that although increasing the net negative charge of cargo proteins is critical for complexation and delivery, it eventually becomes unproductive or even deleterious for delivery efficiency.
Cytosolically Delivered QCRL3 Can Inhibit MRP1, a Drug Efflux Pump.
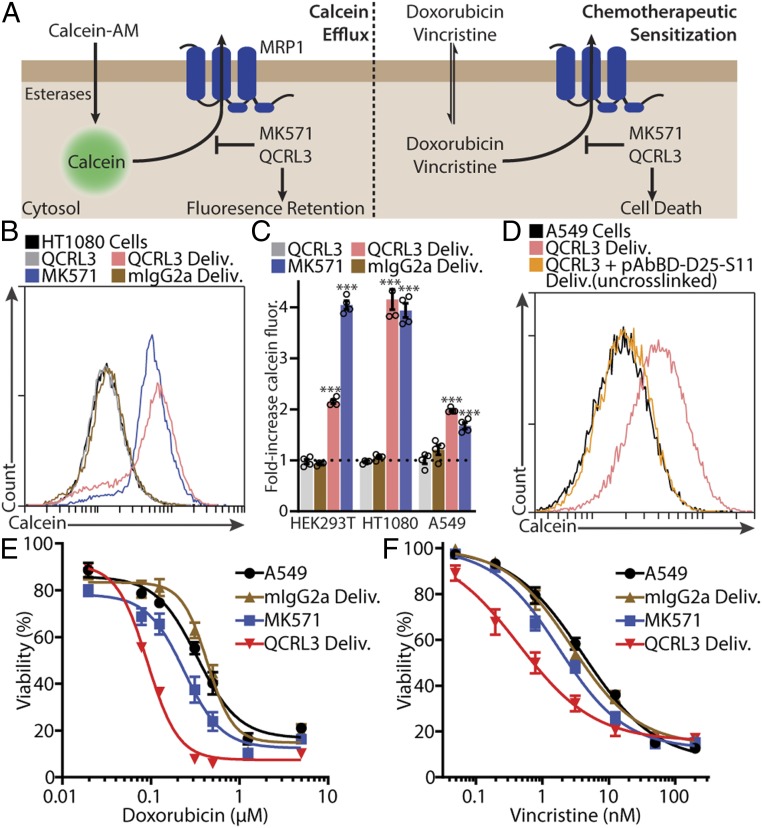
Having shown robust delivery, we next sought to demonstrate the utility of cytosolic IgGs by inhibiting MRP1, a drug-export pump associated with chemotherapy resistance (30). Although it is a transmembrane protein, we chose MRP1 because it can be inhibited by a well-characterized monoclonal IgG, QCRL3, which inhibits via binding to one of MRP1’s cytosolic nucleotide binding domains (30–33).
Initially, we used the calcein-efflux assay to assess MRP1 activity in which MRP1 inhibition results in calcein fluorescence retention (Fig. 5A). A total of 500 nM QCRL3-(pAbBD-D25-S11)2 delivery resulted in calcein retention in HEK293T, HT1080, and A549 cells (Fig. 5 B and C), indicating inhibition of endogenous MRP1 activity. No inhibition was observed following delivery of the mIgG2a-(pAbBD-D25-S11)2 isotype control or simply incubating cells with QCRL3-(pAbBD-D25-S11)2. QCRL3-(pAbBD-D25-S11)2 delivery performed as well as MK571, a nonselective small-molecule MRP1 inhibitor (30), in all except for HEK293T cells (Fig. 5C). We attribute this to the high expression of other efflux pumps that are also inhibited by MK571 in HEK293T cells. Finally, photocrosslinking is necessary for IgG delivery, as delivery of QCRL3 mixed with pAbBD-D25-S11 did not result in calcein retention (Fig. 5D). This suggests that during complexation, noncovalent interactions between pAbBD and IgGs are disrupted.
Fig. 5.
