Significance
The protooncogene KRAS, a GTPase, is responsible for activating the MAPK pathway. Cancer mutations at codons 12, 13, or 61 are thought to stabilize KRAS-GTP primarily by preventing GAP protein-stimulated GTP hydrolysis, thereby stabilizing the active form of KRAS. We report that the tumor suppressor NF1 is comutated in KRAS G13-mutated cells. NF1 GAP protein is inactive against KRAS G12 and Q61-mutated KRAS but stimulates GTP hydrolysis when bound to KRAS G13D. KRAS G13D mutant cells also respond to EGFR inhibitors in a neurofibromin-dependent manner. Crystallographic analysis of wild-type and G13D KRAS complexed with neurofibromin provides the structural basis for neurofibromin-mediated GTP hydrolysis. This suggests that neurofibromin-competent KRAS G13-mutated cancers may respond to EGFR therapies.
Keywords: KRAS, G13D, NF1, EGFR, GTPase
Abstract
KRAS mutations occur in ∼35% of colorectal cancers and promote tumor growth by constitutively activating the mitogen-activated protein kinase (MAPK) pathway. KRAS mutations at codons 12, 13, or 61 are thought to prevent GAP protein-stimulated GTP hydrolysis and render KRAS-mutated colorectal cancers unresponsive to epidermal growth factor receptor (EGFR) inhibitors. We report here that KRAS G13-mutated cancer cells are frequently comutated with NF1 GAP but NF1 is rarely mutated in cancers with KRAS codon 12 or 61 mutations. Neurofibromin protein (encoded by the NF1 gene) hydrolyzes GTP directly in complex with KRAS G13D, and KRAS G13D-mutated cells can respond to EGFR inhibitors in a neurofibromin-dependent manner. Structures of the wild type and G13D mutant of KRAS in complex with neurofibromin (RasGAP domain) provide the structural basis for neurofibromin-mediated GTP hydrolysis. These results reveal that KRAS G13D is responsive to neurofibromin-stimulated hydrolysis and suggest that a subset of KRAS G13-mutated colorectal cancers that are neurofibromin-competent may respond to EGFR therapies.
The RAS family of protooncogenes cycle between active GTP-bound and inactive GDP-bound states in response to mitogenic stimuli. In the GTP-bound state, RAS proteins bind to and activate the RAF/MAPK (mitogen-activated protein kinase) and PI3K pathways to promote cell-cycle progression. The rate of intrinsic GTPase reaction of RAS is slow (1). Thus, the active GTP-bound state of RAS proteins is primarily regulated by RasGAPs (Ras GTPase-activating proteins), which increase the rate of GTP hydrolysis by ∼105-fold (2). Two well-characterized RasGAPs are neurofibromin (the protein is referred to here as NF1, encoded by the NF1 gene) and RASA1 (also called p120GAP) (3). RasGAPs achieve this by providing an arginine residue (also known as an arginine finger) in the nucleotide-binding pocket of RAS, where it stabilizes and orients the catalytic residue, Q61, for an inline nucleophilic attack on the gamma-phosphate of GTP (4–6).
Biophysical analysis of the GAP-mediated GTPase reaction in the RAS–RasGAP complex has suggested 3 key steps for the reaction mechanism (7). In the initial step, RasGAP interacts with active GTP-bound RAS and forms a ground-state complex. During this step, the arginine finger remains exposed to the aqueous environment. The ground state is followed by the transition state, where the arginine finger positions itself in the active site, triggering the cleavage of GTP and formation of protein-bound Pi intermediates. The last and rate-limiting step of the GAP-mediated GTPase reaction involves the release of Pi from the active site. So far, the structural information on the RAS–RasGAP complex is limited to the transition-state structure of the HRAS complexed with the GAP-related domain (GRD) of RASA1 (HRAS–RASA1GRD) in the presence of GDP and AlF3, where AlF3 mimics the cleaved gamma-phosphate during the cleavage reaction (4).
GAP-mediated GTP hydrolysis is frequently disrupted in human cancers by activating point mutations of RAS genes. KRAS is the most frequently mutated of the 3 RAS isoforms. Mutations are commonly observed near the nucleotide-binding pocket at glycine 12, glycine 13, or glutamine 61 (8). Codon 12 mutations predominate across lung, pancreas, and colon, while codon 13 mutations largely appear in colorectal cancers (CRCs). All mutations in this region are thought to prevent formation of the RasGAP transition state by blocking the arginine finger from accessing the GTP terminal phosphate, thereby preventing RasGAP-mediated GTP hydrolysis. NF1 mutations are also frequent in malignant peripheral nerve sheath tumors (9) and in a fraction of lung and colorectal tumors but are thought to be functionally redundant and mutually exclusive with activating KRAS mutations.
It is unknown why the KRAS G13D mutation appears almost exclusively in gastrointestinal cancers and is rare in lung and pancreas. This may be due in part to allelic differences in signal transduction (10–12). It has also been proposed that KRAS G13D colorectal cancers may respond to inhibition of upstream signaling (13, 14). Epidermal growth factor receptor (EGFR) inhibitors are approved for treatment of KRAS wild-type (WT) colorectal cancers, but KRAS mutations are contraindicated. Retrospective analyses of the EGFR inhibitor cetuximab showed a modest response in KRAS G13D-mutated colorectal cancers while KRAS G12-mutated CRCs were resistant. However, these results were not observed for panitumumab (15), and a subsequent clinical trial showed no response in KRAS G13D-mutated CRC patients treated with cetuximab as a single agent or in combination with irinotecan (16). Clearly, the presence of the KRAS G13D allele alone is not sufficient to identify KRAS-mutated tumors that may respond to anti-EGFR therapies. However, CRC tumor genomes are frequently unstable and diverse (17). The complex underlying genetics and biology of KRAS-mutated CRCs raise the possibility that a subset of KRAS G13D-mutated colorectal cancers may be EGFR-dependent, and identifying a functional biomarker may help to further stratify patients.
Here we report increased comutation of the RasGAP NF1 with KRAS G13-mutated cancers and show that KRAS G13D-mutated cells can respond to EGFR inhibitors in an NF1-dependent manner. In addition, we show how NF1 influences KRAS GTP levels in KRAS G13 mutant cells and that NF1 hydrolyzes GTP bound to KRAS G13D-mutated oncoproteins in biochemical assays. We also show structures of the wild type and G13D mutant of KRAS in complex with the catalytic region (GRD) of NF1. Since these structures were obtained using KRAS bound to GMPPNP (a nonhydrolyzable analog of GTP), they represent a ground-state conformation of the RAS–NF1 complex. Structural comparison with the HRAS–RASA1GRD complex obtained in the transition state provides insights into conformational changes that occur when RAS–RasGAP complexes move from the ground state to the transition state. Structural analysis of the KRASG13D–NF1GRD complex provides a rationale for a significant level of GAP-mediated GTP hydrolysis in this mutant compared with G12 and Q61 mutants.
Results
Mutational Landscape of KRAS G13-Mutated Cancers.
