Significance
Mouse studies demonstrating regression of p53-null tumors following reinstatement of functional p53 have fueled the development of p53 reactivating drugs. However, successful p53 reactivation responses have only been formally demonstrated in tumor models where p53 inactivation served as the initiating event. Our study provides the first proof-of-principle evidence that p53 inactivation at late stages of tumorigenesis can also generate a vulnerability to p53 reactivation. However, this is dependent on intact ARF function highlighting ARF as a potential biomarker for p53 reactivation responses in tumors with late-stage p53 inactivation. It furthermore suggests the use of Mdm2 inhibitors as ARF mimetics for sensitizing ARF-deficient tumors to p53-reactivating drugs.
Keywords: p53, tumor suppressor gene, reactivation therapy, Mdm2, Arf
Abstract
Cancer development is driven by activated oncogenes and loss of tumor suppressors. While oncogene inhibitors have entered routine clinical practice, tumor suppressor reactivation therapy remains to be established. For the most frequently inactivated tumor suppressor p53, genetic mouse models have demonstrated regression of p53-null tumors upon p53 reactivation. While this was shown in tumor models driven by p53 loss as the initiating lesion, many human tumors initially develop in the presence of wild-type p53, acquire aberrations in the p53 pathway to bypass p53-mediated tumor suppression, and inactivate p53 itself only at later stages during metastatic progression or therapy. To explore the efficacy of p53 reactivation in this scenario, we used a reversibly switchable p53 (p53ERTAM) mouse allele to generate Eµ-Myc–driven lymphomas in the presence of active p53 and, after full lymphoma establishment, switched off p53 to model late-stage p53 inactivation. Although these lymphomas had evolved in the presence of active p53, later loss and subsequent p53 reactivation surprisingly activated p53 target genes triggering massive apoptosis, tumor regression, and long-term cure of the majority of animals. Mechanistically, the reactivation response was dependent on Cdkn2a/p19Arf, which is commonly silenced in p53 wild-type lymphomas, but became reexpressed upon late-stage p53 inactivation. Likewise, human p53 wild-type tumor cells with CRISPR-engineered switchable p53ERTAM alleles responded to p53 reactivation when CDKN2A/p14ARF function was restored or mimicked with Mdm2 inhibitors. Together, these experiments provide genetic proof of concept that tumors can respond, in an ARF-dependent manner, to p53 reactivation even if p53 inactivation has occurred late during tumor evolution.
Cell fusion experiments in the 1960s showed that fusion of normal cells with tumor cells results in a nonmalignant phenotype (1). This not only provided the experimental evidence for the existence of tumor suppressor genes, but also suggested restoration of tumor suppressors as a tumor therapy. Today, stimulated by the clinical success of oncogene-targeted drugs, there is a regained interest in therapeutic targeting of defective tumor suppressors, in particular the most frequently mutated tumor suppressor p53.
The tumorigenic potential of p53 mutations correlates with the loss of p53’s physiological function as a DNA binding transcription factor (loss of function, LOF) (2, 3). In addition, a subset of p53 missense mutations is known to confer neomorphic activities that promote tumor progression and therapy resistance (gain of function, GOF) (4, 5). Strategies aimed at repairing the LOF are technically challenging, but promise to be a universal therapy approach for a broad spectrum of cancer patients. The best-known compound for reactivating mutant p53 (mutp53) is PRIMA-1 which, in the form of PRIMA-1MET/APR-246, is currently evaluated in clinical trials up to phase III (6). Other compounds aim at reactivating specific p53 missense mutants (7, 8) or nonsense mutants by promoting transcriptional readthrough (9). A major concern with all small molecule approaches are off-target effects, which have been documented broadly for mutp53-reactivating compounds, questioning whether observed therapeutic responses are caused by off-target activities rather than mutp53 reactivation (5, 10, 11).
As there is currently no sufficiently specific compound available to reactivate a p53 LOF mutant to a fully functional wild type, genetically defined models for reactivation therapy have proven essential to establish proof-of-principle evidence for mutp53 as a suitable target for therapeutic reactivation. For example, mutp53 reactivation was modeled by fusing the ligand binding domain of a modified estrogen receptor to the C terminus of p53 (p53ERTAM), thereby rendering p53 switchable at the protein activity level with tamoxifen (TAM) (12, 13). In the absence of tamoxifen (TAM), p53ERTAM is inactive (OFF state) and accumulates similarly to cancer-derived p53 mutants (12). TAM switches p53ERTAM to the ON state, resulting in p53 target gene activation (12). TAM treatment thereby models therapy of p53-mutated tumor cells with p53-reactivating small molecule compounds (12–14). Lymphomas that developed in Eµ-Myc transgenic p53ERTAM mice in the absence of tamoxifen, i.e., in the p53 OFF state, rapidly regressed upon activation of p53ERTAM with tamoxifen. Together with other studies in independent mouse models (15–18), this firmly established the therapeutic potential of p53 reactivation and provided critical support for further research into the development of mutp53-reactivating drugs.
