Significance
Bacterial infection has been extensively investigated; however, little is known about how bacterial pathogens timely shut down infecting machinery after successful infections. Here, a previously unknown sucrose–SghR/SghA–SAG–SA signaling axis was identified which controls the timing to shut off bacterial virulence expression and fine-tune host immune response. Sucrose, salicylic acid (SA), and its storage form SAG are small chemicals produced in plants whereas SghR is a bacterial sensor of sucrose and SghA is a bacterial enzyme that releases SA from SAG. Given that SA is an imperative signaling molecule in defense against a variety of microbial pathogens, these results depict a previously unknown 2-way chemical signaling cross-talk during microbe–host coevolution and shed mechanistic insights into host–bacteria interaction.
Keywords: chemical signaling, cost–pathogen interaction, Agrobacterium, sucrose, glucosidase
Abstract
It is highly intriguing how bacterial pathogens can quickly shut down energy-costly infection machinery once successful infection is established. This study depicts that mutation of repressor SghR increases the expression of hydrolase SghA in Agrobacterium tumefaciens, which releases plant defense signal salicylic acid (SA) from its storage form SA β-glucoside (SAG). Addition of SA substantially reduces gene expression of bacterial virulence. Bacterial vir genes and sghA are differentially transcribed at early and later infection stages, respectively. Plant metabolite sucrose is a signal ligand that inactivates SghR and consequently induces sghA expression. Disruption of sghA leads to increased vir expression in planta and enhances tumor formation whereas mutation of sghR decreases vir expression and tumor formation. These results depict a remarkable mechanism by which A. tumefaciens taps on the reserved pool of plant signal SA to reprogram its virulence upon establishment of infection.
Bacterial pathogens commonly deploy an array of virulence factors to establish infections in various host organisms. For example, major virulence genes of Agrobacterium tumefaciens are carried by a large plasmid (over 200 kb), and infection requires a range of regulatory and structural proteins and a DNA fragment, which are transported from bacterial cells into host plant cells (1). These virulence factors are energy-costly to synthesize, and therefore bacteria might have evolved mechanisms to reprogram expression of virulence genes to survive in changed environmental conditions. Pseudomonas aeruginosa is known to switch from acute infection to chronic persistence by turning off expression of the genes encoding the type III secretion system (T3SS) through the Gac/Rsm regulatory pathway at the later stage of infection (2). Contrarily, Salmonella enterica escapes from its intracellular niche and spreads to a secondary infection site by inducing the expression of invasion-associated T3SS genes (3). However, how bacterial pathogens sense and perceive environmental changes to reprogram virulence gene expression remains elusive.
A. tumefaciens is a renowned plant pathogen for causing crown gall diseases on more than 140 plant species (4). Infection of agrobacteria is modulated by various plant-derived chemical signals (5). Initially, wound-associated acidic conditions induce the expression of the chromosomal ChvG/I 2-component system, which activates the expression of transcriptional regulator VirG. VirA senses acetosyringone in the wounding site, phosphorylates VirG, and activates the expression of vir regulon that encodes the type IV secretion system and accessory proteins for processing and transferring transfer DNA (T-DNA) into plant cells. After integration, T-DNA genes encode biosynthesis of auxin, cytokinin, and opines. Plant hormones auxin and cytokinin promote plant cell proliferation and formation of crown gall tumors whereas opines are utilized by A. tumefaciens as specific nutrients. The bacterial pathogen thus creates an ecological niche that provides a selective advantage over other bacterial species, and this phenomenon is known as genetic colonization (6).
Transformation of host plant cells and building up of competitive advantages in ecological systems make A. tumefaciens an excellent model for exploring various features of pathogen–host interactions (7). A. tumefaciens infection is sensitive to high temperature, and therefore crown gall disease rarely happens in tropical regions. Previous findings suggest that Agrobacterium-mediated infection requires only a short period of time (8). This raises an intriguing question how pathogen can turn down vir expression after the infection to reduce energy cost. In this regard, it is interesting to note that exogenous application of plant defense signal salicylic acid (SA) can inhibit vir gene expression and virulence of A. tumefaciens (9, 10). In plants, a proportion of SA conjugates with glucose to prepare the storage form SA β-glucoside (SAG) (11, 12). Thus, it would be fascinating to explore if Agrobacterium has evolved a mechanism to hijack SA of host plants to switch from infection mode to free living style after the initiation of crown gall formation.
In this study, we report identification and characterization of a gene, sghA, encoding a hydrolytic enzyme. This enzyme releases SA from SAG to elevate its levels in planta. Transcription of sghA is tightly repressed by a repressor (SghR), and plant metabolite sucrose specifically releases this repression. Study further revealed that exogenous addition of SA decreased vir gene expression in A. tumefaciens. This phenomenon confirmed that the pathogen could use SA to reset its virulence via SghR/SghA after completing their infection. SA is an essential signal molecule for plant local defense and systemic resistance against numerous plant pathogens. These results indicate a broad implication in controlling crown gall disease and increasing plant transformation efficiency in agriculture. This study also unravels a previously unknown sophisticated strategy of microbial infection evolved during the long history of pathogen–host interaction.
Results
Null Mutation of Repressor SghR Increases the β-Galactosidase Activity of A. tumefaciens.
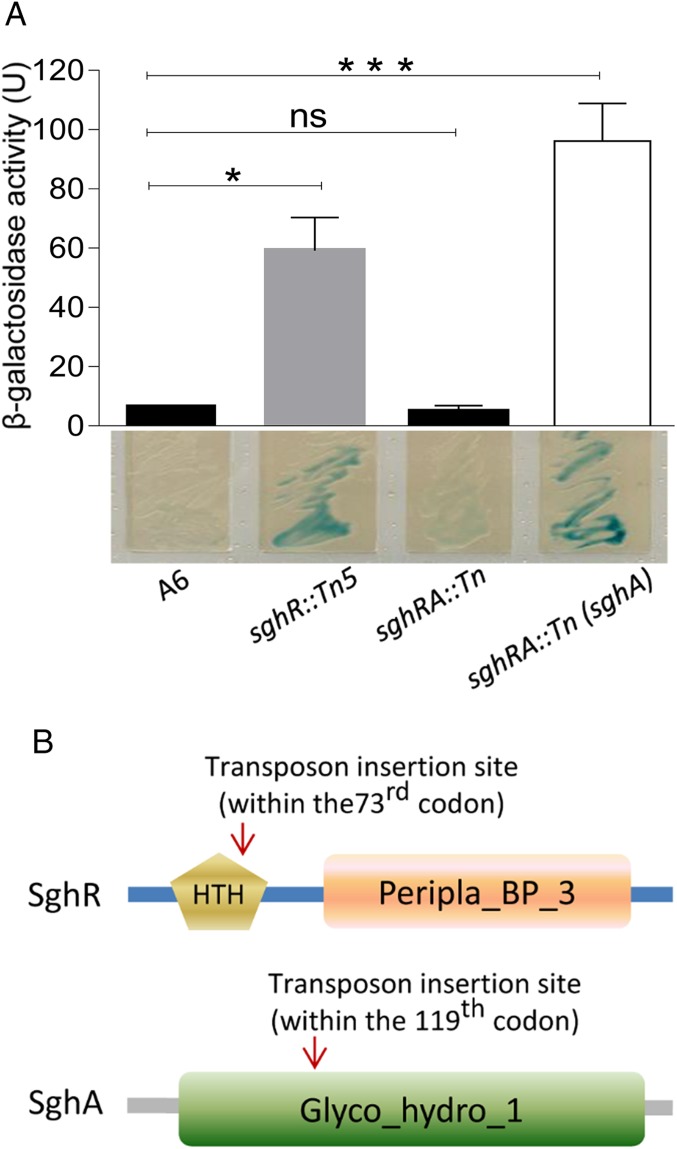
SAG, a glucose-conjugated salicylic acid in plant hosts, shares a similar structure with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) and ortho-nitrophenyl-β-d-galactopyranoside (ONPG) (SI Appendix, Fig. S1). This tempted us to search for potential SAG-hydrolyzing enzymes by using agar plates supplemented with X-gal. Wild-type A. tumefaciens strain A6 did not show obvious β-galactosidase activity as the color of bacterial colonies remained unchanged in the X-gal plate (Fig. 1A). Assuming that the bacterium may contain a repressed gene encoding SAG hydrolytic enzyme, we generated a library of transposon mutants and screened for enhanced β-galactosidase activity. After screening over 20,000 mutants, one mutant was identified that exhibited a dark blue color on basic medium (BM) plates supplemented with X-gal. Sequence analyses showed that Tn5 was inserted in A. tumefaciens strain C58 homolog atu1522 sharing 92% amino acid sequence identity (GenBank no. KU512833). Its translational product is a 350-amino acid (aa) protein containing a helix-turn-helix (HTH)-type DNA-binding domain at its N terminus and a periplasmic binding protein-like domain (Peripla_BP_3) at its C terminus (Fig. 1B). These findings suggest that atu1522 homolog gene might encode a SAG hydrolase gene repressor and was named as sghR. Fig. 1A shows blue color morphology of sghR mutant (sghR::Tn5) on an X-gal agar plate. ONPG-based quantitative analysis indicated that sghR mutation increased β-galactosidase–like activity by about 10-fold as compared to parental strain A6 (Fig. 1A)
Fig. 1.