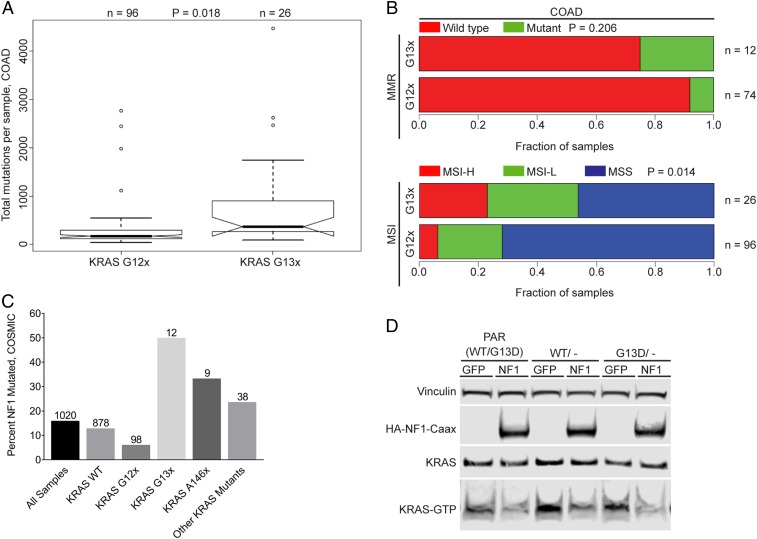
It has been reported that gastric tumor types are associated with high overall mutational burdens (17). To determine if KRAS G13D-mutated cancers appear in a distinct genetic background compared with other KRAS-mutated cancers, a systematic class comparison was performed between KRAS G12 mutant (G12x) samples and KRAS G13 mutant (G13x) samples using The Cancer Genome Atlas (TCGA) data (Fig. 1A). Only sample sets with more than 10 G13-mutated samples were included in the analysis, which included colon adenocarcinoma (COAD), stomach adenocarcinoma (STAD), and rectal adenocarcinoma (READ) samples. We found that G13x-harboring COAD samples had a significantly (t test, P = 0.018; Fig. 1A) higher number of mutations when compared with KRAS G12x samples. Increased mutational burden for the G13x STAD samples did not reach statistical significance (t test; P = 0.053; SI Appendix, Fig. S1A) and no difference was observed for READ (SI Appendix, Fig. S1B). High mutation rate in colorectal tumors is often associated with both microsatellite instability (MSI) and loss of mismatch repair (MMR) gene expression. Therefore, we also compared these clinical terms between the 2 mutant classes. For COAD, there was a significant difference (P = 0.014) between the G13x and G12x mutant classes for the clinical term associated with microsatellite instability (Fig. 1B). Loss of MMR and increased MSI was also observed for G13x STAD samples, but both terms failed to reach statistical significance (MSI: P = 0.095) and no difference was observed for G13x vs. G12x READ samples (SI Appendix, Fig. S1 C and D).
Fig. 1.
KRAS G13D CRC cells are sensitive to NF1-mediated GTP hydrolysis. (A) Total number of mutations per sample in KRAS G12- and G13-mutated COAD from TCGA cancer samples (P = 0.018). (B) Mutational burden comparing KRAS codon 12 and codon 13 mutations with mismatch repair mutations and microsatellite instability (MSI-H, MSI high; MSI-L, MSI low; MSS, MS stable) in TCGA COAD samples. (C) Comparison of NF1 mutations and KRAS mutation status in CCLE cell lines. Category sample sizes are indicated above each bar. (D) NF1 overexpression in HCT-116, a KRAS G13D/NF1-mutated cell line, leads to reduced KRAS-GTP levels.
These observations led us to look more closely at the mutational spectra present in the G12x and G13x sample classes. A screen to query the top 100 mutated genes (collapsing all per-gene mutations for each sample into a single instance) was performed in each sample class and then evaluated for differences in mutational frequencies between the classes. All of the identified statistically significant differentially mutated genes showed an enrichment in mutation in the G13-mutated sample class relative to the G12-mutated sample class (as this class shows significantly higher mutational frequencies) (SI Appendix, Fig. S1 E–G). This pattern was observed for COAD, STAD, and READ samples. Interestingly, there were several shared differentially mutated genes between the COAD and STAD tumor groups (PCLO, KMT2D, MBD6, CACNA1E, MDN1, ANK3, and ZDBF2). Importantly, among the genes showing mutational enrichment in the G13x KRAS mutant samples in COAD, there were several receptor tyrosine kinases—FGFR2, EGFR, INSR, and ALK. Lists of statistically significant differentially mutated genes identified for each tumor type are provided in SI Appendix.
Cell-line data were also used to perform a mutational enrichment analysis. Mutational frequencies of genes within the RAS pathway were compared among G12x and G13x KRAS mutant cell lines using the COSMIC cell line collection. Enrichment analysis revealed that ARHGAP35, CDC25A, CNKSR1, DAB2IP1, EIF4EBP1, IRS2, PAK3, PRKAA1, RASA3, SPRED2, and NF1 were enriched in KRAS G13x-mutated cell lines. NF1 was the most common comutation identified, and many of the cell lines with multiple comutations frequently carried NF1 mutations as well (Dataset S1).
Comutational Frequencies of NF1 with Different KRAS Mutant Alleles.
To confirm that observation, the frequency of NF1 mutations was determined across several KRAS mutant alleles. Silent NF1 mutations were removed for this analysis, although the results are still significant if synonymous point mutations are included. Indeed, the G13-mutated samples showed a higher comutation frequency of NF1 than the G12-mutated cell lines (0.5 vs. 0.06; Fig. 1C). Interestingly, the comutational frequency in the Q61 mutant class was zero, while the A146 mutant samples were elevated similar to the G13-mutated class (Fig. 1C). Together, these observations are consistent with a model whereby both the KRAS G12 and Q61 mutant alleles are able to operate independent of NF1 mutational status while both G13 and A146 mutations appear to benefit from an NF1 comutation. Finally, the cBioPortal was used to identify KRAS and NF1 comutation status in a larger sample size of patient tumor samples from 155 genome-sequencing studies across multiple cancer indications (18, 19). Consistent with the enriched comutations found in the COSMIC cancer cell lines, ∼12% of KRAS G13x-mutated samples also have NF1 mutations (n = 371), compared with 5.5% of NF1 comutations in KRAS G12x-mutated samples (n = 2,695).
KRAS G13D Is Sensitive to NF1-Stimulated GTP Hydrolysis.
To test if NF1 expression functionally alters KRAS activity in a KRAS G13D CRC cell line, HCT-116 cells were transfected with the catalytic domain of NF1, which binds to the KRAS WT and catalyzes GTP hydrolysis. Cellular levels of KRAS GTP were reduced after NF1 expression, indicating that NF1 is functionally active and promotes GTP hydrolysis in a KRAS G13D-mutated CRC cell line, and KRAS G13D protein may be sensitive to NF1-stimulated GTP hydrolysis (Fig. 1D).