An important caveat to these experiments is their focus on tumors that have developed in a p53-compromised background where p53 loss served as an initiating driver of tumorigenesis (13, 15–18). p53 mutations are certainly driver mutations in Li-Fraumeni syndrome patients, who suffer from hereditary cancer susceptibility because of germline p53 mutations (19), and in certain sporadic cancer types such as high-grade serous ovarian cancer where p53 mutations are found already in the earliest premalignant cells (20). In other cancer entities, the timing of p53 mutations during tumor evolution is highly variable and often occurs only at later stages of tumor development (21, 22). For example, according to the prevailing multistep progression model for colorectal cancer, p53 mutations occur only late at the adenoma-to-carcinoma transition (23). Furthermore, recent tumor genome sequencing studies identified widespread subclonal p53 mutations in p53 wild-type tumors that expand during metastatic progression or therapy relapse, implicating them as a cause of therapy failure (21, 24–26). It is therefore of considerable clinical interest to explore whether tumors that have initially evolved in the presence of wild-type p53—but inactivated p53 at later stages of tumorigenesis—are similarly dependent on persistent p53 loss as tumors originating from a p53-compromised precursor cell.
This question is far from trivial as p53 wild-type tumors usually acquire aberrations in the p53 pathway. For example, TCGA tumors without p53 mutations are significantly enriched for copy-number deletions affecting the CDKN2A alternative reading frame (ARF) encoding the p14ARF protein (27). This observation on cancer patients is experimentally well recapitulated by the Eµ-Myc lymphoma model in mice: p53 wild-type lymphomas frequently lack expression of the murine CDKN2A/p14ARF homolog Cdkn2a/p19Arf and, vice versa, enforced Cdkn2a/p19Arf loss protects from p53 inactivation (28, 29). Importantly, several mouse models identified Cdkn2a/p19Arf to be essential for tumor regression upon p53 reactivation (13, 17, 18). If a p53 wild-type tumor cell with inactive p14ARF/p19Arf (in short ARF) acquires a secondary p53 mutation at a late stage of tumorigenesis, p53 reactivation would therefore be expected to be ineffective. In other words, p53 reactivation as a therapeutic strategy would only be effective if the p53 mutation has been the initiating driver lesion, but not in cases where the p53 mutation has occurred at later stages of tumor evolution on the background of other p53 pathway aberrations.
To explore whether tumors with late stage p53 inactivation respond to p53 reactivation, we genetically modeled late-stage p53 inactivation and therapeutic reactivation in mice and human tumor cells using the reversibly switchable p53ERTAM (12). Our results confirm that ARF alterations prevent a p53ERTAM reactivation response in tumor cells with late-stage p53ERTAM inactivation as predicted. However, we also observed that Myc-driven lymphomas with active p53ERTAM down-regulated ARF, but restore ARF expression upon p53ERTAM inactivation and thereby become susceptible to p53ERTAM reactivation. ARF expression in tumors with late stage p53 mutations could therefore serve as a potential biomarker for predicting p53 reactivation responses and Mdm2 inhibitors could be exploited as ARF mimetics to sensitize ARF-deficient p53-mutated tumor cells to p53 reactivation.
Results
Generation of Tamoxifen-Switchable p53ERTAM Lymphomas.
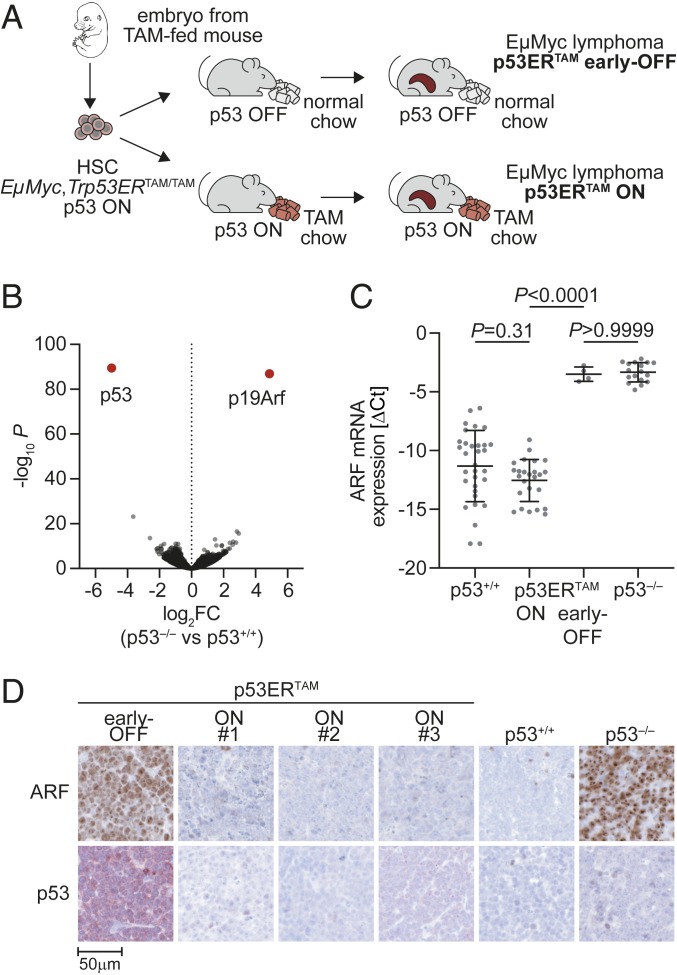