β-galactosidase activity of A. tumefaciens strain A6 and derivatives. (A) Analysis of β-galactosidase activity of A. tumefaciens A6 and its derivatives in liquid culture (Top) and on solid agar plates (Bottom). U, Miller Unit. (B) Domain structures of SghR and SghA. Glyco_hyddro_1, glycoside hydrolase family 1 domain; HTH, HTH-type DNA binding domain; Peripla_BP_3, periplasmic binding protein-like domain 3. Experiments in A were repeated 5 times, and each time in duplicate. A nonparametric Kruskal–Wallis 1-way ANOVA was performed in GraphPad for statistical analyses. Values and error bars represent means and SD. *P < 0.05; ***P < 0.0001; ns, not significant.
To identify the SghR-repressed gene encoding putative SAG hydrolase, another round of mutagenesis was conducted with transposon Mariner by using mutant sghR::Tn5 as the parental strain. Screening of the resultant mutant library identified 1 mutant with much reduced blue color on X-gal agar plates than the parental strain. Sequence analysis showed that Mariner was inserted in an ORF sharing about 94% identity to the atu4485 gene localized on the linear chromosome of A. tumefaciens strain C58. Sequence analysis showed that homolog atu4485 encodes a 467-aa protein (GenBank no. KU512832), and this putative SAG hydrolase gene was designated as sghA. An X-gal plate assay showed that disruption of sghA by sghR mutant abolished its blue color morphology (Fig. 1A), suggesting that sghA is the sole SghR-repressed gene encoding putative SAG hydrolase. In trans expression of sghA in double mutant sghRA::Tn restored blue color morphology on the X-gal plate and significantly increased β-galactosidase activity against ONPG (Fig. 1A). In silico analysis was conducted with the online tool SMART (http://smart.embl-heidelberg.de), and results revealed that SghA contains a glycoside hydrolase family 1 (Glyco_hydro_1) domain (Fig. 1B). This is a typical feature of the glycoside hydrolase family comprising numerous enzymes, including β-glucosidase, β-galactosidase, 6-phospho-β-galactosidase, 6-phospho-β-glucosidase, and β-mannosidase.
SghA as SA-Releasing Enzyme.
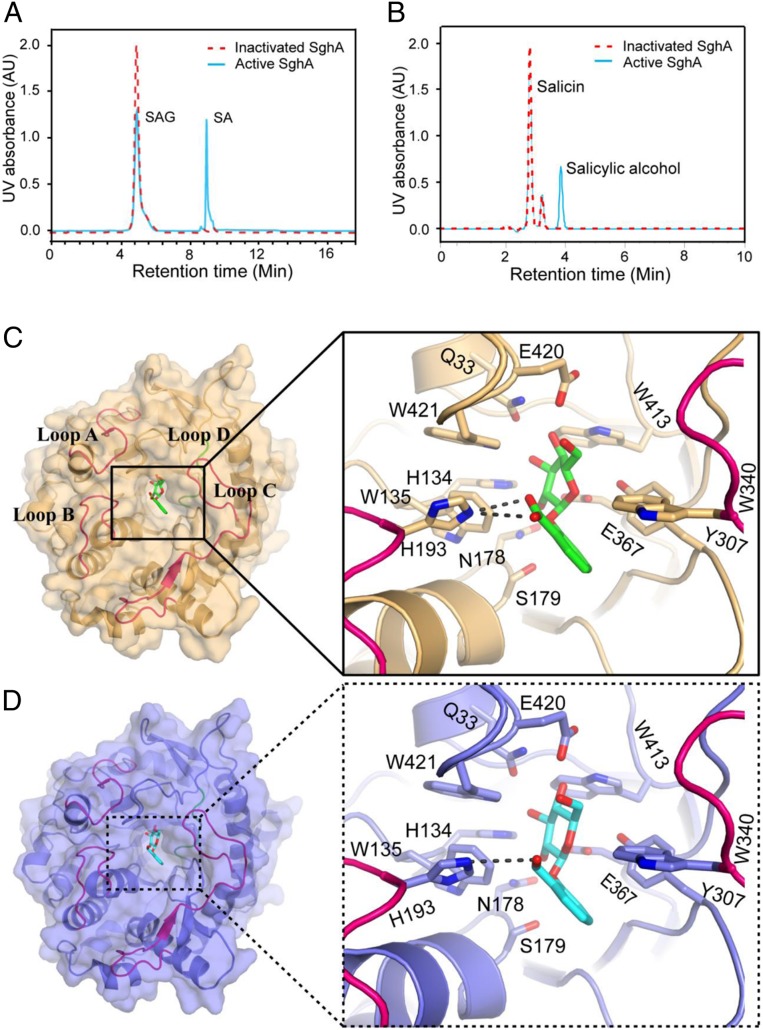
To test whether SghA could hydrolyze SAG to release SA, SAG was synthesized by using a modified protocol (SI Appendix, Fig. S2). After structural verification with mass spectrometry (MS) and NMR, SAG molecules were incubated with recombinant SghA and boiling-denatured SghA, respectively. High performance liquid chromatography (HPLC) analysis of the denatured SghA reaction mixture detected only 1 SAG peak at a retention time of 5.0 min (Fig. 2A) and was confirmed by electrospray ionization mass spectrometry (ESI-MS) analysis (SI Appendix, Fig. S3A). In contrast, HPLC analysis resolved 2 peaks from the reaction mixture of active SghA: i.e., SAG peak at 5 min and a new peak at a retention time of 9.0 min (Fig. 2A). ESI-MS analysis of the new peak showed a strong quasimolecular (M-H) ion with an m/z of 137.00 (SI Appendix, Fig. S3B) corresponding to the molecular mass of SA. Glucose, another predicted enzymatic reaction product, was not detected in this HPLC analysis as the molecule lacks detectable UV absorbance. These results unequivocally demonstrated that SghA is a β-glucosidase that hydrolyzes SAG to release SA.
Fig. 2.
SghA releases SA from SAG. (A) HPLC profile of reaction mixtures containing SAG treated with SghA (blue) and denatured SghA (red). (B) HPLC profile of reaction mixtures containing salicin treated with SghA (blue) and denatured SghA (red). HPLC experiments were repeated at least 3 times, and 1 set of representative data is presented. AU, arbitrary unit. (C) Catalytic site of SghA in the presence of SAG. SAG is shown as a stick, and carbon and oxygen atoms are colored green and red, respectively. Residues involved in SAG binding are shown as a stick model. Hydrogen bonds between His216 of SghA and SAG are indicated by a dashed line. Loops A to D (residues 54 to 69, 189 to 200, 314 to 344, and 365 to 384, respectively) are colored red and blue (loop D only) (same as in D). (D) Binding pocket of SghA in the presence of salicin. Salicin is shown as a stick, and carbon and oxygen atoms are colored cyan and red, respectively. Residues interacting with salicin are shown as a stick model. The hydrogen bond between His216 of SghA and salicin is indicated by a dashed line.
To determine the enzyme specificity of SghA, its catalytic activity was compared against SAG analogs. Results showed that SghA also hydrolyzed salicin to produce a product peak at around 4.0 min (Fig. 2B). ESI-MS analysis of this new peak fraction showed a strong quasimolecular (M-H) ion at an m/z of 123.00, similar to salicin-hydrolyzed product salicylic alcohol (SI Appendix, Fig. S3C). However, SghA could not hydrolyze other SAG structural analogs coniferin and sucrose (SI Appendix, Fig. S4), indicating the substrate selectivity of SghA. Consistent with this finding, SghA digested SAG at a much faster rate than salicin and ONPG under the same reaction conditions (Fig. 2 A and B). Michaelis constant (Km) of SghA was determined as 0.47 mM, 2.81 mM, and 5.91 mM for SAG, salicin, and ONPG, respectively (SI Appendix, Fig. S5). SghA of agrobacterium cells in culture in medium readily cleaved exogenous SAG to its metabolic product SA, as detected in the culture supernatant (SI Appendix, Fig. S6). These results suggest that SAG entered the bacterial cells and its cleaved product was exported out.