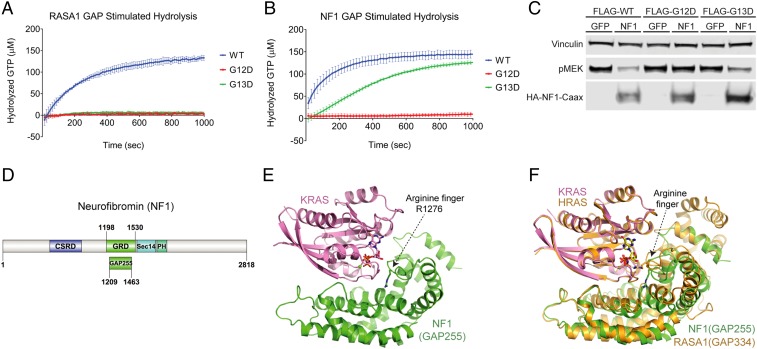
The ability of NF1 to catalyze GTP hydrolysis directly in complex with KRAS G13D was then tested in vitro using purified KRAS and NF1 proteins. Consistent with previously published data (11, 20), RASA1 GAP is active only against wild-type KRAS, and KRAS proteins with activating mutations at codons 12, 13, and 61 are unaffected by RASA1 GAP. RASA1 GAP accelerated GTP hydrolysis of wild-type KRAS GTP to ∼100-fold above the reported rate of intrinsic GTP hydrolysis for wild-type KRAS, whereas RASA1 GAP did not significantly alter the rate of KRAS G12D or G13D (Fig. 2A and Table 1) (11, 21). Similar to RASA1, NF1 stimulated GTP hydrolysis for wild-type KRAS and did not affect GTP hydrolysis for KRAS G12D (Fig. 2B). However, unlike RASA1, NF1 stimulated GTP hydrolysis of KRAS G13D to a rate much closer to that of NF1 with wild-type KRAS (Fig. 2B). The ability of NF1 to alter KRAS signal transduction was also confirmed in cells. KRAS WT, G13D, or G12D constructs were cotransfected with NF1 or GFP control plasmids. MEK phosphorylation levels were measured as an indicator of MAPK pathway activity because MEK is the first kinase activated in the MAPK pathway, and may be less prone to attenuation through DUSP-mediated negative feedback mechanisms that target pERK (22, 23). All KRAS constructs are capable of activating the MAPK pathway leading to elevated pMEK. However, pMEK levels were markedly reduced for KRAS wild type and KRAS G13D when cotransfected with NF1 but unaltered in cells cotransfected with KRAS G12D and NF1, suggesting that NF1 inhibits KRAS signal transduction in both KRAS wild type- and KRAS G13D-expressing cells but NF1 has no effect on cells expressing KRAS G12D (Fig. 2C). Together, these results suggest that NF1 directly regulates KRAS GTP levels in KRAS G13D-mutated cells due to a unique structural conformation for KRAS G13D that distinguishes it from other KRAS oncogenic mutations at codons 12 or 61.
Fig. 2.
RASA1 and NF1 GAP-mediated GTP hydrolysis with multiple KRAS oncoproteins. Intrinsic and GAP-stimulated KRAS GTPase activity was measured using EnzChek. (A) RASA1-stimulated hydrolysis of KRAS WT/G12D/G13D (represents 1 EnzChek assay with 3 replicates of each KRAS mutant all on the same plate, error bars represent standard deviation). (B) NF1-stimulated hydrolysis of KRAS WT/G12D/G13D (represents 1 EnzChek assay with 6 replicates of each KRAS mutant all on the same plate, error bars represent standard deviation). (C) Decreased pMEK levels in WT and G13D when cotransfected with FLAG-KRAS WT or G13D and NF1 in NCI-H1355 cells. (D) Domain organization of human NF1. (E) The overall structure of GMPPNP-bound wild-type KRAS in complex with NF1GRD. KRAS and NF1GRD are shown in cartoon representation and colored pink and green, respectively. GMPPNP and the arginine finger (NF11276) are shown in stick representation where oxygen, nitrogen, and phosphorus atoms are colored red, blue, and orange, respectively. (F) Structural comparison of the ground-state KRAS–NF1GRD complex with the transition-state HRAS–RASA1GRD complex. Here KRAS and NF1GRD are color-coded as in E whereas HRAS and RASA1GRD shown in cartoon representation are colored orange. Secondary structural elements around the arginine finger show rotational displacement of GAP-related domains during the conversion from the ground state to the transition state.
Table 1.
Biochemical comparison of KRAS WT and oncogenic mutants
| KRAS | RASA1-stimulated rate of hydrolysis kobs 10−5, s−1 | NF1-stimulated rate of hydrolysis kobs 10−5, s−1 | NF1 binding affinity KD, µM |
| WT | 2599.00 ± 398.46 | 5027.80 ± 2241.63 | 1.2 ± 0.2 |
| G12D | ND | 194.70 ± 85.42 | 8.1 ± 0.4 |
| G13D | ND | 3457.00 ± 1789.87 | 6.4 ± 0.7 |
| G13C | ND | 3098.60 ± 1024.47 | 11.0 ± 0.0 |
| Q61R | ND | ND | 18.4 ± 4.4 |
The hydrolysis rates listed are derived from averaging the hydrolysis rates of each KRAS mutant from multiple EnzChek hydrolysis runs. ND, not determined (hydrolysis activity too low to measure).
Crystal Structure of Wild-Type KRAS in Complex with NF1GRD.
To better understand the underlying mechanism of hydrolysis observed for KRAS G13D proteins, we sought to determine the structures of the wild type and G13D mutant of KRAS in complex with NF1. NF1 is a large protein comprising 2,818 residues and the GAP-related domain is located between residues 1198 and 1530 (Fig. 2D). Although the apo structure of NF1GRD(GAP333) (residues 1198 to 1530) was solved more than 2 decades ago (5), no structural information is available on how NF1 interacts with RAS and carries out NF1-mediated GTP hydrolysis. Despite extensive efforts, we could not obtain crystals of KRAS in complex with NF1GRD(GAP333). Previous studies have shown that the central part of NF1GRD is sufficient to carry out GAP-mediated GTPase activity (24). Therefore, a smaller NF1GRD construct was made (GAP255; residues 1209 to 1463) containing the minimum catalytic region by excluding C- and N-terminal regions that have been shown to form the extra domain (Fig. 2D). Using an isothermal titration calorimetry (ITC) experiment, NF1-GAP255 showed similar binding affinity to KRAS when compared with NF1-GAP333 (SI Appendix, Fig. S2 A and B). Crystallization of NF1GRD(GAP255) in complex with GMPPNP-bound wild-type KRAS (hereafter referred to as the KRAS–NF1GRD complex) was successful and we solved the structure of this protein–protein complex at 2.85 Å (SI Appendix, Table S1). The domain organization showing the location of GAP255 in the NF1 protein and the overall structure of KRAS–NF1GRD are shown in Fig. 2 D and E, respectively. Despite having a relatively low amino acid sequence identity of 30% between the GRD of RASA1 and NF1, they form a similar tertiary structure (with an rmsd of 1.8 Å for Cα atoms) and interact with KRAS in a similar way (Fig. 2F). In the KRAS–NF1GRD structure, NF1GRD forms a crescent-shaped structure and KRAS binds in the central groove via residues of the switch regions.
