T lymphocytes are essential for adaptive immune responses; most recognize peptides bound to cell-surface major histocompatibility complex (MHC) molecules. The T cell receptor for antigen (TCR; on most T cells, a dimer of an α and a β chain associated with invariant signaling subunits) binds both peptide and MHC determinants (1–3). Accordingly, for any given T cell, the TCR specificity is determined by both the antigenic peptide and the particular MHC molecule the peptide binds. This property, called MHC restriction, is a critical feature of T cell antigen recognition. It has major functional and clinical implications in settings such as organ transplantation and T cell-mediated therapies. MHC restriction results from, and is often used to refer to, the ability of TCRαβ complexes to interact with MHC molecules. A new study by Krovi et al. in PNAS (4) clarifies the controversial question of whether TCR gene sequences have been skewed during evolution toward MHC recognition.
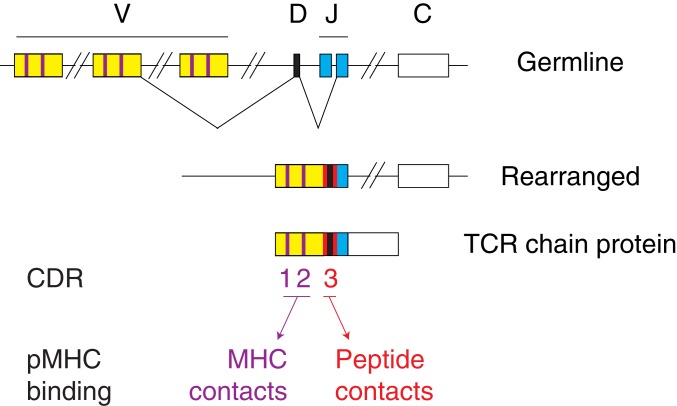
The dominant constraint governing TCR interactions with peptide-MHC complexes (pMHC) is the diversity of both components (2). In addition to being clonotypic (one cell expressing a single TCR specificity), the mammalian TCR repertoire is highly diverse, with a potential for ∼1015 specificities (1). Such diversity is generated in developing T cells through rearrangement of genomic regions of similar organization for both α and β loci (Fig. 1). Rearrangement generates de novo diversity within both α and β chains in a segment called complementarity determining region 3 (CDR3), through both deletion and nontemplated addition of nucleotides. Two additional segments, CDR1 and CDR2, carry diversity among variable (V) domains of TCR chains. All 3 CDRs form loops protruding from the core Vα or Vβ domain and mediate pMHC binding. Mirroring TCR diversity, allelic diversification has made MHC molecules highly polymorphic both at the species and individual level, a property essential to counteract pathogen evasion of MHC presentation and T cell defenses.
Fig. 1.
Schematic showing the genomic regions generating domains of TCR chains before (Top) or after (Middle) TCR gene rearrangement, and the resulting protein products (Bottom). CDR1 and CDR2 are shown as purple boxes. CDR3 takes sequences from V, D, or J genomic regions (as color-coded) and untemplated sequences (red boxes). TCRα genes have no D segments.
The combined diversity of TCR and MHC molecules makes most TCRs generated by a given individual unable to interact with the specific allelic set of MHC molecules carried by that individual. T cell precursors carrying such “useless” TCRs undergo programmed cell death during their development in the thymus (5). At the individual level, this ensures that only “useful” self−MHC-restricted TCRs contribute to the mature T cell repertoire, a process called positive selection. But this leaves unanswered the question of whether MHC restriction is “inscribed” in germline TCR gene sequences, that is, whether the genomic sequences encoding TCR V regions generate a preselection repertoire of TCRs skewed toward MHC recognition.
The structure and organization of TCRs is quite similar to that of immunoglobulins, which serve as receptors for antigens in B lymphocytes and are secreted as antibodies (2). Furthermore, immunoglobulin diversity is generated in B cell precursors through a process highly similar to that operating in T cell precursors at TCR gene loci. However, unlike TCR, immunoglobulins recognize 3D structures of diverse chemical composition, independently of their spatial context, and such binding does not require third-party molecules, whether MHC-related or not. Thus, if T and B cell precursors carry similar receptors generated along similar processes, why would the preselection TCR repertoire be MHC-restricted, whereas that of B cells is not?