To further understand the molecular bases of SghA substrate specificity and catalytic mechanism, 2 substrates SAG and salicin were used to determine SghA structures in apo and in complex. The apo-SghA structure showed a typical (β/α)8-barrel fold in a single domain (SI Appendix, Fig. S7A), which is commonly found in β-glycoside hydrolase family 1 (GH1). The asymmetric unit contains 2 SghA molecules and forms a homodimer whereas dimerization does not influence the free access to its active site (SI Appendix, Fig. S7 B and C). Based on the comparison of the apo-SghA structure with other reported homolog structures, we identified that E179 of SghA plays a critical role in its activity. Substitution of E179 with S179 completely abolished enzymatic activity, as reflected by ONPG hydrolysis.
SghA–SAG and SghA–salicin complexes were cocrystallized by generating a hydrolase-dead mutant SghA(E179S) (SI Appendix, Fig. S8). Similar to other GH1 family proteins (13), SghA also has a bell-shaped binding cavity where glycone (sugar) buries inside and aglycone is located at the wide entrance gate (Fig. 2 C and D). To form the SghA–SAG complex, highly conserved residues (Q33, H134, N178, E367, and E420) formed hydrogen bonds with hydroxyl groups of sugar. Sugar binding was stabilized by the interacting of SAG with Y307 and W413 side chains on one side and indole groups of W135 and W421 from the other. SghA interaction with the aglycone group of SAG involved H193 in loop B and W340 in loop C to form a sandwich arrangement. In particular, 2 hydrogen bonds were established between the residue H193 and carboxylic acid group of SAG. In the case of SghA–salicin, salicin orientation was similar to SAG, and structural comparison revealed that there was little change of the active site except for H193. H193 appeared to form a hydrogen bond with the hydroxymethyl group of salicinaglycone; however, its geometrical preference was less perfect than SghA–SAG.
In contrast to the compact and conserved bottom half of (β/α)8 barrel (β-strand N terminus side), the top half (β-strand C terminus side) of SghA was surrounded by 4 flexible loops that gate the exit of the bell-shaped cavity. It was proposed that these loops are involved in the formation of the aglycone-binding site that determines substrate preference (14). In particular, loops B and C of the 4 loops A to D (Fig. 2 C and D) are mainly responsible for aglycone binding. In line with this notion, residues H193 and W340, respectively, located in loop B and C established direct interactions with the SAG salicylic acid group or salicin. Residue W340 formed an imperfect π–π interaction with the benzene ring, considering that W340 is quite conserved in the binding pocket among SghA homologs whereas H193 is only found in the binding pocket of SghA (SI Appendix, Fig. S9). The unique residue H193 in agrobacterial SghA interacting with SAG only suggests that H193 might play a more crucial role in substrate recognition of SghA than other residues. H193 made strong hydrogen-binding interactions with the SAG salicylic acid group that rationalizes our biochemical data about the substrate preference of SghA to SAG, rather than salicin.
Compared with the structure of Neotermes koshunensis β-glucosidase (13), SghA exhibits a conserved substrate-binding site (SI Appendix, Fig. S9). It is common for GH1 family proteins to have subtle differences in substrate specificity. In addition to the native substrate, these proteins could hydrolyze a broad spectrum of artificial substrates. Similarly, SghA hydrolyzed ONPG and X-gal, which are synthetic analogs of SAG. The GH1 protein family hydrolyzes glycosidic bonds through a retention mechanism that retains the overall anomeric configuration of the saccharide substrate. The combined data of structural and biochemical analysis proposed that a catalytic mechanism of SghA on substrate SAG:E367 makes a nucleophilic attack on the anomeric carbon C1 in SAG and E179 acts as a proton donor to release SA (SI Appendix, Fig. S10).
SghR Suppresses sghA Expression by Binding to Its Promoter.
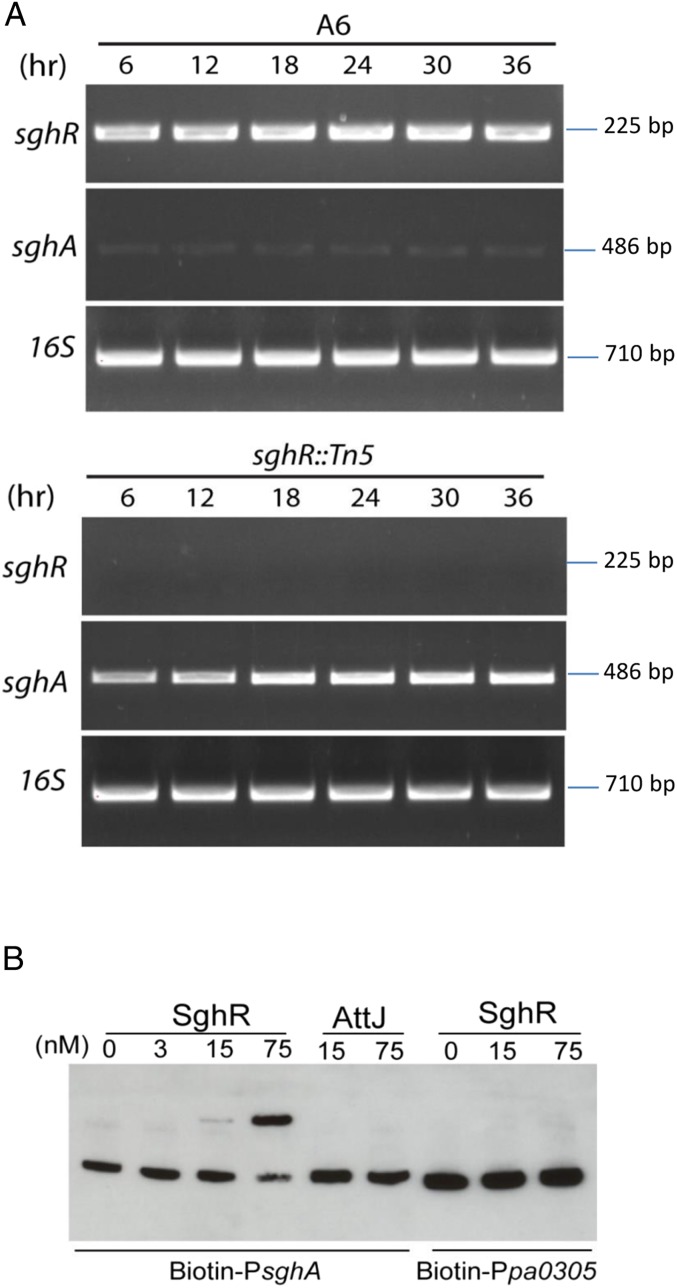
Results of RT-PCR and real time RT-PCR analyses revealed that the level of sghA messenger RNA (mRNA) was much higher in sghR mutant than wild-type strain A6 at different intervals (Fig. 3A and SI Appendix, Fig. S11). Gel shift analysis showed that SghR directly and specifically bound with the sghA promoter to shift it in a dosage-dependent manner, but the control promoter (pa0305 from P. aeruginosa PAO1) was unaffected by SghR (Fig. 3B). An unrelated IclR-type regulator (AttJ or BIcR) of A. tumefaciens also failed to shift the sghA promoter (15). Taken together, these results indicate that SghR suppresses sghA transcription via direct binding to its promoter in A. tumefaciens.
Fig. 3.
SghR controls the transcriptional expression of sghA. (A) RT-PCR analysis of the sghA expression patterns in wild-type strain A6 and its sghR mutant at different time intervals postinoculation in BM medium with mannitol as the sole carbon source. (B) Electrophoretic mobility-shift assay (EMSA) analysis of SghR binding to the sghA promoter. The unrelated transcriptional regulator AttJ from A. tumefaciens and the promoter Ppa0305 from P. aeruginosa were used as specificity controls. Purified proteins of SghR and AttJ were mixed with the biotin-labeled probes (1 nM) at various concentrations, and reaction mixtures were separated by electrophoresis. Experiments were repeated at least 3 times with similar results, and 1 set of representative data is presented.