Measurement of binding affinity using ITC agrees with previous observations that NF1GRD binds to RAS with high affinity (SI Appendix, Fig. S2A) (20). Previous studies have shown that RASA1 binds to RAS with a 50-fold lower affinity than NF1 (20, 25, 26). Structural analysis of the protein–protein interface in previously solved HRAS–RASA1GRD and KRAS–NF1GRD complexes described here shows similar buried surface areas and hydrogen-bonding patterns between these 2 complexes (SI Appendix, Fig. S3). At the RAS–RasGAP interface, nearly half the residues that form the RasGAP interface are different between NF1 and RASA1 (SI Appendix, Fig. S4). However, the structural comparison showed the presence of 4 strong salt-bridge interactions in the KRAS–NF1GRD complex spanning the protein–protein interface whereas the HRAS–RASA1GRD complex has only 2 such interactions at its interface (SI Appendix, Fig. S3 C and D). It is likely that the additional salt bridges present at the KRAS–NF1GRD interface provide a higher-affinity interaction of KRAS with NF1 compared with RASA1 GAP.
NF1-Mediated GTP Hydrolysis in KRAS and Conformational Changes in RAS–RasGAP Complexes.
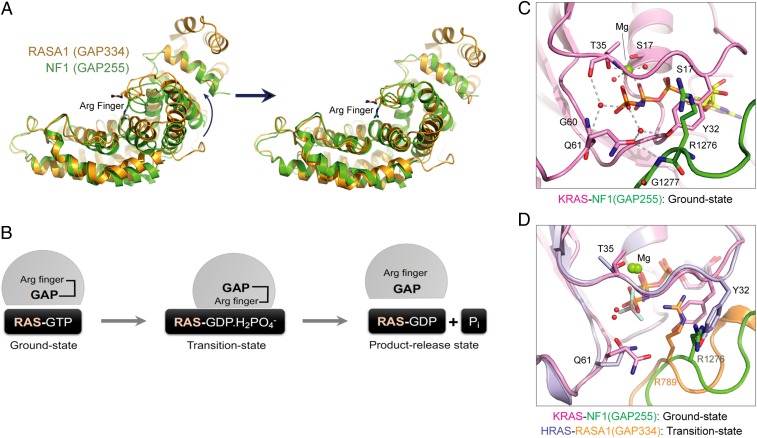
The structure of GMPPNP-bound wild-type KRAS complexed with NF1GRD shows the ground-state conformation with the switch I region in the active conformation. Despite our extensive efforts, we could not obtain well-diffracting crystals of KRAS–NF1GRD in the transition state (using GDP and AlF3). Thus, we compared our ground-state structure with the previously reported transition-state structure of the HRAS–RASA1GRD complex to gain insights into NF1-mediated GTP hydrolysis and conformational changes that occur when the RAS–RasGAP complex shifts from the ground state to the transition state (4). The structural superposition of HRAS–RASA1GRD and KRAS–NF1GRD using RAS proteins shows an ∼8° rotation and 1.8-Å translation of the GAP domain toward RAS when it moves from the ground state to the transition state (Fig. 3A). This rotation brings the GAP protein close to RAS and places the arginine finger in the active site, where it stabilizes and orients KRASQ61 for an inline nucleophilic attack on the gamma-phosphate of GTP. Calculation of the interaction interface shows an increase of 500 Å2 for both KRAS and NF1GRD as they shift from the ground state to the transition state. Similar rotation, although to a large extent (∼20°), has been seen in the case of RHO proteins, where the structures of RHO–RhoGAP complexes are available in both ground and transition states (27).
Fig. 3.
Structural analysis of the active-site pocket and protein–protein interaction interface in the KRAS–NF1GRD and HRAS–RASA1GRD complexes. (A) Conformational changes in NF1GRD during the conversion from the ground state to the transition state. (B) Schematic of the GAP-mediated GTP hydrolysis reaction in RAS. (C) Active-site pocket in the KRAS–NF1GRD complex. (D) Structural superposition of the active-site pocket in the KRAS–NF1GRD and HRAS–RASA1GRD complexes.
In the ground-state structure of KRAS–NF1GRD, the side-chain atoms of KRASY32 occupy the position of the arginine finger (NF1R1276) as seen in the transition-state structure of the HRAS–RASA1GRD complex (Fig. 3 B–D). Side-chain atoms of KRASY32 and KRASQ61 interact with each other via a bridging water molecule. Unlike the transition-state structure, where HRASQ61 forms a hydrogen bond with the main-chain carbonyl oxygen of the arginine finger, in the ground-state structure KRASQ61 forms a hydrogen bond with the main-chain amide nitrogen of NF1G1277, a residue following the arginine finger (Fig. 3D). Structural superposition of ground- and transition-state structures suggests that to facilitate NF1-mediated GTPase activity in wild-type KRAS, a rotation of NF1 toward KRAS would allow placement of the arginine finger in the nucleotide-binding pocket by swapping its position with KRASY32. This rotation would now bring NF1R1276 (instead of NF1G1277) present in the finger loop close to KRASQ61, allowing a new hydrogen bond to form between the amide nitrogen of the KRASQ61 side chain and main-chain carbonyl oxygen of the arginine finger. These conformational changes in the active site would stabilize and orient KRASQ61 and NF1R1276 in the active site for the GTP hydrolysis reaction via formation of the transition state, as observed in the structure of the HRAS–RASA1GRD complex.
Structure of KRASG13D Complexed with NF1GRD Provides a Rationale for NF1-Mediated GTPase Activity.
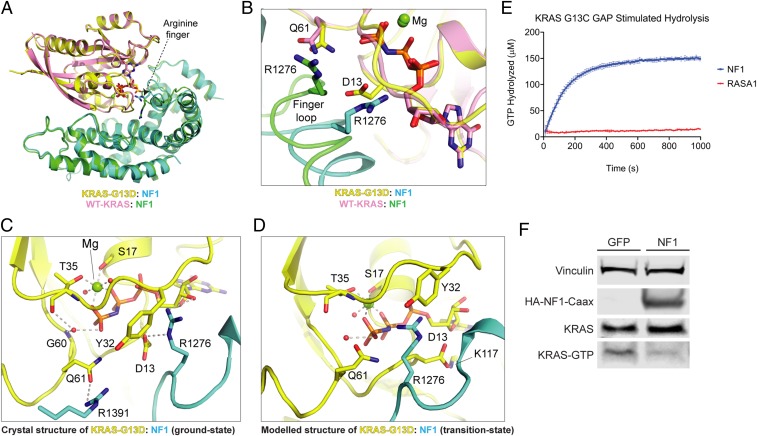
As predicted earlier, unlike wild-type KRAS, the oncogenic mutants G12D, G13D, and Q61R of KRAS show relatively weak affinity for NF1GRD. Among these 3 mutants, G13D and G12D mutants of KRAS bind to NF1GRD with a relatively higher affinity than Q61R mutants (Table 1 and SI Appendix, Fig. S2 C–F). As mentioned above, the rate of NF1-mediated GTP hydrolysis for KRASG13D is surprisingly similar to wild-type KRAS (Table 1). To understand the structural basis of NF1-mediated GTPase activity in the G13D mutant of KRAS, we solved the structure of GMPPNP-bound KRASG13D complexed with NF1GRD(GAP255) at 2.1 Å (SI Appendix, Table S1). The overall structure and the interaction interface of the KRASG13D–NF1GRD complex are similar to those of the wild-type KRAS–NF1GRD structure (Fig. 4A). In the KRASG13D–NF1GRD structure, the presence of side-chain atoms of KRASD13 displaces KRASY32 away from the nucleotide-binding pocket (SI Appendix, Fig. S5) and this results in the displacement of the finger loop containing the arginine finger and surrounding residues in NF1GRD at the KRAS–NF1 interface. This vertical displacement, which is close to 4 Å for the Cα atom of NF11276, allows partial entry of the arginine finger into the nucleotide-binding pocket, where it interacts with KRASD13 and occupies the position of KRASY32 observed in the wild-type KRAS–NF1GRD structure (Fig. 4B). Unlike the KRAS–NF1GRD structure, one of the side-chain carbonyl oxygen atoms of KRASD13 of the KRASG13D–NF1GRD structure interacts with the main-chain amide hydrogen of NF1G1277, and the side chain of KRASQ61 forms a hydrogen bond with the side chain of NF1R1391 (Fig. 4C). The local conformational changes observed in the finger loop of the KRASG13D–NF1GRD structure suggest that it has inherent flexibility that could allow it to place the arginine finger in the active-site pocket, even in the case of minor steric hindrance due to the G13 mutation.