Cytosolic QCRL3 delivery inhibits MRP1. (A) Schematic of assays that assess MRP1 inhibition. In the calcein efflux assay, cells are first loaded with calcein, a fluorescent membrane-impermeable MRP1 substrate. Cells with high MRP1 activity will rapidly export calcein, whereas MRP1 inhibition results in calcein fluorescence retention. In the chemotherapeutic sensitization assay, MRP1 inhibition results in greater intracellular accumulation of doxorubicin or vincristine, resulting in sensitization to both compounds. (B) Representative flow cytometry histograms of calcein fluorescence after 16 h of export in calcein-loaded HT1080 cells treated with 20 μM MK571, 500 nM QCRL3-(pAbBD-D25-S11)2, 500 nM cytosolically delivered mIgG2a-(pAbBD-D25-S11)2, or 500 nM cytosolically delivered QCRL3-(pAbBD-D25-S11)2. (C) Calcein-efflux assay quantification across HEK293T, HT1080, and A549 cell lines. Only QCRL3 delivery and MK571 treatment resulted in calcein fluorescence retention. Data are mean ± SEM, n = 4, ***P < 0.001 (1-sided 1-sample t test of log ratios). (D) Same as for B, but in calcein-loaded A549 cells treated with cytosolic delivery of 500 nM QCRL3 with or without photocrosslinking to pAbBD-D25-S11. Calcein fluorescence retention is only seen with photocrosslinked QCRL3, indicating that photocrosslinking is necessary for delivery. (E and F) A549 cell sensitivity to doxorubicin (E) or vincristine (F) following treatment with 20 μM MK571, 500 nM cytosolically delivered mIgG2a-(pAbBD-D25-S11)2, or 500 nM cytosolically delivered QCRL3-(pAbBD-D25-S11)2. Data are mean ± SEM, n = 4.
Next, we attempted to sensitize A549 cells to doxorubicin and vincristine, which are chemotherapeutic drugs known to be MRP1 substrates (30, 34). Cytosolic delivery of 500 nM QCRL3-(pAbBD-D25-S11)2 was able to sensitize A549 cells to doxorubicin by 3.7 ± 0.45 fold and vincristine by 9.0 ± 2.0 fold (Fig. 5 E and F). In comparison, MK571 treatment resulted in only moderate sensitization, and delivery of the mIgG2a isotype control resulted in no sensitization (Fig. 5 E and F). Thus, cytosolically delivered IgGs remain functional following delivery and can inhibit biologically interesting proteins.
Cytosolically Delivered IgGs Can Inhibit NFκB.
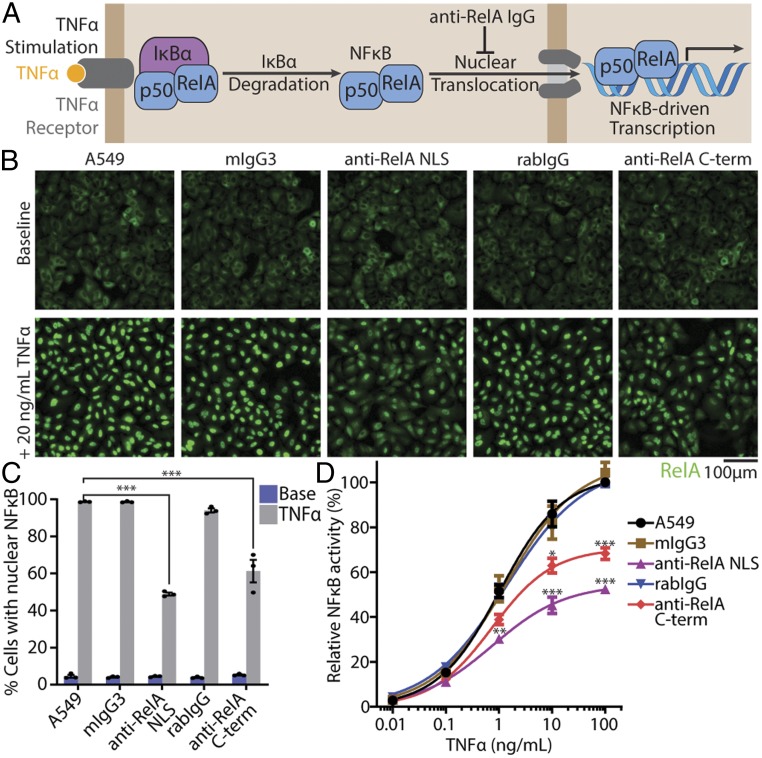
Finally, we investigated whether cytosolically delivered IgGs could inhibit protein–protein interactions, which are particularly difficult to perturb with small molecules. We targeted the transcription factor NFκB, which is a heterodimer between p50 and RelA (p65) whose nuclear localization signals (NLSs) are normally masked by IκBα, sequestering NFκB in the cytosol. With TNFα stimulation, IκBα is degraded, allowing NFκB to enter the nucleus to stimulate transcription (35). We hypothesized that an anti-RelA NLS IgG could sterically block RelA from engaging with its cognate nuclear import factor (NIF) to prevent RelA nuclear translocation and NFκB-mediated transcription (Fig. 6A). We also tested an anti-RelA C terminus IgG that bound to an epitope distinct from the NLS.