The p53ERTAM knockin mouse provides a unique opportunity to reversibly switch p53 at the protein activity level with TAM (12). To engineer lymphomas with early or late p53 inactivation, hematopoietic stem cells were obtained from the liver of Eµ-Myc;Trp53ERTAM/TAM embryos at embryonic day 13.5 (E13.5), transduced with luciferase for monitoring lymphomagenesis and transplanted into lethally irradiated recipient mice. Transplanted mice were fed either normal or TAM-supplemented chow, giving rise to Eµ-Myc lymphomas with inactive (early OFF) or active (p53 ON) p53ERTAM, respectively (Fig. 1A). Comparative gene expression profiling of Eµ-Myc;Trp53+/+ (p53+/+) and Eµ-Myc;Trp53−/− (p53−/−) lymphomas identified ARF to be the most differentially expressed gene (Fig. 1B). Early-OFF lymphomas expressed equally high ARF mRNA and protein levels as p53−/− lymphomas (Fig. 1 C and D). In contrast, ARF was undetectable in both p53-ON and p53+/+ lymphomas, indicating that in the presence of TAM p53ERTAM lymphomas bypass p53-mediated tumor suppression just like p53+/+ lymphomas via ARF inactivation (Fig. 1 C and D and SI Appendix, Fig. S1A). Of note, p53ERTAM protein was almost undetectable in p53-ON lymphomas, but strongly expressed in early-OFF lymphomas (Fig. 1D and SI Appendix, Fig. S1A), reminiscent of the stabilization of mutant p53 proteins in human cancer tissues (4).
Fig. 1.
Generation of Eµ-Myc;p53ERTAM lymphomas with active p53. (A) Scheme illustrating the experimental outline. HSC, hematopoietic stem cells from fetal liver. (B) Volcano plot depicting fold change in gene expression (log2FC) and significance (−log10P) for Eµ-Myc;p53+/+ and Eµ-Myc;p53−/− lymphomas (n = 4 each). (C) ARF mRNA expression of lymphomas with indicated p53 genotype and activity status. Expression of individual lymphomas is shown normalized to β-actin as ΔCt (mean ± SD, 1-way ANOVA with Tukey’s multiple comparisons test). (D) Immunostaining of ARF and p53 (CM5) for lymphomas with indicated p53 genotype and activity status.
Late-Stage p53 Inactivation and Reactivation Therapy.
To model late-stage p53 inactivation, p53-ON Eµ-Myc lymphomas from moribund TAM-fed mice were transplanted into normally fed recipients, switching p53ERTAM to the OFF state and thereby generating lymphomas with late-stage p53 inactivation (late OFF) (Fig. 2A). Survival of mice with late-OFF lymphomas was significantly shorter than for p53-ON lymphomas and similar to early-OFF lymphomas (Fig. 2B), indicating that late-stage p53 inactivation can render Eµ-Myc lymphomas more aggressive.
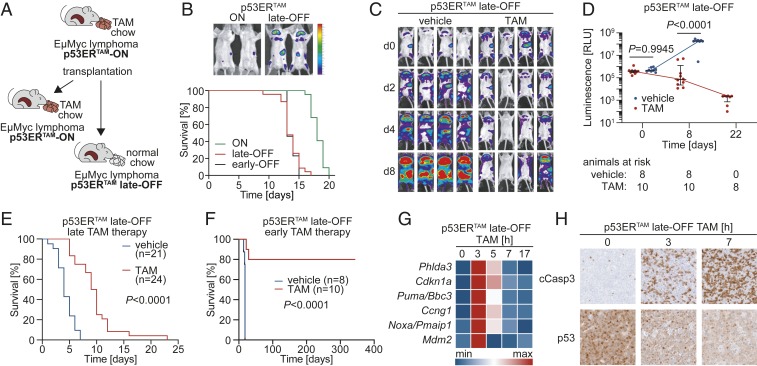
Fig. 2.
Tumor regression following p53 reactivation in late-OFF Eµ-Myc lymphomas. (A) Scheme depicting generation of late-OFF lymphomas by transplantation of p53-ON lymphomas into normal chow-fed mice. (B, Top) Bioluminescence images of representative p53-ON and late-OFF lymphoma mice at day 9. (B, Bottom) Kaplan–Meier survival plots for mice with p53-ON (n = 22), late-OFF (n = 23), and early-OFF (n = 13) Eµ-Myc lymphomas. Median survival: ON, 18 d; early OFF, 13 d; and late OFF, 13 d. Log-rank test: ON vs. late OFF P < 0.0001; ON vs. early OFF P < 0.0001; late OFF vs. early OFF P = 0.9043. (C) Bioluminescence images of representative mice with late-OFF lymphomas treated with either TAM or vehicle. (D) Quantification of whole-body bioluminescence. Shown is the photon flux for individual mice and the mean ± SD for both cohorts at indicated time points. Multiple t test corrected with the Holm–Sidak method. (E and F) Kaplan–Meier survival plots for mice with late-OFF Eµ-Myc lymphomas treated as indicated. Treatment started when mice showed full-blown lymphoma (E) or 4 d after transplantation (F). Shown is time after start of treatment. (G) p53 target gene expression in late-OFF lymphomas at indicated time points after TAM administration was quantified by RTqPCR and depicted as a heatmap of the row-wise min-max scaled mean mRNA expression (n = 3). (H) Immunostaining of late-OFF lymphomas for cleaved caspase-3 and p53 (CM5) at indicated time points after TAM administration.