Fig. 4.
RASA1 and NF1 GAP-mediated GTP hydrolysis with multiple KRAS oncoproteins. (A) Structural superposition of NF1(GAP255) complexed with WT KRAS and KRASG13D. (B) Enlarged view of the active-site pocket in the superposed structures of NF1(GAP255) complexed with WT KRAS and KRASG13D (ground state). (C) The active-site pocket in the crystal structure of the KRASG13D–NF1GRD complex in the ground state. (D) The active-site pocket in the modeled structure of the KRASG13D–NF1GRD complex in the transition state. (E) GTP hydrolysis of KRAS G13C mediated by RasGAP proteins (error bars represent standard deviation of 3 replicates). (F) KRAS-GTP levels after NF1 expression in NCI-H1355, a KRAS G13C-mutated CRC cell line.
Structural comparison of the KRASG13D–NF1GRD complex with the KRAS–NF1GRD and HRAS–RASA1GRD complexes suggests that to enable NF1-mediated GTPase activity in KRASG13D, besides the above-mentioned rotation and translation of NF1GRD toward KRASG13D, it would require that KRASD13 adopts another rotamer conformation which points away from the gamma-phosphate so that the arginine finger can occupy its place and interact with phosphate moieties of GTP. Examination of other possible side-chain rotamer conformations of KRASD13 in the KRASG13D–NF1GRD complex structure reveals a plausible rotamer conformation of the KRASD13 side chain pointing toward KRASK117 (away from the gamma-phosphate) and forming a salt-bridge interaction with it. Thus, it is likely that the conversion of the ground state to the transition state would be accompanied by a change to this rotamer conformation of the KRASD13 side chain in the KRASG13D–NF1GRD complex. This would allow the arginine finger to enter the active-site pocket and orient KRASQ61 as in the wild-type KRAS complex for inline nucleophilic attack on the gamma-phosphate of GTP. These conformational changes incorporated in the modeled structure of the KRASG13D–NF1GRD complex in the transition state provide a structural basis for the observed NF1-mediated GTP hydrolysis in this mutant (Fig. 4D). This model also predicts that other oncogenic KRAS mutations at codon 13 may also be sensitive to NF1-stimulated GTP hydrolysis.
KRAS G13C Is Also Sensitive to NF1-Stimulated GTP Hydrolysis.
To determine if NF1-mediated GTPase activity is specific for KRAS G13D or if other codon 13 mutations are also NF1-sensitive, NF1 biochemical activity was tested with purified KRAS G13C protein. Although it only accounts for a fraction of a percent of human cancers, KRAS G13C is the second most common KRAS codon 13 mutation observed in human tumors. As predicted and consistent with our model, the NF1 GRD stimulated GTP hydrolysis in KRAS G13C with a kobs rate similar to KRAS G13D hydrolysis stimulation by NF1 (Fig. 4E and Table 1). To determine if NF1 can affect KRAS G13C activity in cells, NF1 was transfected into NCI-H1355 cells, which carry an endogenous KRASG13C mutation. Consistent with the in vitro biochemical data, KRAS GTP levels were reduced in the presence of NF1 (Fig. 4F).
NF1 Alters Response to EGFR Inhibitors in KRAS WT and G13D-Mutated Cells.
It has been suggested that a subset of KRASG13D-mutated colorectal cancers may respond to EGFR-inhibitor therapies (13, 14). However, clinical responses to cetuximab have not been statistically significant, and no underlying mechanism has been proposed. We postulated that NF1 activity may alter drug sensitivity in KRAS G13x-mutated colorectal cancer cells. Because many KRASG13D-mutated CRC cell lines also harbor an NF1 mutation, we chose to test this in SW48 cells, a CRC cell line with a KRAS and NF1 wild-type genetic background. SW48 cells engineered to carry either a KRASG12D or KRASG13D mutation in the endogenous locus were treated with the EGFR inhibitor gefitinib. The parental KRAS wild-type cells had an EC50 of 7.26 nM, while the KRASG12D SW48 cells were resistant up to 10.49 µM. The KRASG13D SW48 cells had a moderate sensitivity to gefitinib with an EC50 of 35.54 nM (Fig. 5A). Similar results were obtained with erlotinib, another small-molecule EGFR inhibitor (SI Appendix, Fig. S6). In addition, the SW48 isogenic cell lines had comparable doubling times while maintaining the cells in culture, and thus response to EGFR inhibitors is unlikely to be caused by differences in rates of proliferation. To determine if EGFR-inhibitor sensitivity in the KRASG13D SW48 cells is NF1-dependent, NF1 expression was depleted using small interfering RNA (siRNA) transfection. mRNA levels were depleted by 70 to 80% and the NF1 protein was not detectable by Western blot (Fig. 5D and SI Appendix, Fig. S7). After NF1 depletion, KRASG13D SW48 cells became resistant to gefitinib, whereas KRASG12D SW48 cells remained resistant to gefitinib without NF1 (Fig. 5 B and C). Consistent with these data, Western blots showed reduced pMEK levels after gefitinib treatment in the parental KRAS wild-type SW48 cells, while pMEK levels remained unchanged in the KRASG12D SW48 cells even at the highest concentrations tested. NF1 knockdown partially restored pMEK levels in the KRAS wild-type and G13D SW48 cells but remained unchanged in the KRASG12D SW48 cells (Fig. 5D). Together, these data suggest that, in an isogenic system, the presence of a KRASG13D mutation may confer sensitivity to EGFR inhibitors, but only in the context of functional NF1 protein.
Fig. 5.