A conceptually appealing answer is that, despite their diversity, the germline-encoded CDR1 and CDR2 of TCRs, but not those of immunoglobulins, have been under evolutionary pressure to bind MHC molecules (3, 6). Although MHC and TCR diversity are not compatible with binary, sterically fixed interactions as for most ligand−receptor pairs (7), the idea of a TCR bias for MHC is supported by several lines of evidence. Analyses of TCR-pMHC structures have shown that most use a similar (although not identical) docking mode (2, 3): TCR α and β V domains are obliquely positioned over the 2 MHC α-helices and peptide-containing groove that make up the pMHC interaction interface. Most peptide contacts come from CDR3, whereas most MHC contacts are made by CDR1 and CDR2 residues. Although there is no general pattern of pairing between MHC allelic isoforms and specific TCR V chains, comparison of multiple crystallographic structures suggests a loose correspondence between MHC and conserved CDR1 and CDR2 residues that are needed for TCR-pMHC interactions (8, 9). This supports the hypothesis that CDR1 and CDR2 have evolved to be MHC-skewed, so that the TCR repertoire is intrinsically MHC-biased at the species level, prior to thymic selection. Indeed, earlier studies supported the idea of an MHC-biased preselection TCR repertoire (10, 11).
In apparent contrast with this idea, genetic analyses in mice have identified αβ TCRs that recognize MHC-independent 3D structures, including CD155, the mouse ortholog of the human poliovirus receptor (12, 13). Cells expressing such MHC-independent TCRs are functionally similar to MHC-restricted T cells; as a population, they exhibit a broad TCR repertoire, although with a trend to lower diversity than MHC-restricted cells. MHC-independent reactivity requires CDR3 and conserved CDR2 residues previously reported to contribute to MHC reactivity (14–16). Thus, TCR gene rearrangement can generate MHC-independent specificities, akin to those of immunoglobulins. Importantly, while such TCRs are generated in the preselection repertoire, they are normally absent from the mature T cell repertoire (consistent with the broader concept of MHC restriction) (13). Instead, the development of MHC-independent cells requires 2 conditions: absence of MHC molecules from the thymus and disruption of genes encoding 2 surface “coreceptors,” CD4 and CD8. CD4 and CD8 normally facilitate the selection of MHC-restricted T cell precursors through 2 mechanisms (17): 1) binding of invariant regions of MHC molecules, thereby promoting TCR tethering to MHC, and 2) recruitment via their intracellular domain of a tyrosine kinase needed for TCR signal transduction. That the development of MHC-independent cells requires CD4 and CD8 deletion raises the tantalizing possibility that coreceptors actually prevent the selection of TCRs with MHC-independent reactivity, and thereby are the primary enforcers of MHC restriction. In that perspective, the preselection repertoire generated by TCR gene rearrangement does not need not to be skewed toward MHC reactivity.
Distinguishing between these 2 possibilities requires evaluating the respective frequencies of cells reacting with MHC vs. non-MHC determinants in the preselection repertoire, which Krovi et al. (4) do using a high-throughput single-cell reporter assay. The basic approach is to reproduce TCR diversity in a cell line expressing a fluorescent reporter measuring TCR responsiveness. To this end, libraries encoding TCR Vα domains (including all 3 CDRs) are generated from mouse T cell precursors lacking the constant (C) region of the TCRα gene (Fig. 1), which express no surface TCR despite normal TCRα gene rearrangement. Thus, these libraries sample the preselection Vα repertoire. After verification of their diversity by deep sequencing, the libraries are transfected into hybridoma cells that carry a reporter for TCR engagement and express either a single TCRβ chain or a library of TCRβ chains obtained through the same approach. The experimental design is tailored to express one TCR specificity per cell, and transfected cells are evaluated for reactivity against MHC-expressing cell lines. Reactivity is assessed in both the absence and presence of anti-MHC antibodies that prevent TCR-MHC interactions; the latter assay detects reactivity against non-MHC antigens expressed by the MHC-expressing cells or by the hybridoma itself, and therefore estimates MHC-independent binding.
The study by Krovi et al. builds a strong case that T cell MHC restriction is largely supported by a built-in bias in TCR germline sequences.
TCR generated by pairing fixed TCRβ chain with members of 5 diverse TCRα libraries showed little or no MHC-independent reactivity in these assays, whereas the frequency of MHC-reactive cells depended on which specific TCRβ was expressed. In this setting, the fixed TCRβ specificity conceivably restrained assay sensitivity. Thus, Krovi et al. (4) generated hybridomas coexpressing TCRβ and TCRα libraries, therefore not limiting Vα pairing to a few specific Vβ. Remarkably, even though these TCRαβ pairs were randomly generated from preselection chains, 5 to 10% of them were MHC-reactive; in contrast, little or no MHC-independent reactivity was detected. While the sensitivity of the assay could be limiting for the detection of MHC-independent reactivity, this should also affect the detection of MHC-dependent reactivity; thus, the data strongly support the idea of a germline skewing of TCR toward MHC reactivity.