Fig. 6.
Cytosolic anti-RelA IgG delivery inhibits NFκB. (A) Schematic of NFκB inhibition. Anti-RelA IgGs inhibit NFκB transcriptional activity by preventing its nuclear translocation following TNFα stimulation. (B and C) Representative immunofluorescence images (B) and quantification (C) of RelA nuclear translocation following delivery of the indicated 150 nM IgG-(pAbBD-D25-S11)2 antibody and TNFα treatment. Only delivery of anti-RelA IgGs reduced RelA nuclear translocation. Data are mean ± SEM, n = 3, ***P < 0.001 (1-way ANOVA). (D) A549 cells were transiently transfected with a NFκB-driven firefly luciferase reporter plasmid. NFκB transcriptional activity was detected by luminescence following delivery of the indicated 150 nM IgG-(pAbBD-D25-S11)2 antibody and TNFα treatment. Only delivery of anti-RelA IgGs inhibited NFκB transcriptional activity. Data are mean ± SEM, n = 3; *P < 0.05, **P < 0.01, ***P < 0.001 (1-way ANOVA).
Initially, we delivered 150 nM anti-RelA NLS-(pAbBD-D25-S11)2, anti-RelA C-term-(pAbBD-D25-S11)2, or their isotype controls (mIgG3 for anti-RelA NLS, rabIgG for anti-RelA C-term) into A549 cells and assessed for RelA nuclear translocation following TNFα stimulation. Immunofluorescence revealed that both anti-RelA NLS and anti-RelA C-term delivery reduced RelA nuclear translocation to 48.0 ± 0.8% and 60.1 ± 5.9% of that of normal cells, respectively (Fig. 6 B and C and SI Appendix, Fig. S8). Next, by using a NFκB-driven luciferase reporter plasmid, we showed that delivery of both anti-RelA NLS and anti-RelA C-term reduced NFκB transcriptional activity to 52.4 ± 1.1% and 68.3 ± 2.6% of that of normal cells, respectively (100 ng/mL TNFα) (Fig. 6D). This degree of inhibition is excellent considering the delivery efficiencies with 150 nM anti-RelA NLS and anti-RelA C-term were 70.9 ± 0.8% and 29.7 ± 4.5%, respectively (SI Appendix, Fig. S9). Because anti-RelA C-term IgG delivery was capable of inhibiting NFκB, antibody binding to non-NLS epitopes may be sufficient to sterically block nuclear translocation of many target proteins. We anticipate that cytosolic IgG-dependent cytoplasmic sequestration can be easily adopted to inhibit other transcription factors or nuclear proteins.
Discussion
Efficient cytosolic delivery of proteins, particularly antibodies, has long been sought for expanding the toolbox that one can use for perturbing biological systems. By leveraging a modular antibody functionalization technology (21), we were able to easily append ApPs onto off-the-shelf IgGs to enable their complexation with a variety of commercially available cationic lipids and efficiently deliver them into the cytosol of cells. Cytosolically delivered IgGs remain functional and are capable of inhibiting diverse proteins such as MRP1 and NFκB.