Sucrose Inactivates SghR and Induces sghA Expression.
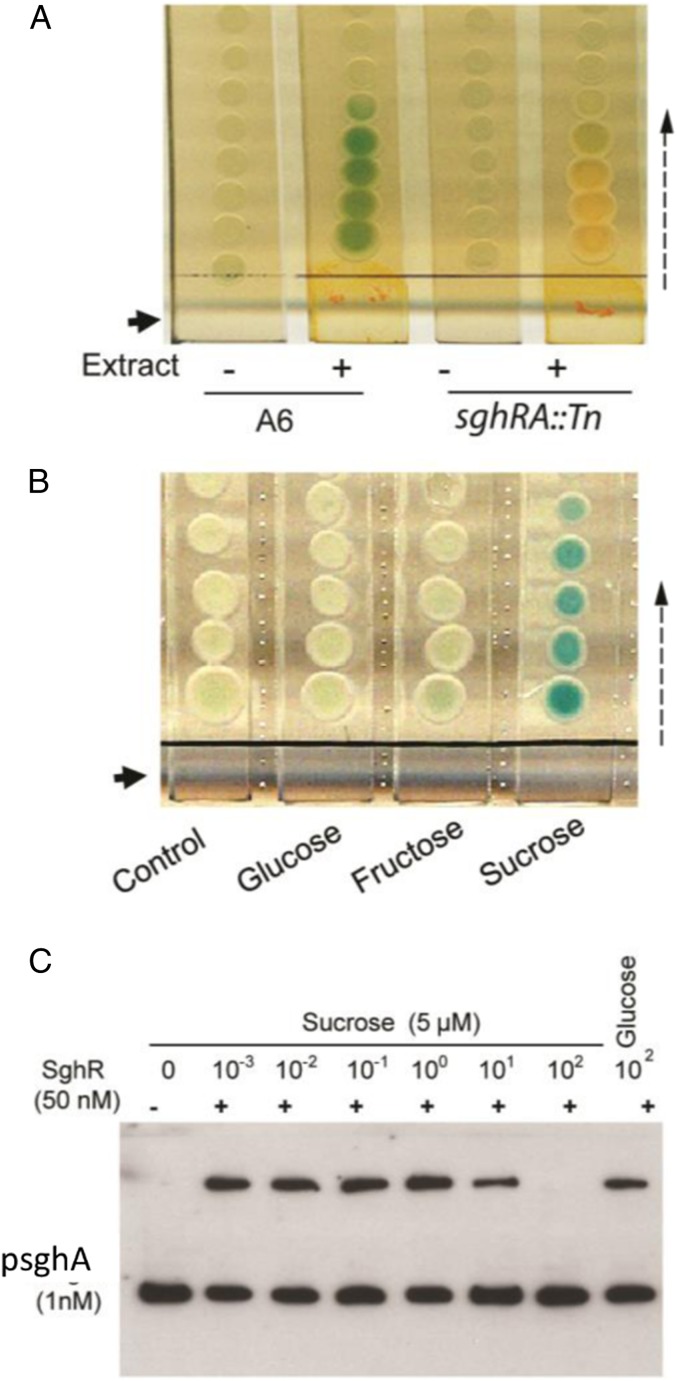
Given that SghA is tightly suppressed by SghR under in vitro conditions, we hypothesized that there might be a signal ligand interacting with SghR to induce sghA expression in planta. An extract of carrot tubers was prepared to test its activity in the induction of β-galactosidase encoded by sghA, and results depicted that sghA expression was induced with the addition of the extract (Fig. 4A). To purify the active signal, dry powders of carrot extract were further extracted with methanol, and samples were separated by silica column chromatography. Active fractions were pooled, and a compound was crystallized during the evaporation of solvent. This crystallized molecule displayed a strong activity to induce sghA expression when dissolved in water (SI Appendix, Fig. S12A). MS analysis showed that the molecular weight of this compound is 342.20, and, together with the results of NMR analysis, the active compound was identified as disaccharide sucrose (SI Appendix, Fig. S12 B–F). Commercial sucrose induced β-galactosidase activity in A. tumefaciens A6 on the X-gal plate (Fig. 4B). Consistent with the above results, gel shift analysis demonstrated that sucrose dissociated repressor SghR from the sghA promoter. SghR binding to the sghA promoter was substantially reduced and completely abolished when sucrose was added at a final concentration of 50 μM and 500 μM, respectively (Fig. 4C). This is within the range of extracellular sucrose concentration in planta. Quantitative measurement of intercellular sucrose levels in carrot tissues showed that sucrose concentrations were higher than 40 mM in both uninoculated control and tissues infected with bacterial cells after deletion of sghA and sghR (SI Appendix, Fig. S13). These findings demonstrate that plant metabolite sucrose is a cognate signal deactivating SghR and induces transcriptional expression of sghA.
Fig. 4.
Sucrose is the host signal that induces sghA expression. (A) X-gal plate assay of strain A6 sghA-encoded β-galactosidase activity in the presence or absence of carrot extract. A6, wild-type A. tumefaciens A6; sghRA::Tn, negative control of A. tumefaciens A6 with mutations in SghA and SghR. (B) Sucrose induced the expression of sghA in A6 on solid plates containing X-gal where A. tumefaciens A6 was spotted as the indictor strain on separated agar slices. The solid arrow in A and B represents the loading location of carrot extract and related compounds (5 µL of solution with a concentration of 1 M) on separated BM agar slices. The dashed arrow indicates the diffusion direction of the compounds. (C) EMSA analysis of sucrose activity in dissociation of SghR-promoter (PsghA) complex. Experiments were repeated at least 3 times with similar results, and 1 set of representative data is presented.
SghA Increases Plant SA Level during A. tumefaciens Infection.
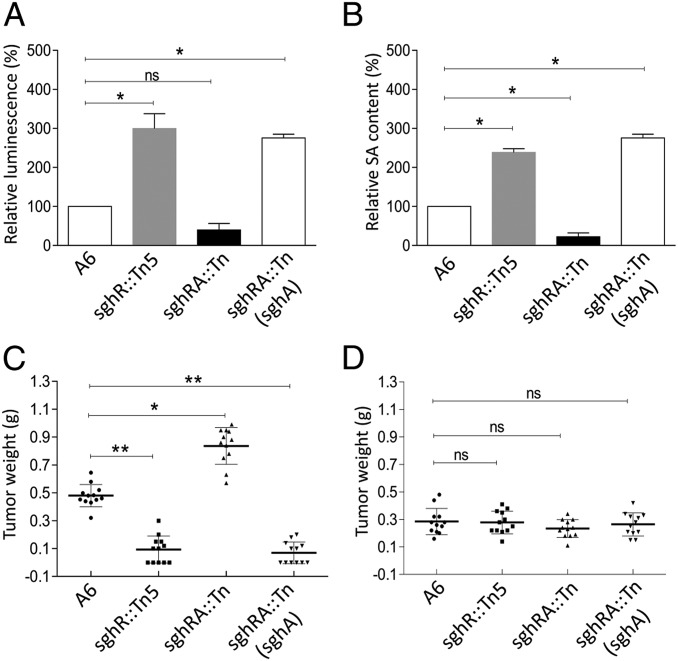
To confirm that SghA hydrolyzes SAG to SA during agrobacteria plant interaction, the level of free SA in infected plants was determined by using SA biosensor Acinetobacter sp. strain ADPWH_lux (16). Results revealed that inoculation of sghR mutant significantly enhanced free SA concentration in Arabidopsis at 20 h postinfection. Contrarily, challenge of the sghA/sghR double mutant led to lower SA level than plants inoculated with wild-type strain A6 (Fig. 5A). As expected, overexpression of sghA in the mutant sghA::Tn5 resulted in substantially increased free SA level. Similar results were also obtained with HPLC by directly measuring free SA concentration in the roots of Arabidopsis infected with A. tumefaciens A6 and its derivatives (Fig. 5B). Data indicated that, during infection, A. tumefaciens used SghA to release SA from SAG in the host plant.
Fig. 5.