To explore whether late-OFF tumors respond to p53 reactivation, p53-ON Eµ-Myc lymphomas were retransplanted into cohorts of normal-fed mice and monitored by bioluminescence imaging (BLI). When the animals showed first signs of disease in BLI, they were treated for 1 wk with daily injections of TAM to switch p53ERTAM to the ON state as a model for treatment with a p53 reactivating drug. Already 2 d after initiation of treatment, stagnation of lymphoma growth was detectable (Fig. 2C). Photon flux as a surrogate marker of lymphoma burden progressively decreased over the following days (Fig. 2 C and D). When TAM was administered to animals that showed full-blown lymphoma, the treatment more than doubled median survival (vehicle: 4 d, TAM: 9.5 d, P < 0.0001, Fig. 2E) similar to what has been reported for lymphomas that had evolved in the absence of active p53 (13). We observed equivalent reactivation responses for late-OFF lymphomas generated from 3 independent primary p53-ON lymphomas (SI Appendix, Fig. S2A). p53−/− lymphomas failed to profit from TAM treatment (SI Appendix, Fig. S2B), confirming that the TAM effect in late-OFF lymphomas is p53 mediated and therefore on target. When mice were treated already 4 d after transplantation, TAM even cured the majority (80%) of animals (Fig. 2F). In contrast, control mice that received oil injections rapidly progressed and reached a median survival of just 18 d following transplantation (Fig. 2F). TAM-treated late-OFF lymphomas showed induction of canonical p53 target genes peaking at 3 h (Fig. 2G) and massive and progressive apoptosis over 3 to 7 h following TAM injection (Fig. 2H). In parallel, p53ERTAM protein levels decreased, consistent with the lower half-life of active p53 (Fig. 2H). We conclude, that late-OFF Eµ-Myc lymphomas, which have developed in the presence of active p53ERTAM and were later switched to a p53-inactive state, regress when p53ERTAM is reactivated, indicating that they have rapidly become addicted to p53 inactivation.
Late-OFF Lymphomas Reactivate p19ARF.
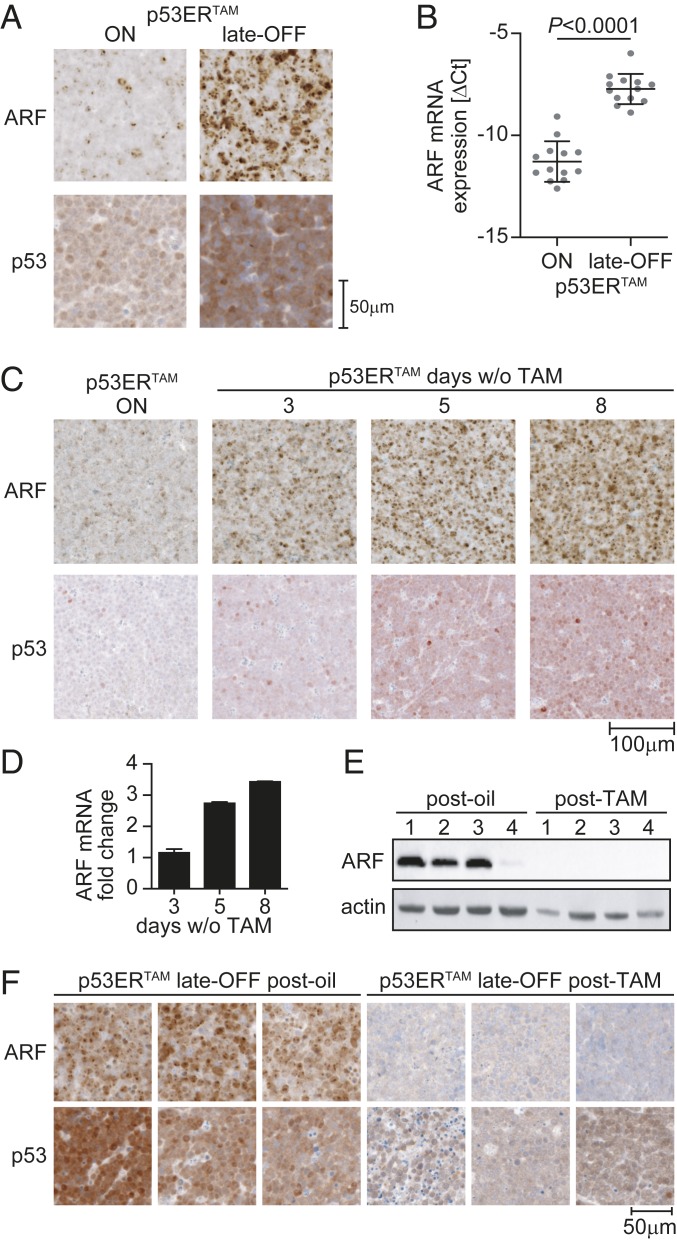
Regression of late-OFF lymphomas upon TAM treatment was rather unexpected, considering that Eµ-Myc lymphomas, which have originated in the presence of active p53, commonly blunt p53-mediated tumor suppression by losing ARF expression (Fig. 1B) (28, 29). As ARF is induced by oncogenes such as Myc and stabilizes p53 by inhibiting Mdm2-mediated ubiquitination (30), ARF loss promotes p53 degradation and enables tumor cells to survive and expand in the presence of wild-type p53 despite high oncogenic signaling flux. Intriguingly, although p53ERTAM and ARF were expressed at low levels in p53-ON lymphomas, late-OFF lymphomas showed ARF up-regulation at the protein and mRNA level along with p53ERTAM accumulation (Fig. 3 A and B and SI Appendix, Fig. S1B). In fact, ARF up-regulation and p53ERTAM stabilization were evident in p53-ON lymphomas already 3 d after TAM withdrawal and increased further within the next days (Fig. 3 C and D). Consistent with an absence of ARF gene deletions or promoter methylation (SI Appendix, Figs. S3 and S4), this indicated that the defect in the Myc-ARF-Mdm2-p53 pathway of p53-ON Eµ-Myc lymphomas is reversible, which allows tumor cells to rapidly rewire the signaling