When compared to CPP-mediated delivery of small peptides or proteins, our approach enables cytosolic delivery of a much larger IgG cargo with similar or greater efficiencies at an ∼100-fold lower concentration (25–28). It is difficult to directly compare our technology to previous carrier-mediated approaches due to the use of different reporter systems. We note, however, that previous studies have relied on either a reporter that only detects total cellular uptake (17) or by delivering proteins capable of greatly amplifying their signal, such as Cre-recombinase or enzymes (16, 18–20). In contrast, our functional assays—MRP1 and NFκB inhibition—are more stringent and require delivery of close to stoichiometric amounts of IgG relative to their target, which is more representative of most protein inhibition assays.
Intrabodies have long been used to modulate cell biology, ranging from simply inhibiting target proteins (8–10, 36, 37) to marking target proteins for degradation (38). However, those approaches require either expertise in in vitro display technologies or antibody engineering to create fragments that can fold properly in the cytoplasmic environment, which can be major barriers for easy adoption by researchers. Because IgG antibodies have been invaluable research tools for decades, there exists large and well-validated antibody collections (39, 40) that, using our modular IgG functionalization and cytosolic delivery approach, can now be repurposed for perturbing the activity of intracellular proteins. Although IgG binding is not guaranteed to inhibit all target proteins, we believe that the large size of IgGs is sufficient to sterically block many biological interactions.
Cytosolic delivery of inhibitory antibodies also offers unique advantages over traditional genetic or small-molecule approaches for modulating protein function. Because genetic approaches for protein knockdown or knockout do not directly act on target proteins, they perform poorly against proteins with long half-lives (41) and can induce significant cellular compensatory responses (42). Small-molecule inhibitors and modulators can avoid these limitations, but many proteins are not druggable by small molecules and for those that are, identifying potent compounds and then validating their selectivity can be challenging (43). In contrast, inhibitory antibodies directly bind to target proteins, act on fast time scales, and can be generated far more easily than small molecules. Finally, cytosolic inhibitory antibodies also offer the opportunity to discriminate between and modulate the activity of proteins with specific posttranslational modifications or certain isoforms of a protein, which are generally not possible with traditional approaches.
Cytosolic IgGs have previously been reported to be capable of engaging the TRIM21 E3 ubiquitin ligase to degrade their target proteins (7), but pAbBD photocrosslinking sterically blocks the TRIM21 binding site (21, 44, 45) and prevents our cytosolically delivered IgGs from harnessing any endogenous protein degradation machinery. Future studies could address this by engineering alternative antibody-binding domains (46) to photocrosslink to IgGs outside of the Fc region. In this study we complexed IgGs with commercially available cationic lipids. Although the resulting complexes perform efficiently in cultured cells, their poor pharmacokinetics render them unsuitable for in vivo studies. Future studies should test alternative delivery formulations for in vivo delivery, such as those containing ionizable and poly(ethylene glycol)-conjugated lipids (14), which have shown promise for the in vivo delivery of siRNA. However, it is not clear whether these formulations, when prepared with antibodies, will have adequate pharmacokinetics, be capable of extravasating from the vasculature to induce uptake into desired cell populations, or be capable of promoting escape from the endosome–lysosome system with adequate efficiency to impart a therapeutic effect in vivo.
In summary, we have designed and rigorously validated a modular cytosolic IgG delivery platform that allows previously developed IgG collections to now be used as intracellular tools. We believe that this capability will significantly expand the ways in which intracellular pathways can be perturbed and will shed unique insight into cell function in both health and disease.
Methods
pAbBD Expression and Purification.
All pAbBD variants were purified via proximity-based sortase-mediated ligation (PBSL) (47). Using PBSL to purify proteins consists of 2 steps: 1) PBSL resin preparation and 2) target protein purification. PBSL resin was prepared as previously described with 2 minor modifications due to the use of SpyCatcher-SrtA-His12 (47). Pellets were lysed by resuspending in lysis buffer (PBS + 1% wt/vol N-octyl-β-d-1-thioglucopyranoside [OTG] + 200 μg/mL lysozyme + 4 μg/mL DNaseI + EDTA-free protease inhibitor mixture [Roche]) and rotating for 30 min at room temperature (RT). Following binding, the resin was washed with PBS + 20 mM imidazole 3 times followed by PBS once.