SghA releases SA from SAG during bacterial infection and is involved in the modulation of tumor development. Biosensor analysis (A) and HPLC analysis (B) of SA in Arabidopsis roots infected with A. tumefaciens strain A6 and its derivatives. Experiments were repeated 5 times in duplicate, and relative fold change was calculated by normalizing against wild-type A6, arbitrarily set as 100%. (C) Tumor sizes incited by strain A6 and derivatives in A. thaliana wild-type Wassilewskija. (D) Tumor sizes incited by strain A6 and derivatives in the SAG-defective mutant of A. thaliana Ugt74f1. Experiments in C and D were repeated 3 times, and each time with 4 plants. Statistical analyses were performed with a nonparametric Kruskal–Wallis 1-way ANOVA test in GraphPad. Values and error bars represent means and SD. *P < 0.05; **P < 0.001; ns, not significant.
The tumor mass on Arabidopsis stems was measured to examine the impact of SghA-mediated SA increment in the host plant on bacterial infection (17, 18). Results showed that the tumor mass was substantially reduced in wild-type Arabidopsis ecotype WS-2 plants inoculated with the sghR mutant, as compared to those inoculated with wild-type A. tumefaciens A6. In contrast, disruption of sghA from the sghR mutant background led to significantly enhanced tumor growth, and tumor weight was almost 2 times that resulting from strain A6. Consistent with this trend, overexpression of sghA substantially attenuated bacterial tumorigenicity (Fig. 5C). These results were further consolidated by examining tumor weights on carrot discs (SI Appendix, Fig. S14A) and validated by quantification of tumor numbers after inoculation of carrot discs with serial dilutions of bacterial inoculum (SI Appendix, Fig. S14B).
Tumorigenicity of strain A6 and its derivatives was tested on Arabidopsis Ugt74f1 as control. Arabidopsis Ugt74f1 is a mutant derived from the Arabidopsis thaliana ecotype WS-2. In this mutant, SA glucosyltransferase was mutated to substantially reduce SAG formation, leading to a significantly higher SA level than in wild-type Arabidopsis (19, 20). As shown in Fig. 5D, strain A6 and its mutants caused a similar level of tumor growth on Arabidopsis Ugtf741, but the tumor mass was substantially less than that incited by strain A6 on wild-type Arabidopsis. In controls, no significant growth difference was recorded among wild-type and mutants in lysogeny broth (LB) and BM media, or with glucose and sucrose as the sole carbon source (SI Appendix, Fig. S15 A–D). sghR- and sghA-mutants produced similar levels of biofilms, and mRNA levels of virE3 in mutants were comparable with wild-type strain A6 in virulence-inducible or noninducible media (SI Appendix, Fig. S15 E and F). Data are consistent with the notion that an enhanced free SA level in host plants is positively associated with increased disease resistance and SghR/SghA play an important role in the modulation of A. tumefaciens virulence.
SghA-Released SA Reprograms A. tumefaciens Virulence.
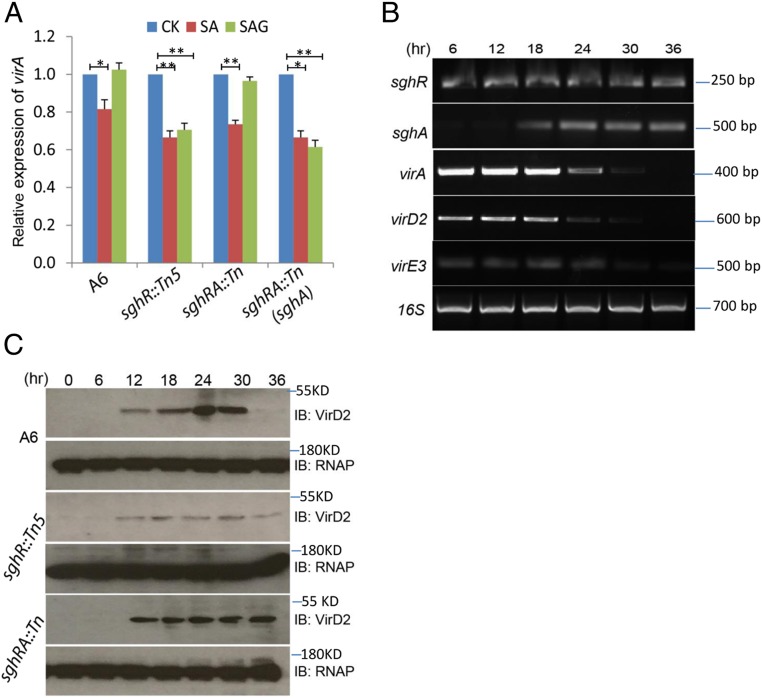
To understand the role of SghR/SghA in A. tumefaciens tumorigenicity, SA and SAG effects on the expression patterns of sgh and vir genes were analyzed. With a basal level of gene expression of virE3 in BM minimal medium, real time RT-PCR results showed that SA and SAG treatments had no detectable effect on the expression of sghR and sghA in the presence or absence of functional SghA (SI Appendix, Fig. S16). SA significantly inhibited virA expression in all tested strains whereas SAG affected virA expression only in the presence of functional SghA (Fig. 6A). Results suggest an essential role of SghA in the modulation of vir gene expression by releasing SA from the inactive SAG.
Fig. 6.
SghA-releasing SA down-regulates vir gene expression of A. tumefaciens during plant infection. (A) Real time RT-PCR analysis of virA expression in the presence of SA and SAG, respectively. The virA expression level in blank control without treatment (CK) was arbitrarily set as 1. The error bar denotes the SD of 3 repeats. Experiments were repeated 3 times in duplicate. Statistical analyses were performed with a nonparametric 1-way ANOVA test in GraphPad. *significant < 0.05; **significant < 0.01; ns, not significant. (B) In planta expression patterns of sgh and vir genes of A. tumefaciens strain A6. The 16S rRNA was amplified as an internal control. (C) Western blot analysis of VirD2 in planta expression patterns using VirD-specific antibody. Expression levels of bacterial RNA polymerase (RNAP) were determined with beta subunit RNAP-specific antibody as a loading control. IB, immunoblot. KD, kilodalton.
RT-PCR and real-time RT-PCR were used to determine in planta expression patterns of sgh and vir genes in wild-type strain A6 at different intervals after the inoculation of A. thaliana. SghA and vir genes displayed differentially overlapping expression patterns while sghR constitutively expressed at different postinoculation intervals (Fig. 6B and SI Appendix, Fig. S17). The 3 tested vir genes including virA, virD2, and virE3 significantly expressed during early stages of infection at 6, 12, and 18 h postinoculation, but their expressions were inhibited at the later stages. In contrast, hardly any sghA transcript was found at 6 and 12 h, and maximum expression occurred at 24 h postinoculation. Western blot analysis also showed that VirD2 protein expression in wild-type strain A6 occurred at around 12 h postinoculation, peaked at 24 h, and decreased afterward (Fig. 6C). Contrarily, sghR mutant resulted in a significantly decreased level of VirD2 protein (Fig. 6C), but sghA mutant caused higher expressions of VirD2, even at the later stage of infection. Taken together, these results suggest that SghR/SghA-mediated release of SA plays a key role in reprograming the virulence gene expression in A. tumefaciens.