This conclusion fits with earlier reports that coreceptors (notably CD4) are not required to generate an MHC-restricted repertoire (18). However, consistent with an important role of coreceptors in establishing MHC restriction, Krovi et al. (4) find that MHC-specific reactivity was enhanced by expression of an improved CD4 (with higher affinity for its MHC target). Because coreceptors only bind MHC and because positive selection is a competitive process, the help provided by coreceptors would contribute to expunging the few MHC-independent TCRs generated by the rearrangement process. On the other side of the spectrum, precursors with high affinity for intrathymic ligands are eliminated by TCR-induced death or redirected toward lineages with regulatory functions, and it would be interesting to see whether those are enriched for MHC-independent specificities (5).
Thus, the study by Krovi et al. (4) builds a strong case that T cell MHC restriction is largely supported by a built-in bias in TCR germline sequences. Even though numerous MHC-independent T cells can be generated in MHC-deficient animals, the preselection repertoire comprises a greater frequency of MHC-reactive than of MHC-independent TCRs, and the MHC bias of the mature repertoire is further enhanced by coreceptor expression (which could also enhance the evolutionary pressure toward the selection of MHC-reactive TCR CDRs). Future studies will explore which TCR determinants mediate such MHC reactivity, since, in addition to CDR1 and CDR2 sequences, there is evidence that MHC recognition is also constrained by CDR3 attributes (4, 14). Last, despite their rarity, MHC-independent T cells are interesting on their own merit. Because of the many functions T cells can carry out, notably cytotoxic activity, MHC-independent T cells may offer alternative strategies, notably against tumor cells.
Acknowledgments
I thank J. Ashwell and P. Love for reading the manuscript. Research in my laboratory is supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health.
Footnotes
The author declares no competing interest.
See companion article on page 22252.
References
- 1.Davis M. M., Bjorkman P. J., T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Rossjohn J., et al. , T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Marrack P., Scott-Browne J. P., Dai S., Gapin L., Kappler J. W., Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu. Rev. Immunol. 26, 171–203 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krovi S. H., Kappler J. W., Marrack P., Gapin L., Inherent reactivity of unselected TCR repertoires to peptide-MHC molecules. Proc. Natl. Acad. Sci. U.S.A. 116, 22252–22261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogquist K. A., Jameson S. C., The self-obsession of T cells: How TCR signaling thresholds affect fate ‘decisions’ and effector function. Nat. Immunol. 15, 815–823 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia K. C., Reconciling views on T cell receptor germline bias for MHC. Trends Immunol. 33, 429–436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadinski B. D., et al. , A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity 35, 694–704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng D., Bond C. J., Ely L. K., Maynard J., Garcia K. C., Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon.’ Nat. Immunol. 8, 975–983 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Scott-Browne J. P., White J., Kappler J. W., Gapin L., Marrack P., Germline-encoded amino acids in the αβ T-cell receptor control thymic selection. Nature 458, 1043–1046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zerrahn J., Held W., Raulet D. H., The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell 88, 627–636 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Merkenschlager M., et al. , How many thymocytes audition for selection? J. Exp. Med. 186, 1149–1158 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siliciano R. F., et al. , Direct evidence for the existence of nominal antigen binding sites on T cell surface Ti α-β heterodimers of MHC-restricted T cell clones. Cell 47, 161–171 (1986). [DOI] [PubMed] [Google Scholar]
- 13.Van Laethem F., et al. , Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity 27, 735–750 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Lu J., et al. , Molecular constraints on CDR3 for thymic selection of MHC-restricted TCRs from a random pre-selection repertoire. Nat. Commun. 10, 1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tikhonova A. N., et al. , αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity 36, 79–91 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Laethem F., et al. , Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell 154, 1326–1341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Laethem F., Tikhonova A. N., Singer A., MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection. Trends Immunol. 33, 437–441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locksley R. M., Reiner S. L., Hatam F., Littman D. R., Killeen N., Helper T cells without CD4: Control of leishmaniasis in CD4-deficient mice. Science 261, 1448–1451 (1993). [DOI] [PubMed] [Google Scholar]