Plasmids encoding for various pAbBD variants were transformed in conjunction with pEVOL-pBpF into T7 express competent Escherichia coli cells (New England Biolabs). Starter cultures were grown in LB + 100 μg/mL ampicillin (amp) + 25 μg/mL chloramphenicol (cam) at 37 °C with shaking until OD600 ∼ 0.6. The starter culture was added at a 1:1,000 dilution to autoinduction media (Formedium AIMLB0210 autoinduction media LB broth base including trace elements supplemented with 0.6% vol/vol glycerol and 100 μg/mL amp) further supplemented with 25 μg/mL cam + 0.1% wt/vol arabinose + 3.33 μM 4-benzoyl-l-phenylalanine (BPA, Bachem). All pAbBD variants were grown at 37 °C with shaking for 24 h, except for pAbBD-D30-S11 and pAbBD-E30-S11, which were grown at 25 °C with shaking for 48 h. Expression cultures were then pelleted and stored at −20 °C.
Frozen pellets were lysed by resuspending in lysis buffer for 30 min at RT. Afterward, lysates were frozen at −80 °C and then thawed in a 37 °C water bath. The lysates were clarified by centrifuging for 15 min at ≥14,000 × g and discarding the pellet. Clarified lysates were incubated with the SpyCatcher-SrtA-His12 resin prepared above while rotating for 25 min at RT. Following binding, the resin was transferred to a Poly-Prep chromatography column (Bio-Rad) and washed with 1 column volume (CV) of PBS, 1 CV of PBS + 20 mM imidazole, and 1 CV of PBS + 1 M NaCl + 20 mM imidazole. pAbBD variants were then eluted from the resin by adding PBS + 250 μM CaCl2 + 2 mM Gly-Gly-Gly (triglycine) and incubating at 25 °C for 3 h. Following elution, pAbBD variants were buffer exchanged into PBS and concentrated to ≥0.5 mg/mL via a 10k MWCO Amicon Ultra centrifugal filter (MilliporeSigma). The final protein was analyzed by SDS/PAGE for purity, tested for splitGFP complementation, stored at −80 °C, and tolerated freeze–thaw cycles well.
See SI Appendix, Supplementary Methods, for details on plasmid generation, splitGFP(1–10) purification, and splitGFP complementation assays.
Photocrosslinking pAbBD Variants to IgGs.
For photocrosslinking, pAbBD variants were added to IgGs at a 2:1 molar ratio in PBS. IgG concentration was kept at ≤5 μM and the pAbBD-IgG uncrosslinked mixture was aliquoted in 2 mL clear polypropylene microcentrifuge tubes. The mixture was then placed in an ice bath and irradiated for 3 h with 365 nm UV light using a UVP CL-1000L UV crosslinker placed in a 4 °C cold room. After photocrosslinking, IgG-pAbBD2 conjugates were washed with PBS 3 times and then concentrated to ≥10 μM via a 100k MWCO Amicon Ultra centrifugal filter to remove any uncrosslinked pAbBD. SDS/PAGE was used to confirm that >95% of IgG heavy chains were photocrosslinked and that any excess pAbBD was removed. The final protein was then tested for splitGFP complementation and stored at 4 °C for short durations (<1 wk) and at −80 °C for long durations. See SI Appendix, Supplementary Methods, for details on IgG antibodies used in this study.
Protein Delivery.
Cells were seeded to 70 to 80% confluency in a 48-well flat-bottom tissue culture plate. Just prior to delivery, the media was replaced with 180 µL fresh media. The protein to be delivered was diluted in OptiMEM I (Thermo Fisher 31985-070) to 20× final protein concentration in 10 µL total volume. The indicated transfection reagent was diluted in OptiMEM to 10 µL total volume and incubated at RT for 10 min. The diluted protein and diluted cationic lipid was then mixed and incubated at RT for 10 min. Finally, the mixture was immediately added to reporter cell lines to a final volume of 200 µL (180 µL media + 20 µL protein/cationic lipid mixture). Reporter cell lines were incubated at 37 °C for 6 h before splitGFP complementation was determined by live-cell fluorescence microscopy or flow cytometry. Each delivery condition was paired with the same concentration of pAbBD-S11 or Ritux-(pAbBD-S11)2 with the same amount of the indicated cationic lipid as a negative control. Viability was determined using the lactate dehydrogenase (LDH) assay (Dojindo Molecular Technologies) per the manufacturer’s recommended protocol.