Discussion
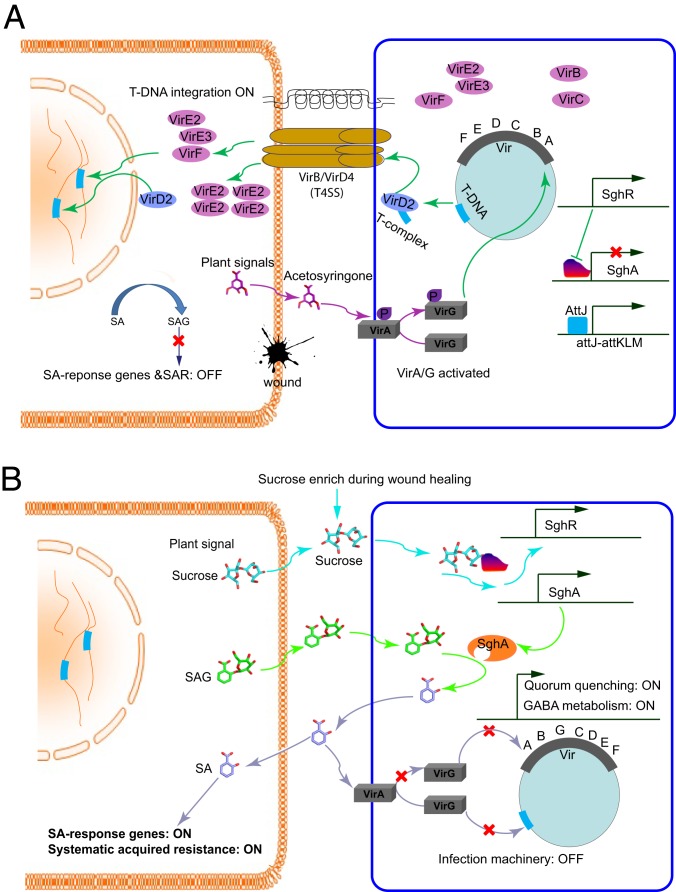
At least 2 lines of evidence indicate that the time required for tumor induction by A. tumefaciens is less than 10 h (21, 22), but expression patterns of the bacterial virulence gene after establishing infections is not clear. This study showed that, upon inoculation, vir genes of A. tumefaciens maintained a high level of expression in planta at the early stage of infection but their transcript levels were substantially decreased after 18 h. The key mechanism that reprograms vir gene expression in A. tumefaciens consists of a regulator and enzyme pair SghR/SghA that timely releases SA from SAG in a working model, as presented in Fig. 7. In this model, we proposed that plant SAG enters in agrobacterial cells for hydrolysis rather than the secretion of bacterial SghA to act extracellularly. Secretion-specific signal peptide was not identified in the SghA amino acid sequence; however, it is well-known that SAG analogs having high structural similarity with X-gal, ONPG, and salicin can enter bacterial cells. However, further studies using isotope-labeled SAG are required to verify this working model. SAG has been reported to exclusively deposit in the vacuole via ATP-dependent transportation (23), but it is currently unknown whether plant cells can also actively export SAG for hydrolysis. Generally, agrobacteria initiate infection at the wound site of the host plant, and SAG could be one of the wound-released chemical signals perceived by bacterial cells.
Fig. 7.
Schematic presentation of entry and exit of A. tumefaciens infection. (A) Infection entry at the beginning of bacterial infection. The plant signal acetosyringone is released upon wounds to bind with sensor VirA and thereby activates VirG by phosphorylation. Activated VirG triggers vir gene expression and initiates T-DNA synthesis in A. tumefaciens. Bacterial type IV secretion system (T4SS) transfers T-DNA into plant cells and integrates into the plant chromosome. At this stage, salicylic acid (SA) is inactivated by conjugation with glucose to produce SAG in plant cells, SA-responsive genes are not induced, and genes encoding systematic acquired resistance (SAR) are turned off. In the bacterial cell, quorum quenching gene attM and SA hydrolase gene sghA are suppressed by AttJ and SghR, respectively. (B) Infection exit at the later stage of infection. Sucrose enhances at the wounded site during healing and is transported into bacterial cells to dissociate SghR from its complex with the promoter DNA inducing sghA expression. Accumulated SghA hydrolyzes SAG to release SA, which, on one hand, triggers the expression of the AttJ-AttKLM operon, thus initiating quorum quenching and γ-aminobutyric acid (GABA) catabolism systems inside the bacterial cells. On the other hand, released SA inactivates VirA to shut down infection machinery. Additionally, released SA could also activate plant SA responsive genes and SAR.
SghA is known to hydrolyze different compounds, such as X-gal, ONPG, and salicin. Biochemical and structural analyses in this study suggested that SAG can serve as a bona fide substrate of SghA because 1) SghA failed to hydrolyze coniferin and its inducer sucrose, 2) SghA mutant grows readily with sucrose as the only carbon source, and 3) SghA releases SA from SAG in a highly efficient manner, as compared to other substrates. SA is a key signal that activates both local and systemic acquired resistance to regulate disease resistance. Under normal growth conditions, SA conjugates with β-glucosides and is stored in the inactive form to minimize its cytotoxicity, maximize SA stability, and facilitate its transportation. Among SA conjugates, SAG has been identified as a predominant and stable metabolite (24, 25). A previous study showed that plant growth hormone auxin influences A. tumefaciens virulence by inhibiting vir gene induction and bacterial growth (26). However, this effect was only observed at higher concentrations of auxin (25 to 250 μM), marginally matching with its level in Arabidopsis crown galls (17.3 ± 8.8 µM) and taking multiple days from agrobacteria inoculation to the formation of visible crown galls (27). During this study, inactivation of SghA promoted tumor formation in plants whereas it failed to improve agrobacterial growth in vitro. Future work is required to examine whether specific release of SA from SAG by SghA benefits bacteria during in vivo infection. SA is considered a systemic signal triggering a defense response in uninfected host cells. Initiation of SA synthesis in host cells by SghA lacking pathogens suggests that SghA-based SA release is not the only mechanism to manipulate the host SA signaling pathway during microbial infection. SghA may also metabolize additional substrates, and the fact that SghA releases SA might be coincidental although it exhibits no enzymatic activity toward sucrose and coniferin.
Transcriptional repressor SghR and its cognate signal ligand sucrose control the expression of sghA. Mutation of sghR led to constitutional expression of sghA, suggesting that SghR is the key regulator that governs transcriptional expression of sghA. Our data revealed that sghA is specifically induced by the plant metabolite sucrose, which interacts to inactivate repressor SghR at a concentration lower than 0.5 mM. Sucrose is a major photosynthesis product (>95%) in plants that could reach to a concentration of 1 M in conducting vascular cells and 2 to ∼7 mM in extracellular spaces (28). Consistent with this report, soluble sucrose concentration in carrot tissues was noted in the range of 47 to 100 mM, depending on postinoculation intervals. Extracellular concentration of sucrose can dramatically increase after tissue disruption. In addition to a carbon source, sucrose also acts as a signal molecule to regulate plant growth, development, differential gene expression, and stress-related responses (29). Furthermore, evidence supporting sucrose as a potent signal to induce plant defense responses has been reported (30, 31). However, the precise mechanism of sucrose-induced immunity and the related signaling pathway remains a mystery. Results of this study depict that sucrose specifically induces the expression of hydrolase SghA by deactivating repressor SghR, which degrades the SAG conjugate to increase the cellular level of plant signal SA. Therefore, identification of SghR/SghA depicts a mechanism that corroborates sucrose-induced immunity and explains that the SA level in plant tissues is not elevated by the initiation of agrobacterial infection, but dramatically enhances at the later infection stage (32).
Characterization of SghR/SghA unveils a mechanism that allows the bacterial pathogen to tap on the reserved pool of plant defense signal SA to reprogram its virulence gene expression after establishing infection. Bioinformatics analysis showed that SghA is highly conserved in A. tumefaciens and other members of the Rhizobiaceae family of α-proteobacteria. SghA shares moderate similarities with the homologs of other bacterial species, such as Dickeya and Pectobacterium. It is not clear whether these homologs can hydrolyze SAG or pair with an SghR-like regulator to release SA in a controlled manner. In this regard, it is interesting to note that SA could directly influence virulence gene expression in A. tumefaciens and other bacterial pathogens. At lower concentrations, SA was found to impair bacterial attachment and biofilm formation and down-regulate fitness and production of virulence factor in P. aeruginosa (33). Similarly, SA inhibited biofilm formation, motility, and N-acyl homoserine lactone quorum sensing signal production in Pectobacterium carotovorum and Pseudomonas syringae pv. Syringae (34). Moreover, the amazing sensing abilities of microbes and the response to changing environmental conditions require further investigations to unveil mechanisms that allow pathogens to reprogram virulence gene expression after establishing infections.
Methods
Bacterial Strains, Plasmids, and Growth Conditions.
SI Appendix, Table S1 lists bacterial strains and plasmids used in this study. A. tumefaciens strains were grown at 28 °C in BM medium (pH 7.0, 0.2% mannitol) or in VIB medium (NH4CI, 1 g/L; MgSO4 • 7H20, 0.3 g/L; KCl, 0.15 g/L; CaCl2, 0.01 g/L; FeSO4 • 7H20, 2.5 mg/L; K2HPO4, 0.06 g/L; NaH2PO4, 0.023 g/L; pH 5.5, 0.2% arabinose and 100 µM acetosyringone) (10). One liter of BM medium contained K2HPO4 (10.5 g), KH2PO4 (4.5 g), MgSO4 • 7H2O (0.2 g), (NH4)2SO4 (2.0 g), FeSO4 (5 mg), CaCl2 (10 mg), MnCl2 (2 mg), and mannitol (2.0 g). LB medium was used for general cultivation of Escherichia coli strains, and antibiotics were added at the following concentrations when required: kanamycin, 100 µg/mL; tetracycline, 5 µg/mL; gentamycin, 100 µg/mL; ampicillin, 200 µg/mL.