network and reengage this pathway as soon as p53ERTAM is inactivated by TAM withdrawal. Mechanistically, wild-type p53 is known to repress the p19Arf promoter by recruitment of Polycomb group (PcG) proteins as part of a negative regulatory feedback loop (31), that has also been described for human p14ARF (32). In line with disruption of negative feedback upon loss of p53, we observed significantly reduced PcG protein-mediated histone modifications at the ARF gene locus of late-OFF versus p53-ON Eµ-Myc lymphomas as an explanation for the observed increase in ARF mRNA levels and consequent stabilization of p53ERTAM (SI Appendix, Fig. S5).
Fig. 3.
Late-OFF lymphomas reactivate p19ARF. (A) Immunostaining of p53-ON and late-OFF lymphomas for ARF and p53 (CM5). Images are representative of more than 10 animals of each group. (B) ARF mRNA expression normalized to β-actin shown as ΔCt (mean ± SD, 2-sided unpaired t test). (C and D) ARF and p53 protein (CM5) (C) and ARF mRNA levels (D) were analyzed in p53-ON lymphomas before (n = 4) and at indicated time points after tamoxifen withdrawal (n = 2). (E and F) p53 late-OFF lymphomas from mice that had relapsed after therapy were analyzed for ARF and p53 (CM5) expression by (E) Western blot and (F) immunohistochemistry. Late-OFF lymphomas from vehicle-treated mice are shown for comparison. Images are representative of multiple samples from 3 mice of each treatment group.
p19Arf Loss Causes Resistance to p53 Reactivation.
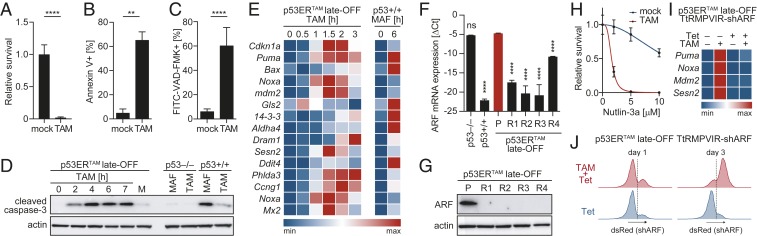
These observations suggested that the reengagement of the Myc-ARF-Mdm2-p53 pathway in late-OFF Eµ-Myc lymphomas is responsible for the therapeutic effect of acute p53ERTAM reactivation. In support of this, late-OFF lymphomas that had relapsed after TAM treatment displayed loss of ARF expression along with reduced p53ERTAM levels (Fig. 3 E and F). To formally test the role of ARF for the reactivation response, a late-OFF lymphoma was explanted in culture where the cells responded to TAM treatment with loss of viability (Fig. 4A), induction of apoptosis (Fig. 4 B–D), and transactivation of bona fide p53 target genes similar to p53+/+ lymphoma cells under chemotherapy (Fig. 4E). The few late-OFF lymphoma cells that survived TAM treatment in vitro could be expanded and yielded reactivation/TAM-resistant late-OFF lymphoma cell populations. Similar to late-OFF lymphomas that have relapsed in vivo (Fig. 3 E and F), TAM-resistant lymphoma cell cultures showed significantly reduced ARF expression at the mRNA and protein levels (Fig. 4 F and G). Genetic analysis revealed a deletion in the ARF gene locus comprising exons 2 and 3 that was not detectable before TAM treatment (SI Appendix, Fig. S3B). As all TAM-resistant cultures contained the same deletion, we concluded that an ARF-deleted subclone was present in the original lymphoma and selected under TAM treatment. This illustrates that a genetic, nonreversible loss of ARF results in resistance to p53ERTAM reactivation. Moreover, when we “repaired” the ARF-Mdm2-p53 pathway in TAM-resistant cells using the Mdm2 inhibitor Nutlin-3a to mimic the Mdm2-inhibitory function of ARF, sensitivity to TAM was restored (Fig. 4H), formally proving that ARF loss was responsible for resistance to TAM-induced p53ERTAM reactivation.