See SI Appendix, Supplementary Methods, for details on cell line generation and maintenance, flow cytometry, live-cell fluorescence microscopy, MRP1 functionality assays, and NFκB functionality assays.
Supplementary Material
Acknowledgments
We thank Philip Zoltick for providing splitGFP(1–10) lentivirus and HEK293T splitGFP(1–10) cells. We are grateful for assistance from the University of Pennsylvania Flow Cytometry and Cell Sorting Facility and Cell Center cores. This work was supported in part by the NIH (NCI P30 CA016520, NCI R01CA241661, and F30 CA221385), the Breakthrough Bike Challenge, and the Penn-Health Tech Pilot Award.
Footnotes
Competing interest statement: H.H.W. and A.T. have a pending patent on this technology. A.T. is a founder and owns equity in AlphaThera, a biotechnology company that sells pAbBD-based products.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913973116/-/DCSupplemental.
References
- 1.Bradbury A. R. M., Sidhu S., Dübel S., McCafferty J., Beyond natural antibodies: The power of in vitro display technologies. Nat. Biotechnol. 29, 245–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter P. J., Lazar G. A., Next generation antibody drugs: Pursuit of the ‘high-hanging fruit’. Nat. Rev. Drug Discov. 17, 197–223 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Mulcahy L. S., Smith M. R., Stacey D. W., Requirement for ras proto-oncogene function during serum-stimulated growth of NIH 3T3 cells. Nature 313, 241–243 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Riabowol K. T., Mizzen L. A., Welch W. J., Heat shock is lethal to fibroblasts microinjected with antibodies against hsp70. Science 242, 433–436 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Yeung K., et al. , Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401, 173–177 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Habedanck R., Stierhof Y. D., Wilkinson C. J., Nigg E. A., The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140–1146 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Clift D., et al. , A method for the acute and rapid degradation of endogenous proteins. Cell 171, 1692–1706.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marschall A. L. J., et al. , Delivery of antibodies to the cytosol: Debunking the myths. MAbs 6, 943–956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marschall A. L. J., Frenzel A., Schirrmann T., Schüngel M., Dübel S., Targeting antibodies to the cytoplasm. MAbs 3, 3–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slastnikova T. A., Ulasov A. V., Rosenkranz A. A., Sobolev A. S., Targeted intracellular delivery of antibodies: The state of the art. Front. Pharmacol. 9, 1208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akishiba M., et al. , Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide. Nat. Chem. 9, 751–761 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Herce H. D., et al. , Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem. 9, 762–771 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Shin S. M., et al. , Antibody targeting intracellular oncogenic Ras mutants exerts anti-tumour effects after systemic administration. Nat. Commun. 8, 15090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanasty R., Dorkin J. R., Vegas A., Anderson D., Delivery materials for siRNA therapeutics. Nat. Mater. 12, 967–977 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Adams D., et al. , Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Zuris J. A., et al. , Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y., et al. , Efficient delivery of bioactive antibodies into the cytoplasm of living cells by charge-conversional polyion complex micelles. Angew. Chem. Int. Ed. Engl. 49, 2552–2555 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Eltoukhy A. A., et al. , Nucleic acid-mediated intracellular protein delivery by lipid-like nanoparticles. Biomaterials 35, 6454–6461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postupalenko V., et al. , Protein delivery system containing a nickel-immobilized polymer for multimerization of affinity-purified his-tagged proteins enhances cytosolic transfer. Angew. Chem. Int. Ed. Engl. 54, 10583–10586 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Mout R., et al. , General strategy for direct cytosolic protein delivery via protein-nanoparticle co-engineering. ACS Nano 11, 6416–6421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui J. Z., Tamsen S., Song Y., Tsourkas A., LASIC: Light activated site-specific conjugation of native IgGs. Bioconjug. Chem. 26, 1456–1460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deprey K., Becker L., Kritzer J., Plückthun A., Trapped! A critical evaluation of methods for measuring total cellular