Genetic Manipulation of A. tumefaciens.
Transposon mutagenesis was performed in E. coli BW020767 containing a mini Tn5 or E. coli SM10 (pBT20) carrying a mariner transposon, and disrupted genes were identified (35). A. tumefaciens mutants were screened on BM solid plates containing X-gal (50 µg/mL) and relevant antibiotics. To conduct complementation analysis, a DNA fragment containing the putative promoter and full-length sghA was amplified from A. tumefaciens A6 by using PCR primers (SI Appendix, Table S2). The PCR product was digested by BamHI and cloned in pLAFR3, and the resultant construct pLA-sghA was verified by sequencing before transformation into A. tumefaciens by electroporation (15). Deletion of SghR, SghA, and its double deletion were performed according to Wang et al. (36).
Recombinant Proteins.
The coding regions of target genes were amplified from the genomic DNA of A. tumefaciens A6 and fused into 6×His-tagged expression vector pET14b (Novagen). After verification by DNA sequencing, resultant constructs were separately transformed into strain BL21(DE3), and recombinant proteins were purified (37).
Enzyme Assay.
SghA β-galactosidase activity was indicated by the appearance of a blue color on BM agar plates supplemented with X-gal (50 µg/mL) according to Zhang et al. (38). Briefly, solidified BM medium in Petri dishes was cut into separate slices, and plant extracts or chemical compounds were loaded at the end of the agar slice, which generated gradient concentrations of the tested sample by diffusing along the separated agar slices. Specified bacterial cultures were progressively spotted along the agar slice, and results were recorded after incubation at 28 °C for 48 h.
Quantification of β-galactosidase activity was carried out by gently shaking at 37 °C for 3 h in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-ME, pH 7.0) containing 1 mM ONPG or salicin or SAG and 10 mM SghA. The same amount of boiled SghA was added to the reaction mixture as a negative control. Samples were introduced onto a symmetry reverse-phase column (4.6 × 250 mm) for HPLC. Fractions were eluted with 50/50 methanol/water (vol/vol) at a flow rate of 1 mL/min and detected by a Waters 996 photodiode detector. ESI-MS was performed on a Finnigan/MAT ion-trap mass spectrometer following standard procedures.
RT-PCR and Western Blot Analysis.
To study sghR and sghA expression levels during bacterial growth, strain A6 and its mutant sghR::Tn5 were cultivated in BM medium and harvested at specific time intervals. Total RNAs were extracted by using the RNeasy Mini Kit (Qiagen). To treat SAG and SA, specified bacterial cells were grown overnight in LB medium and then subcultured in 5 mL of induction medium (with 100 μM acetosyringone) at an initial OD600 of 0.1, with or without SAG (10 μM) and SA (5 μM). After 5 h of shaking (rpm 200) at 28 °C, 1 mL of cell cultures was harvested for RNA extraction with the RNAprotect Reagent Kit (Qiagen). Residual DNAs were digested by RNase-free DNase I (Promega), and its absence was validated by PCR with purified RNA templates. RNA integrity was assessed by agarose gel electrophoresis, and RNA concentration was measured by Nanodrop ND-1000 (Nanodrop Technologies). To minimize data variation, total RNAs were prepared from 3 independent repeats and pooled together for RT-PCR analysis.
The protocol of the 1-step strategy (Qiagen) was followed to conduct RT-PCR. An aliquot of 0.2 µg of total RNAs served as the template for RT-PCR to amplify a portion of target genes with primers (SI Appendix, Table S2). A fragment of 16S ribosomal RNA (rRNA) was also amplified in each RT-PCR as an internal control. An aliquot of 0.1 mg of total RNAs served as the template for real-time RT-PCR, and real-time reverse transcription-PCR analysis was performed in Rotor-Gene Q (Qiagen) by using the SYBR Green RT-PCR Kit (Qiagen). Housekeeping gene rpoC encoding the β subunit of RNA polymerase was included as an internal control. Results were presented as the ratio of target gene expression versus control gene. Real-time RT-PCR primers are listed in SI Appendix, Table S2.
To determine the expression levels of sghR, sghA, and vir genes during Agrobacterium infection, strain A6 cells grown in LB medium at late exponential phase were harvested, washed 3 times with DPBS (Dulbecco’s phosphate-buffered saline) buffer and resuspended in the same buffer for carrot infection. Carrots were washed 3 times with 75% ethanol, sliced into 1.5-cm-thick disks, and inoculated with 200 µL of bacterial cells. Infected carrot disks were placed into Petri dishes moisturized with wet filter papers and incubated at 28 °C. Bacterial cells were collected at specific time intervals by washing with DPBS buffer containing Triton X-100 (0.1%), treated with RNAprotect Bacteria Reagent (Qiagen), and kept at −80 °C until RNA extraction. RT-PCR was conducted following the protocol of the 1-step strategy (Qiagen) or 2-step strategy (Promega). An aliquot of 0.2 µg of total RNAs was used as template to amplify a portion of the target genes with primers listed in SI Appendix, Table S2.
Western blotting was carried out by harvesting bacterial cells from infected plant tissues as mentioned above and lysed in urea buffer (100 mM NaH2PO4, 10 mM Tris⋅HCl, 8 M Urea, 1 mM Na3VO4, 20 mM NaF, 0.1 mM β-glycerophosphate, and 20 mM sodium pyrophosphate) supplemented with a complete Protease Inhibitor Mixture tablet (Roche, pH 8.0). Lysates were cleared by centrifugation (14,000 rpm, 10 min, 4 °C), and protein concentration in supernatants was determined with the BCA Protein Assay Kit (Pierce). Equal amounts of total proteins were used for Western blot with VirD2-specific (catalog no. MBS5304219; Mybiosource) and RNA polymerase (RNAP)-specific antibodies (catalog no. 663903; Biolegend,).
Electrophoretic Mobility-Shift Assay.
The SghA promoter region (−345 bp to +43 bp) was amplified from genomic DNA of A. tumefaciens strain A6 by using 5′-biotinylated primers (SI Appendix, Table S2). The fragment was designated as PsghA and used as a probe for gel retardation analysis to determine potential interaction with transcriptional repressor SghR. As a specificity control, PCR was used to generate the Ppa0305 probe (−281 bp to +81 bp) from genomic DNA of P. aeruginosa with 5′-biotinylated primers (SI Appendix, Table S2). A gel retardation assay was performed with the LightShift Chemiluminescent EMSA Kit by following the recommended protocol (Pierce Biotechnology).
SA Measurement.
A. thaliana ecotype WS-2 seeds were sterilized with a solution (50% bleach and 0.1% SDS) for 10 min and rinsed in sterile water several times. Seeds were placed on MS agar plates (Sigma), incubated at 4 °C for 2 d, and transferred to a growth chamber with a short-day (8 h light) photoperiod at 23 °C for root culture. After 2 wk, roots were axenically cut into ∼3-cm segments on a sterile filter paper and transferred to MS medium for agrobacterial infection. Root segments were collected at 20 h postinfection for SA extraction and quantified with HPLC (39) and biosensor Acinetobacter sp. ADPWH_lux (16).
Protein Preparation, Crystallization, and Structural Determination.
The SghA coding region was amplified by PCR and cloned into vector pET-14b. The resultant construct was verified by DNA sequencing and transformed into E. coli BL21 CodonPlus-(DE3) RIL for protein expression. SghA purification was carried out according to Ye et al. (37). SghA derivative E179S was generated by QuikChange (Stratagene) and purified by following the same procedure as SghA.
Crystallization of SghA has been previously reported (37). To crystallize SghA–SAG, –salicin, and –glucose protein–ligand complexes, the ligands were mixed with SghA derivative E179S at a final concentration of 20 mM and incubated for 30 min at room temperature, followed by a similar crystallization procedure for the apo form.
Diffraction data were collected at 100 K on beam lines I04 (Diamond) or X06SA (Swiss Light Source). Data were processed in XDS (40), and structures were determined with Phaser (41) using homolog structure (PDB ID code 1NP2) as a search model for SghA. Automatic model building was performed in ARP/wARP (40) and improved by building a manual model with COOT (42). Models were refined with Phenix (43) and checked by PROCHECK (44). Crystallographic data and refinement statistics are listed in SI Appendix, Table S3, and figures were generated with Pymol (Delano Scientific).
Quantitative Analysis of Soluble Sucrose in Carrot Tissues.