KRAS G13D is sensitive to EGFR inhibition in an NF1-dependent manner. SW48 isogenic cell line dose–response to gefitinib. (A) Dose–response of WT, G12D, and G13D isogenic cell lines without siRNA treatment (error bars represent standard deviation). (B) G12D siRNA treated shows no change in response to gefitinib (error bars represent standard deviation). (C) G13D shows resistance to drug when NF1 is knocked down (error bars represent standard deviation). (D) G13D shows pMEK resistance when NF1 is knocked down, whereas the WT shows sensitivity with or without NF1 and G12D remains resistant in both situations. Drug treatment was for 48 h without EGF stimulation.
Discussion
KRAS residues G12, G13, and Q61 are all important for coordinating GTP binding, and mutation at these residues has been suggested to disrupt GAP-stimulated GTP hydrolysis and constitutively activate the RAF/MAPK pathway (28). Mutations at these sites occur at different frequencies in different cancer indications, and KRASG13D mutations appear almost exclusively in gastrointestinal cancers (16, 28–30). This observation raises the intriguing possibility that although all KRAS mutations stabilize KRAS GTP levels, the different KRAS oncogenic alleles may have distinct biological functions. Indeed, the biochemistry of KRAS proteins harboring G13 mutations appears distinct from codon 12 and 61 variants. KRASG13-mutated proteins have been described with reduced but measurable GAP-stimulated GTPase activity, suggesting that KRASG13x may be less oncogenic than KRASG12x or KRASQ61x cells (11). Here we report that within gastric cancers, tumor genomes with a KRASG13x mutation are associated with higher genetic instability, and frequently exhibit comutations within the RAS pathway. Furthermore, ∼12% of KRASG13x-mutated patient samples and 50% of KRASG13x-mutated cell lines also carry an NF1 comutation (Fig. 1C) while NF1 mutations appear mutually exclusive with KRASG12x- and KRASQ61x-mutated cells, suggesting that KRASG13x cells may benefit from an additional mutation to fully activate the RAS pathway. Interestingly, NF1 was also comutated in KRASA146T-mutated tumor samples and cell lines (Fig. 1C), again suggesting that low-frequency, weakly oncogenic KRAS-mutated cells may require an additional comutation in the RAS signal-transduction pathway.
Although all oncogenic KRAS alleles appear resistant to GTP hydrolysis by RASA1 GAP (11), the 2 most common codon 13 variants, KRASG13D and KRASG13C, are sensitive to NF1-stimulated GTPase activity in vitro and in cells. Restoration of NF1 activity in NF1-mutated cells stabilizes KRASG13D GTP levels and activates downstream signal transduction. Crystal structures of the wild type and G13D mutant of GMPPNP-bound KRAS in complex with the GAP-related domain of NF1 described here provide the structural basis of NF1-mediated GTP hydrolysis in the wild type as well as G13 mutants of KRAS. Structural comparison between the KRAS–NF1GRD complex and HRAS–RASA1GRD complex suggests that, like the Rho–RhoGAP complex, a rotation of 8° of NF1GRD would enable conversion from the ground state to the transition state, resulting in placement of the catalytic arginine finger inside the KRAS active site. Structural results described here validate that residue R1276 acts as an arginine finger in the NF1-mediated GTP hydrolysis reaction.
Comparison of the wild type and G13D mutant of KRAS (GMPPNP-bound) in complex with NF1GRD suggests that G13D/C mutants can allow placement of the arginine finger (present in the flexible finger loop) in the active-site pocket by adopting a side-chain rotamer conformation, which points away from the gamma-phosphate. In the case of KRASG13D, interaction of D13 with K117 is likely to stabilize its alternate rotamer conformation and prevent any steric clash between D13 (KRAS) and the arginine finger (NF1). Previously, it has been suggested that unlike G12, oncogenic mutation at G13 in KRAS is less stringent in the steric requirements and is able to tolerate small perturbations, and thus can allow placement of the arginine finger in the catalytic pocket for NF1-mediated GTPase activity (31). As suggested, loss of NF1-mediated GTP hydrolysis in G12 mutants of KRAS is likely due to steric hindrance, which does not allow placement of the arginine finger in the nucleotide-binding pocket for any possible rotamer conformations the mutated residue at the G12 position adopts. Insensitivity of KRAS Q61 mutants to NF1 and RASA1 GAPs is likely due to its inability to stabilize the arginine finger and orient the catalytic water for nucleophilic attack.
As suggested previously, formation of RAS–RasGAP complexes in the presence of a nonhydrolyzable GTP analog represents a ground-state conformation, whereas formation of RAS–RasGAP complexes in the presence of GDP and aluminum fluoride mimics the transition state of GAP-mediated GTPase activity (31). Previous biochemical studies have shown that G13A and G13S mutants of HRAS bind to NF1 with close to wild-type HRAS affinity and form an HRAS–NF1 complex in the presence of aluminum fluoride, suggesting NF1-mediated GTPase activity in these mutants (31). Unlike G13A/S, G13R showed weaker affinity and a reduced level of complex formation in the presence of aluminum fluoride. Surprisingly, in that study, no interaction was observed between the G13D mutant of HRAS and NF1GRD (31). We measured the affinity of KRAS mutants and NF1GRD using isothermal titration calorimetry, and the dissociation constants (Kds) of G13D/C mutants were close to that of wild-type KRAS. Biochemical and structural data presented here suggest that the mutation of G13 in RAS to other amino acids with a relatively small side chain (Ala, Ser, Cys, or Asp) is likely to be sensitive to NF1, and this sensitivity would decrease when G13 is mutated to a larger side-chain residue such as arginine.
Interestingly, unlike NF1, G13D/C mutations in KRAS are insensitive to a RASA1-mediated GTP hydrolysis reaction (Figs. 2A and 4E). Analysis of biochemical and structural differences between NF1 and RASA1 interactions with RAS provides a possible rationale for this observation. Previous studies have shown that the binding affinity of RAS for NF1GRD is 50-fold higher than RASA1GRD (20, 25, 26). The association and dissociation rate constants for RASA1 have been shown to be at least 2- and 100-fold faster than NF1 (20) and, because of this, it is difficult to measure binding affinity between KRAS and RASA1 using ITC. In NF1, the arginine finger is followed by Gly instead of Ala as in the case of RASA1, and it has been suggested that this results in higher flexibility in the finger loop of NF1 (5). It is possible that relatively weaker affinity and faster kinetics between KRAS and RASA1 and the relatively rigid finger loop is responsible for G13 mutants’ insensitivity toward RASA1.
NF1 biochemical activity with KRAS G13-mutated proteins may also partially explain the prevalence of these mutations in colorectal cancers. The high mutation rate found in gastric cancer patient samples includes functionally redundant mechanisms that may allow for less oncogenic KRAS variants to emerge. Comutation of tumor suppressors and signaling modifiers within the RAS pathway may augment activity of KRAS G13-mutated oncoproteins. It is also important to note that colorectal cancers frequently harbor underlying alterations in the WNT and TGF-β pathways, mutations that are rare in other KRAS-mutated cancer indications but often cross-talk with MAPK signaling. WNT signaling can stabilize oncogenic KRAS protein (32) and TGF-β signaling augments KRAS-driven oncogenesis in colorectal cancers (33). A better understanding of these mechanisms may help explain the high prevalence of KRAS G13 mutations in colon cancers specifically and how WNT and TGF-β signaling affect NF1 in KRAS G13-mutated colon cancer cells.