uptake versus cytosolic localization. Bioconjug. Chem. 30, 1006–1027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith S. A., Selby L. I., Johnston A. P. R., Such G. K., The endosomal escape of nanoparticles: Toward more efficient cellular delivery. Bioconjug. Chem. 30, 263–272 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Cabantous S., Terwilliger T. C., Waldo G. S., Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Milech N., et al. , GFP-complementation assay to detect functional CPP and protein delivery into living cells. Sci. Rep. 5, 18329 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt S., et al. , Detecting cytosolic peptide delivery with the GFP complementation assay in the low micromolar range. Angew. Chem. Int. Ed. Engl. 54, 15105–15108 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Lönn P., et al. , Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci. Rep. 6, 32301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kauffman W. B., Guha S., Wimley W. C., Synthetic molecular evolution of hybrid cell penetrating peptides. Nat. Commun. 9, 2568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timney B. L., et al. , Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 215, 57–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole S. P. C., Targeting multidrug resistance protein 1 (MRP1, ABCC1): Past, present, and future. Annu. Rev. Pharmacol. Toxicol. 54, 95–117 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Loe D. W., Almquist K. C., Deeley R. G., Cole S. P. C., Multidrug resistance protein (MRP)-mediated transport of leukotriene C4 and chemotherapeutic agents in membrane vesicles. Demonstration of glutathione-dependent vincristine transport. J. Biol. Chem. 271, 9675–9682 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Loe D. W., Deeley R. G., Cole S. P. C., Characterization of vincristine transport by the M(r) 190,000 multidrug resistance protein (MRP): Evidence for cotransport with reduced glutathione. Cancer Res. 58, 5130–5136 (1998). [PubMed] [Google Scholar]
- 33.Hipfner D. R., et al. , Monoclonal antibodies that inhibit the transport function of the 190-kDa multidrug resistance protein, MRP. Localization of their epitopes to the nucleotide-binding domains of the protein. J. Biol. Chem. 274, 15420–15426 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Cole S. P. C., et al. , Pharmacological characterization of multidrug resistant MRP-transfected human tumor cells. Cancer Res. 54, 5902–5910 (1994). [PubMed] [Google Scholar]
- 35.Napetschnig J., Wu H., Molecular basis of NF-κB signaling. Annu. Rev. Biophys. 42, 443–468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guillard S., et al. , Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange. Nat. Commun. 8, 16111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta A., et al. , Facile target validation in an animal model with intracellularly expressed monobodies. Nat. Chem. Biol. 14, 895–900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caussinus E., Kanca O., Affolter M., Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–121 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Marcon E., et al. , Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat. Methods 12, 725–731 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Venkataraman A., et al. , A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors. Nat. Methods 15, 330–338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smoak E. M., Stein P., Schultz R. M., Lampson M. A., Black B. E., Long-term retention of CENP-A nucleosomes in mammalian oocytes underpins transgenerational inheritance of centromere identity. Curr. Biol. 26, 1110–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi A., et al. , Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Arrowsmith C. H., et al. , The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–541 (2015).Correction in: Nat. Chem. Biol.11, 541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.James L. C., Keeble A. H., Khan Z., Rhodes D. A., Trowsdale J., Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. U.S.A. 104, 6200–6205 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauer-Eriksson A. E., Kleywegt G. J., Uhlén M., Jones T. A., Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure 3, 265–278 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Choe W., Durgannavar T. A., Chung S. J., Fc-binding ligands of immunoglobulin G: An overview of high affinity proteins and peptides. Materials (Basel) 9, 994 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H. H., Altun B., Nwe K., Tsourkas A., Proximity-based sortase-mediated ligation. Angew. Chem. Int. Ed. Engl. 56, 5349–5352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.