Fig. 4.
p19ARF loss causes resistance to p53 reactivation in vitro. (A) Survival of late-OFF lymphoma cells treated for 24 h with 4-OHT (TAM) relative to mock (n = 4). (B and C) Percentage of (B) Annexin V-positive (n = 3) or (C) active caspase-positive cells treated for 5 h with 4-OHT (TAM). (D) Western blot for cleaved caspase-3 (Asp175) of Eµ-Myc lymphoma cells with indicated p53 status treated with 4-OHT (TAM) or 3 µg/mL mafosfamide (MAF). (E) Expression of p53 target genes measured by RTqPCR in late-OFF lymphoma cells treated with 4-OHT (TAM) compared to Eµ-Myc;p53+/+ lymphoma cells treated with 3 µg/mL MAF. Depicted is the row-wise min-max scaled mean mRNA expression (n = 3). (F) ARF mRNA expression normalized to β-actin shown as ΔCt for late-OFF lymphoma cell lines: parental (P) and TAM-resistant (R1–R4). Eµ-Myc;p53−/− and Eµ-Myc;p53+/+ lymphoma cells are shown for comparison. One-way ANOVA with Tukey’s multiple comparisons test. (G) Western blot of parental and TAM-resistant late-OFF lymphoma cells. (H) Nutlin-3a overcomes TAM resistance of late-OFF lymphoma cells. Shown is survival following 24-h treatment with Nutlin-3a ± TAM relative to untreated. (I and J) Late-OFF lymphoma cells were transduced with a construct for Tet-inducible coexpression of ARF-shRNA and dsRed. (I) Expression of p53 target genes measured by RTqPCR following treatment with Tet and TAM as indicated. Depicted is the row-wise min-max scaled mean mRNA expression (n = 3). (J) Flow cytometry of dsRed expression following 1 to 3 d of treatment with Tet ± TAM. All graphs show mean ± SD and 2-sided, unpaired t tests unless indicated otherwise. **P < 0.01, ****P < 0.0001; ns, not significant.
To further validate the role of ARF for the reactivation response, we experimentally reduced ARF expression in late-OFF Eµ-Myc lymphoma cells by RNA interference. Tet-inducible ARF knockdown blunted the induction of p53 target genes by TAM (Fig. 4I) and conferred a survival advantage resulting in the rapid overgrowth of ARF-depleted (dsRed-positive) cells under TAM treatment (Fig. 4J). In contrast, late-OFF lymphoma cells expressing a control shRNA were completely killed by TAM. Together, this further attests to the critical role of a functional ARF-Mdm2-p53 signaling pathway for a successful therapeutic reactivation response of late-stage p53-inactivated (late OFF) Eµ-Myc lymphomas.
p14ARF-Dependent p53 Reactivation Response in Human p53ERTAM Tumor Cells.
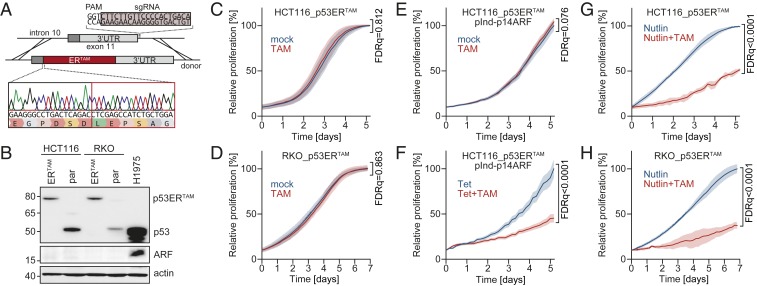
To explore p53 reactivation in human tumor cells with late-stage p53 inactivation, we chose colorectal cancer as a model where, according to the well-accepted multistep progression model, p53 inactivation is most frequently a late event (23). HCT116 and RKO cells are colorectal adenocarcinoma cells that have retained wild-type TP53. Similar to most other TP53 wild-type cell lines, both lack p14ARF expression. In HCT116 cells, one ARF allele is mutated, the other is epigenetically silenced by promoter methylation (33); in RKO cells both alleles are methylated (34). To reversibly inactivate the endogenous TP53 gene in these cell lines, we inserted the tamoxifen-responsive ERTAM cDNA into TP53 exon 11 using CRISPR/Cas9-induced homology-directed repair (Fig. 5A). Successful targeting was confirmed for single-cell clones by sequencing genomic DNA (Fig. 5A) and detection of the p53ERTAM fusion protein instead of the wild-type p53 in Western blots (Fig. 5B). As single cells were clonally expanded in the absence of TAM, the HCT116_p53ERTAM and RKO_p53ERTAM cell lines had undergone at least 20 population doublings in the p53-OFF state. Nevertheless, different from late-OFF lymphomas, HCT116/RKO_p53ERTAM cells showed comparably low p53 protein levels as the parental cell lines and the epigenetically silenced ARF alleles were not reexpressed (Fig. 5B). Of note, mimicking the Mdm2-inhibitory function of ARF with Nutlin-3a strongly stabilized p53ERTAM in both cell lines (SI Appendix, Fig. S6A), supporting that the low p53ERTAM expression level is caused by lack of ARF-mediated Mdm2 inhibition. Consistent with the critical role of p53ERTAM accumulation for the reactivation response of late-OFF lymphomas, TAM failed to inhibit proliferation of HCT116/RKO_p53ERTAM (Fig. 5 C and D). Confirming the lack of ARF as a cause, Tet-inducible reexpression of ARF or treatment with different Mdm2 inhibitors (Fig. 5 E–H and SI Appendix, Fig. S6 B–F) sensitized HCT116/RKO_p53ERTAM cells to TAM. Together these results from human colorectal cancer cells confirm the data obtained on murine Eµ-Myc lymphomas and reinforce the conclusion that a therapeutic reactivation response of tumors with a late-acquired loss of p53 function will depend on ARF status.