Finally, the structural and biochemical properties unique to G13-mutated KRAS proteins may have significant implications for KRAS-mutated colorectal cancer patients. The clinical observation that KRAS G13D CRC may respond to EGFR inhibition (13, 14) may be explained by the increased sensitivity to NF1-stimulated GTP hydrolysis. In the presence of active, functional NF1, KRASG13D cancer cells may have reduced KRAS GTP levels, and be partially dependent on upstream signaling through EGFR or other receptor tyrosine kinases. Although not tested in this study, G13x-mutated cell lines that also harbor FGFR2-, EGFR-, INSR-, or ALK-activating point mutations may similarly respond to direct inhibition of the mutated receptor tyrosine kinase. Conversely, cancers with KRASG12x or KRAS61x mutations are resistant to NF1 activity. Codon 12 or 61 mutations are fully active and independent of NF1 mutation status or NF1 activity, and therefore resistant to inhibition of upstream stimuli. Therefore, identification of the appropriate biomarker for active NF1 could expand the patient population for receptor tyrosine kinase inhibitors to include a subset of KRASG13x-mutated cancers.
Materials and Methods
Bioinformatics.
TCGA mutation and clinical data were downloaded from the Broad Institute GDAC firehose website (https://gdac.broadinstitute.org/). Tumor types with more than 10 G13x and G12x mutant samples included COAD, READ, and STAD. Per-class nonsynonymous mutation counts were compared for each tumor type and evaluated for statistical significance using the t test. Clinical term data were evaluated similarly except that the Fisher exact test was applied. For cell-line analysis, mutation data were downloaded from the COSMIC website’s cell-line project area (https://cancer.sanger.ac.uk/cell_lines). From these data, the top 100 mutated genes showing nonsynonymous comutation with any nonsynonymous KRAS mutation were identified and subsequently evaluated for statistical significance contrasting the G12 and G13 mutant sample classes. Complete lists of the statistically significant differentially mutated genes identified are provided in SI Appendix. For the NF1 mutational analysis, samples were subdivided into classes according to their mutational status and their NF1 comutational frequency status was determined.
Cell Culture and Transfection.
HCT-116 and SW48 isogenic cell lines were licensed from Horizon Discovery. Both sets of cell lines were cultured in RPMI medium (ATCC; 30-2001) supplemented with 10% fetal bovine serum (FBS) (GE Life Sciences; SH30070.03). NCI-H1355 cells were purchased from the ATCC. The cells were cultured in ACL4 media made from DMEM/F12 (ATCC; 30-2006), and FBS was used in place of BSA (SI Appendix, Materials and Methods). SW48 isogenic cell lines were transfected with siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific) as previously described by Vartanian (34). The following sequences were used as the NF1 siRNA: hNF1_558 5′-CUCACUACUAUUUUAAAGAAUA-3′, hNF1_558_as 5′-UUCUUUAAAAUAGUAGUGAGGC-3′. Scramble siRNA (Thermo Fisher Scientific; Ambion Silencer SelectNegative Control 2 siRNA) was used as a control in siRNA transfections. HCT-116 cells and NCI-H1355 cells were transfected with cDNAs using FuGENE HD (Promega) per the manufacturer’s protocol. The following cDNA constructs were used: HA-NF1-Caax STD [CMV51P>HA-TEV-NF1(1198–1509)-CAAX STD] (R711-M06-370), GFP (CMV51p>GFP) (R931-M01-366), KRAS WT (CMV51p>3XFLAG-Hs.KRASopt) (R750-M67-304), KRAS G12D (CMV51p>3xFLAG-HsKRAS4b G12D) (R750-M03-304), and KRAS G13D (CMV51p>3xFLAG-KRAS4b G13D) (R750-M05-304). See SI Appendix, Materials and Methods for sequences and construction. HCT-116 and NCI-H1355 cell lines were stimulated by the addition of 100 ng/mL EGF for 5 min prior to harvesting for Western blot and active RAS pull-down. KRAS GTP levels were measured using the Active Ras Pull-Down and Detection Kit (Thermo Fisher Scientific; 16117) according to the manufacturer’s protocol. CellTiter-Glo (Promega; G7573) was used for proliferation assays per the manufacturer’s protocol. Cells were treated with inhibitors (gefitinib; Tocris Bioscience; 3000) for 72 h unless otherwise indicated.
Biochemical GTP Hydrolysis Assays.
The EnzChek Phosphate Assay Kit (Thermo Fisher Scientific; E6646) was used to measure GTP hydrolysis in vitro. Reactions were performed in 384-well microplates with 30 mM Tris (pH 7.5) with 1 mM DTT. Final reaction concentrations in the wells were 200 μM MESG, 5 U/mL PNP, 200 μM GTP, and 10 mM MgCl2. Final concentrations of KRAS ranged from 80 to 2.5 µM and final GAP concentrations ranged from 80 to 40 µM. Microplates were read every 12 to 18 s at 360 nm. Data were analyzed by subtracting the background (no-substrate control) at each measurement. The amount of GTP hydrolyzed (µM) was determined by converting absorbance to micromolar concentration of GTP hydrolyzed using the linear fit of the KH2PO4 standard curve. Data were fit to a 1-phase association curve. Initial enzyme velocity measurements were used to calculate kobs values.
Structure Determination and Analysis.
The structure of the wild-type KRAS–NF1GRD complex was solved at 2.85 Å by molecular replacement using the program Phaser as implemented in the Phenix/CCP4 suite of programs with a protein-only version of Protein Data Bank ID codes 3GFT (Q61H-KRAS bound to GMPPNP) and 1NF1 (apo structure of NF1GRD) as search models (35, 36). The initial solution was refined using Phenix.refine and the resulting 2Fo − Fc map showed clear electron density for the 2 proteins. The model was further improved using iterative cycles of the manual model building in Coot and refinement using Phenix.refine (35, 37). The structure of GMPPNP-bound KRASG13D complexed with NF1GRD(GAP255) was solved at 2.1 Å using the structure of the wild-type KRAS–NF1GRD complex. Refinement statistics for these 2 crystal structures are summarized in SI Appendix, Table S1. Secondary structural elements were assigned using DSSP (https://swift.cmbi.ru.nl/gv/dssp/). Figures were generated with PyMOL (Schrödinger). Crystallographic and structural analysis software support is provided by the SBGrid Consortium (38).
Isothermal Titration Calorimetry Measurements.