The overnight starter cultures of A. tumefaciens strains were inoculated (1%) in 20 mL of LB medium and grown at 28 °C and 200 rpm until OD600 reached to 1.0. Bacterial cells were collected by centrifuging at 12,000 rpm for 10 min, washed twice with sterile PBS buffer, and resuspended in 2 mL of the same buffer. Carrot samples were sterilized with 80% ethanol and dissected into 1-cm-thick slices. Each carrot slice was evenly spread with 100 μL of bacterial solution, placed on a Petri dish with moisturized filter papers, and incubated at 28 °C for 6 h. Similarly, PBS buffer was added to serve as a control. To measure extracellular sucrose concentration, carrot samples were collected and washed with sterile water to remove bacterial cells. Slices were dried with tissue paper, and a thin layer (2 mm) was cut from the end of inoculation and weighed. Apoplastic fluid was collected with an infiltration technique as previously described (45). After infiltration, carrots were centrifuged at 650 × g for 10 min at 4 °C. Intercellular sucrose content was then determined with sucrose assay kit SCA20-1KT (Sigma-Aldrich) by following the manufacturer’s protocol. Carrot tissue powders were extracted 3 times with 1 mL of sterile water, mixed thoroughly, and placed in a 60 °C water bath for 15 min. Supernatants were collected by centrifuging for 10 min at 12,000 rpm. Supernatants of the same sample were combined, and sterile water was added to a final volume of 11 mL, which was diluted accordingly for sucrose quantification with the SCA20-1KT.
Tumorigenesis Assay.
Arabidopsis infection was carried out according to Deeken et al. (46). Briefly, A. thaliana seeds were cultivated in a growth room under a short-day condition at 23 °C. Tumors were induced by applying A. tumefaciens strains to the base of a wounded young inflorescence stalk. Disease symptoms were recorded on the 28th day of infection. Statistical analyses were conducted with the nonparametric 1-way ANOVA test.
Supplementary Material
Acknowledgments
We thank Dr. Hui Wang (Natural Environment Research Council/Centre for Ecology and Hydrology, Oxford) for providing Acinetobacter sp. strain ADPWH_lux, and Prof. John V. Dean (DePaul University) for critical comments on SA measurement and providing Arabidopsis seeds. We also thank T. Tomizaki and M. Wang (Swiss Light Source) for help in X-ray data collection. This work was supported by National Program on Key Basic Research Project (973 Program) of China 2015CB150600 and the Agency for Science, Technology and Research in Singapore (to L.-H.Z.), Nanyang Technological University Integrated Medical, Biological and Environmental Life Sciences Grant NIM/01/2018 (to Y.-G.G.), and Basic Scientific Research Project of Chongqing Science and Technology Commission CSTC2018JXJL0230 (to X.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structural factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6RJK [Apo], 6RK2 [with SAG], 6RJO [with salicin], and 6RJM [with glucose]).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903695116/-/DCSupplemental.
References
- 1.Zupan J. R., Zambryski P., Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol. 107, 1041–1047 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coggan K. A., Wolfgang M. C., Global regulatory pathways and cross-talk control pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 14, 47–70 (2012). [PMC free article] [PubMed] [Google Scholar]
- 3.Knodler L. A., et al. , Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U.S.A. 107, 17733–17738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore L. W., Chilton W. S., Canfield M. L., Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl. Environ. Microbiol. 63, 201–207 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brencic A., Winans S. C., Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 69, 155–194 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schell J., et al. , Interactions and DNA transfer between Agrobacterium tumefaciens, the Ti-plasmid and the plant host. Proc. R. Soc. Lond. B Biol. Sci. 204, 251–266 (1979). [DOI] [PubMed] [Google Scholar]
- 7.Nester E. W., Agrobacterium: Nature’s genetic engineer. Front. Plant Sci. 5, 730 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun A. C., Mandle R. J., Studies on the inactivation of the tumor-inducing principle in crown gall. Growth 12, 255–269 (1948). [PubMed] [Google Scholar]
- 9.Anand A., et al. , Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 146, 703–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan Z. C., et al. , The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 104, 11790–11795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z., Klessig D. F., Identification of a soluble salicylic acid-binding protein that may function in signal transduction in the plant disease-resistance response. Proc. Natl. Acad. Sci. U.S.A. 88, 8179–8183 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yalpani N., Raskin I., Salicylic acid: A systemic signal in induced plant disease resistance. Trends Microbiol. 1, 88–92 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Jeng W. Y., et al. , High-resolution structures of Neotermes koshunensis β-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates. Acta Crystallogr. D Biol. Crystallogr. 68, 829–838 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Nijikken Y., et al. , Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium. FEBS Lett. 581, 1514–1520 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Wang C., Zhang H. B., Wang L. H., Zhang L. H., Succinic semialdehyde couples stress response to quorum-sensing signal decay in Agrobacterium tumefaciens. Mol. Microbiol. 62, 45–56 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Huang W. E., et al. , Chromosomally located gene fusions constructed in Acinetobacter sp. ADP1 for the detection of salicylate. Environ. Microbiol. 7, 1339–1348 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Chevrot R., et al. , GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 103, 7460–7464 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haudecoeur E., et al. , Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 106, 14587–14592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noutoshi Y., et al. , Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 24, 3795–3804 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean J. V., Delaney S. P., Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 132, 417–425 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Sykes L. C., Matthysse A. G., Time required for tumor induction by Agrobacterium tumefaciens. Appl. Environ. Microbiol. 52, 597–598 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Virts E. L., Gelvin S. B., Analysis of transfer of tumor-inducing plasmids from Agrobacterium tumefaciens to Petunia protoplasts. J. Bacteriol. 162, 1030–1038 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean J. V., Mills J. D., Uptake of salicylic acid 2-O-beta-D-glucose into soybean tonoplast vesicles by an ATP-binding cassette transporter-type mechanism. Physiol. Plant. 120, 603–612 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Lee H. I., Raskin I., Glucosylation of salicylic acid in Nicotiana tabacum Cv. Xanthi-nc. Phytopathology 88, 692–697 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Silverman P., et al. , Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 108, 633–639 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P., Nester E. W., Indoleacetic acid, a product of transferred DNA, inhibits vir gene expression and growth of Agrobacterium tumefaciens C58. Proc. Natl. Acad. Sci. U.S.A. 103, 4658–4662 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gohlke J., Deeken R., Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 5, 155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch K., Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 7, 235–246 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Wind J., Smeekens S., Hanson J., Sucrose: Metabolite and signaling molecule. Phytochemistry 71, 1610–1614 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Bolouri Moghaddam M. R., Van den Ende W., Sugars and plant innate immunity. J. Exp. Bot. 63, 3989–3998 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Hanson J., Smeekens S., Sugar perception and signaling–An update. Curr. Opin. Plant Biol. 12, 562–567 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Lee C. W., et al. , Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21, 2948–2962 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prithiviraj B., et al. , Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans. Infect. Immun. 73, 5319–5328 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagonenko L., Lagonenko A., Evtushenkov A., Impact of salicylic acid on biofilm formation by plant pathogenic bacteria. J. Biol. Earth Sci. 3, 6 (2013). [Google Scholar]
- 35.Larsen R. A., Wilson M. M., Guss A. M., Metcalf W. W., Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178, 193–201 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Wang C., Tang D., Gao Y. G., Zhang L. H., Succinic semialdehyde promotes prosurvival capability of Agrobacterium tumefaciens. J. Bacteriol. 198, 930–940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye F., Wang C., Fu Q., Zhang L. H., Gao Y. G., Cloning, expression, purification and crystallization of a pair of novel virulence factors, SghA and SghR, from Agrobacterium tumefaciens. Acta Crystallogr. F Struct. Biol. Commun. 71, 1139–1145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H. B., Wang L. H., Zhang L. H., Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 99, 4638–4643 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaspar Y. M., et al. , Characterization of the Arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of agrobacterium transformation. Plant Physiol. 135, 2162–2171 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langer G., Cohen S. X., Lamzin V. S., Perrakis A., Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy A. J., Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M., AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Lohaus G., et al. , Solute balance of a maize (Zea mays L.) source leaf as affected by salt treatment with special emphasis on phloem retranslocation and ion leaching. J. Exp. Bot. 51, 1721–1732 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Deeken R., et al. , Tumour development in Arabidopsis thaliana involves the Shaker-like K+ channels AKT1 and AKT2/3. Plant J. 34, 778–787 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.