Fig. 5.
p14ARF-dependent p53 reactivation response in human p53ERTAM tumor cells. (A) Scheme for CRISPR/Cas9-mediated insertion of ERTAM domain into the TP53 gene of human p53 wild-type tumor cells. (Bottom) Validation of correct insertion by Sanger sequencing. (B) Western blot of parental and p53ERTAM-edited HCT116 and RKO cells for ARF and p53 (DO1). H1975 cells with endogenous p53R273H mutation are shown for comparison. (C and D) Proliferation of p53ERTAM-edited cells in response to TAM. (E and F) Proliferation of HCT116_p53ERTAM cells with Tet-inducible expression of ARF in response to TAM in the absence (E) and presence (F) of Tet. (G and H) Proliferation of p53ERTAM-edited cells treated with Nutlin-3a ± TAM. (C–H) In all graphs, day 0 is the start of TAM treatment. All proliferation curves were normalized to the confluence of non–TAM-treated reference cells at the end of the time course and shown as mean ± SD. Statistical significance was assessed with multiple 2-sided t tests in combination with false discovery rates (FDRs). Reported are FDR q-values for relative proliferation at the end of the observation period.
Discussion
In light of the regained interest in therapeutic p53 reactivation, our findings provide proof of principle that tumor cells which have evolved in the presence of nonmutated active p53 can rapidly adapt and become addicted to p53 inactivation. Reinstatement of functional p53 could therefore be an effective approach even if inactivation of p53 is not the initiating driver lesion and has occurred late during tumorigenesis. This was not expected as it is generally believed that p53 wild-type tumors are enriched for p53 pathway alterations that bypass p53-mediated tumor suppression and would similarly blunt a p53 reactivation response. In principle, such aberrations can hit p53 downstream effectors. However, the tumor suppressor response of p53 is highly pleiotropic (35). No single or compound mouse knockout of specific p53 target genes has recapitulated the dramatic tumor predisposition that characterizes p53-null mice (36). Even p53 mutants deprived of most of their transactivation function can retain remarkably potent tumor suppressor activity (36–38). It is therefore unlikely that mutations affecting single arms of the p53 response undermine a p53 reactivation response. Alterations in upstream effectors such as ARF, on the other hand, could be more critical and it has been shown previously for tumors, which have developed in a p53-compromised background, that successful p53 reactivation is dependent on oncogene-driven up-regulation of p19Arf and that p53-null tumors with ARF deletions or insufficient oncogenic signaling are resistant to p53 restoration (17, 18). In line with this, our study also demonstrates that a successful reactivation response depends on ARF expression. Based on the results in the p53ERTAM model, it can be speculated that a p53 loss-of-function mutant might not be sufficiently stabilized if ARF is deleted, mutated, or irreversibly silenced and that Mdm2 inhibitors could serve as ARF mimics to boost the effect of mutant p53 reactivating drugs in this scenario. Consistent with this concept, it was shown that Mdm2 inhibitors synergistically enhance the activity of small-molecule stabilizers of conformationally unstable p53 mutants and readthrough-promoting drugs for p53 nonsense mutants (39, 40). For p53 reactivation to be therapeutically effective, it therefore does not seem to be critical whether p53 was inactivated at the onset of malignant transformation or at later stages, as long as strong oncogenic signals are present and transmitted via ARF to stabilize p53. ARF expression along with stabilization of mutant p53 should therefore be considered as potential biomarkers for tumors susceptible to p53 reactivation approaches.
Materials and Methods
p53ERTAM late-OFF lymphoma cells, mimicking late-stage p53 inactivation, were generated from Eµ-Myc;p53ERTAM/TAM [B6.Cg-Tg(IghMyc)22Bri/JThst; B6;129Sv-Trp53tm1Gev/JThst] fetal liver cells. For p53 reactivation experiments, cohorts of B6-albino mice were transplanted with late-OFF lymphoma cells, treated for 7 d with daily i.p. injections of tamoxifen (100 µL of 10 mg/mL solution in corn oil) or vehicle and monitored by BLI. A detailed description of the materials and methods used in this study is provided in SI Appendix. All animal experiments were performed according to the German Animal Welfare Act and approved by the Regional Board of Giessen.