Binding affinities of the GMPPNP-bound wild type and oncogenic mutants of KRAS (1 to 169) with NF1GRD (GAP333 and GAP255) were measured using isothermal titration calorimetry. Protein samples were prepared by extensive dialysis in a buffer (filtered and degassed) containing 20 mM Hepes (pH 7.3), 150 mM NaCl, 5 mM MgCl2, and 1 mM TCEP. For the ITC experiment, 60 μM KRAS and 600 μM NF1-GRD were placed in the cell and syringe, respectively. ITC experiments were carried out in a MicroCal PEAQ-ITC instrument (Malvern) at 25 °C using an initial 0.4-μL injection and 18 subsequent injections of 2.2 μL each at 150-s intervals. Data analysis was performed based on a binding model containing “one set of sites” using a nonlinear least-squares algorithm incorporated in the MicroCal PEAQ-ITC analysis software (Malvern).
Supplementary Material
Acknowledgments
We thank the following members of the Protein Expression Laboratory (Frederick National Laboratory for Cancer Research) for their help in cloning, protein expression, and purification: Allison Coward, John-Paul Denson, Dominic Esposito, Randy Gapud, Bill Gillette, Jennifer Mehalko, Simon Messing, Shelley Perkins, Lauren Procter, Nitya Ramakrishnan, Jose Sanchez Hernandez, Mukul Sherekar, Troy Taylor, and Vanessa Wall. We would also like to thank Ming Yi for assistance with proliferation data analysis. We are thankful to the staff at the 24-ID-C/E beamlines of the Advanced Photon Source, Argonne National Laboratory, for their help with data collection. This project was funded in whole or in part with federal funds from National Cancer Institute, NIH Contract HHSN261200800001E. Part of this work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from National Institutes of Health Grant P41 GM103403. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the US Government.
Footnotes
Competing interest statement: F.M. is a consultant for the following companies: Aduro Biotech, Amgen, Daiichi Ltd., Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, Inc., PellePharm, Pfizer Inc., PMV Pharma, Portola Pharmaceuticals, and Quanta Therapeutics; has received research grants from Daiichi Ltd. and is a recipient of funded research from Gilead Sciences; is a consultant and cofounder for the following companies (with ownership interest including stock options): BridgeBio Pharma, DNAtrix Inc., Olema Pharmaceuticals, Inc., and Quartz; and is scientific director of the NCI RAS Initiative at the Frederick National Laboratory for Cancer Research/Leidos Biomedical Research, Inc. M.H. is an employee of Quanta Therapeutics.
Data deposition: The atomic coordinates and structure factors of the wild-type KRAS–NF1GRD and KRASG13D–NF1GRD complexes reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6OB2 and 6OB3, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908353116/-/DCSupplemental.
References
- 1.Simanshu D. K., Nissley D. V., McCormick F., RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gideon P., et al. , Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: The C-terminal domain of GAP is not sufficient for full activity. Mol. Cell. Biol. 12, 2050–2056 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vigil D., Cherfils J., Rossman K. L., Der C. J., Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheffzek K., et al. , The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Scheffzek K., et al. , Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 17, 4313–4327 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheffzek K., Ahmadian M. R., Wittinghofer A., GTPase-activating proteins: Helping hands to complement an active site. Trends Biochem. Sci. 23, 257–262 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Kötting C., Kallenbach A., Suveyzdis Y., Wittinghofer A., Gerwert K., The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy. Proc. Natl. Acad. Sci. U.S.A. 105, 6260–6265 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gysin S., Salt M., Young A., McCormick F., Therapeutic strategies for targeting Ras proteins. Genes Cancer 2, 359–372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korf B. R., Malignancy in neurofibromatosis type 1. Oncologist 5, 477–485 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Hobbs G. A., Der C. J., Rossman K. L., RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129, 1287–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter J. C., et al. , Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 13, 1325–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Ihle N. T., et al. , Effect of KRAS oncogene substitutions on protein behavior: Implications for signaling and clinical outcome. J. Natl. Cancer Inst. 104, 228–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Roock W., et al. , Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304, 1812–1820 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Tejpar S., et al. , Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J. Clin. Oncol. 30, 3570–3577 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Peeters M., et al. , Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: Assessment as prognostic and predictive biomarkers of response to panitumumab. J. Clin. Oncol. 31, 759–765 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Segelov E., et al. , Response to cetuximab with or without irinotecan in patients with refractory metastatic colorectal cancer harboring the KRAS G13D mutation: Australasian Gastro-Intestinal Trials Group ICECREAM Study. J. Clin. Oncol. 34, 2258–2264 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Lawrence M. S., et al. , Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E., et al. , The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmadian M. R., Hoffmann U., Goody R. S., Wittinghofer A., Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry 36, 4535–4541 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Bollag G., et al. , Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J. Biol. Chem. 271, 32491–32494 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Kucharska A., Rushworth L. K., Staples C., Morrice N. A., Keyse S. M., Regulation of the inducible nuclear dual-specificity phosphatase DUSP5 by ERK MAPK. Cell. Signal. 21, 1794–1805 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Zeliadt N. A., Mauro L. J., Wattenberg E. V., Reciprocal regulation of extracellular signal regulated kinase 1/2 and mitogen activated protein kinase phosphatase-3. Toxicol. Appl. Pharmacol. 232, 408–417 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmadian M. R., Wiesmüller L., Lautwein A., Bischoff F. R., Wittinghofer A., Structural differences in the minimal catalytic domains of the GTPase-activating proteins p120GAP and neurofibromin. J. Biol. Chem. 271, 16409–16415 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Brownbridge G. G., Lowe P. N., Moore K. J., Skinner R. H., Webb M. R., Interaction of GTPase activating proteins (GAPs) with p21ras measured by a novel fluorescence anisotropy method. Essential role of Arg-903 of GAP in activation of GTP hydrolysis on p21ras. J. Biol. Chem. 268, 10914–10919 (1993). [PubMed] [Google Scholar]
- 26.Eccleston J. F., Moore K. J., Morgan L., Skinner R. H., Lowe P. N., Kinetics of interaction between normal and proline 12 Ras and the GTPase-activating proteins, p120-GAP and neurofibromin. The significance of the intrinsic GTPase rate in determining the transforming ability of ras. J. Biol. Chem. 268, 27012–27019 (1993). [PubMed] [Google Scholar]
- 27.Rittinger K., Walker P. A., Eccleston J. F., Smerdon S. J., Gamblin S. J., Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature 389, 758–762 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Lu S., Jang H., Nussinov R., Zhang J., The structural basis of oncogenic mutations G12, G13 and Q61 in small GTPase K-Ras4B. Sci. Rep. 6, 21949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Normanno N., et al. , Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat. Rev. Clin. Oncol. 6, 519–527 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Stinchcombe T. E., Der C. J., Are all KRAS mutations created equal? Lancet Oncol. 12, 717–718 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Gremer L., Gilsbach B., Ahmadian M. R., Wittinghofer A., Fluoride complexes of oncogenic Ras mutants to study the Ras-RasGap interaction. Biol. Chem. 389, 1163–1171 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Jeong W. J., et al. , Ras stabilization through aberrant activation of Wnt/β-catenin signaling promotes intestinal tumorigenesis. Sci. Signal. 5, ra30 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Grünert S., Jechlinger M., Beug H., Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell Biol. 4, 657–665 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Vartanian S., et al. , Identification of mutant K-Ras-dependent phenotypes using a panel of isogenic cell lines. J. Biol. Chem. 288, 2403–2413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin A., et al. , Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.