Supplementary Material
Acknowledgments
This work was supported by research grants from the Deutsche Forschungsgemeinschaft (TI 1028/2-1 to O.T., TRR81 TPA10 to T.S.), Deutsche José Carreras Leukämie Stiftung (09 R/2018 to O.T. and T.S.), German Center for Lung Research (DZL) to T.S., Deutsche Krebshilfe (70112623 to T.S.), and von Behring-Röntgen Stiftung (65-0004, 66-LV06 to T.S.). We thank Gerard Evan for kindly providing the p53ERTAM mouse strain and the members of the T.S. laboratory for helpful discussion and advice. We acknowledge Sigrid Bischofsberger, Antje Grzeschiczek, Björn Geißert, and Angela Mühling for technical assistance and the Irradiation Core Facility for providing access to the X-RAD 320iX platform.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910255116/-/DCSupplemental.
References
- 1.Harris H., Miller O. J., Klein G., Worst P., Tachibana T., Suppression of malignancy by cell fusion. Nature 223, 363–368 (1969). [DOI] [PubMed] [Google Scholar]
- 2.Kato S., et al. , Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. U.S.A. 100, 8424–8429 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotler E., et al. , A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol. Cell 71, 178–190.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Muller P. A., Vousden K. H., Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joerger A. C., Fersht A. R., The p53 pathway: Origins, inactivation in cancer, and emerging therapeutic approaches. Annu. Rev. Biochem. 85, 375–404 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Bykov V. J. N., Eriksson S. E., Bianchi J., Wiman K. G., Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 18, 89–102 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Yu X., Vazquez A., Levine A. J., Carpizo D. R., Allele-specific p53 mutant reactivation. Cancer Cell 21, 614–625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joerger A. C., et al. , Exploiting transient protein states for the design of small-molecule stabilizers of mutant p53. Structure 23, 2246–2255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Floquet C., Deforges J., Rousset J. P., Bidou L., Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 39, 3350–3362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng X., et al. , APR-246/PRIMA-1MET inhibits thioredoxin reductase 1 and converts the enzyme to a dedicated NADPH oxidase. Cell Death Dis. 8, e2751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D. S., et al. , Inhibiting the system xC-/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat. Commun. 8, 14844 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christophorou M. A., et al. , Temporal dissection of p53 function in vitro and in vivo. Nat. Genet. 37, 718–726 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Martins C. P., Brown-Swigart L., Evan G. I., Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127, 1323–1334 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Sharpless N. E., DePinho R. A., Cancer biology: Gone but not forgotten. Nature 445, 606–607 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Xue W., et al. , Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ventura A., et al. , Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Junttila M. R., et al. , Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 468, 567–571 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldser D. M., et al. , Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malkin D., et al. , Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250, 1233–1238 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Kuhn E., et al. , TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma–Evidence supporting the clonal relationship of the two lesions. J. Pathol. 226, 421–426 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlinger M., et al. , Cancer: Evolution within a lifetime. Annu. Rev. Genet. 48, 215–236 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Rivlin N., Brosh R., Oren M., Rotter V., Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer 2, 466–474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fearon E. R., Vogelstein B., A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Hong M. K., et al. , Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat. Commun. 6, 6605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amin N. A., et al. , A quantitative analysis of subclonal and clonal gene mutations before and after therapy in chronic lymphocytic leukemia. Clin. Cancer Res. 22, 4525–4535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi D., et al. , Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood 123, 2139–2147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mina M., et al. , Conditional selection of genomic alterations dictates cancer evolution and oncogenic dependencies. Cancer Cell 32, 155–168.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Schmitt C. A., McCurrach M. E., de Stanchina E., Wallace-Brodeur R. R., Lowe S. W., INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 13, 2670–2677 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eischen C. M., Weber J. D., Roussel M. F., Sherr C. J., Cleveland J. L., Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13, 2658–2669 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zindy F., et al. , Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12, 2424–2433 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng Y., Kotake Y., Pei X. H., Smith M. D., Xiong Y., p53 binds to and is required for the repression of Arf tumor suppressor by HDAC and polycomb. Cancer Res. 71, 2781–2792 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stott F. J., et al. , The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17, 5001–5014 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burri N., et al. , Methylation silencing and mutations of the p14ARF and p16INK4a genes in colon cancer. Lab. Invest. 81, 217–229 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Esteller M., et al. , Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 60, 129–133 (2000). [PubMed] [Google Scholar]
- 35.Kastenhuber E. R., Lowe S. W., Putting p53 in context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bieging K. T., Attardi L. D., Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 22, 97–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brady C. A., et al. , Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T., et al. , Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M., et al. , Synergistic rescue of nonsense mutant tumor suppressor p53 by combination treatment with aminoglycosides and Mdm2 inhibitors. Front. Oncol. 7, 323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., et al. , Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 41, 6034–6044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.