As we move into the era of personalized medicine, it may not be too far off that a visit to our doctor includes a number of genome-scale “seq” experiments to diagnose disease and estimate our likelihood of response to an array of possible treatments. One such experiment measures the cell’s total RNA abundance (its transcriptome) which provides a snapshot of a cellular activity or state. For decades global gene expression levels and changes in expression have been used to identify disease and drug response markers. This has been enabled by technological advances such as “whole-exome” sequencing which can home in on biomarker genes that define cellular states. These approaches were first designed to survey protein-coding gene (PCG) levels of expression and have uncovered numerous PCG biomarkers, led to advances in how drugs are screened, and generated insights into how gene pathways are misregulated in disease.
Despite this progress, we are still limited in our ability to diagnose cancer subtypes or predict how an individual patient will respond to a particular therapy. This raises the question: Is there valuable information hidden in the rest of the genome outside of PCGs? Considering that PCGs represent a small fraction of the transcriptome, it seems likely that there could be other RNA genes, such as long noncoding RNAs (lncRNAs), that could also be biomarkers of drug response.
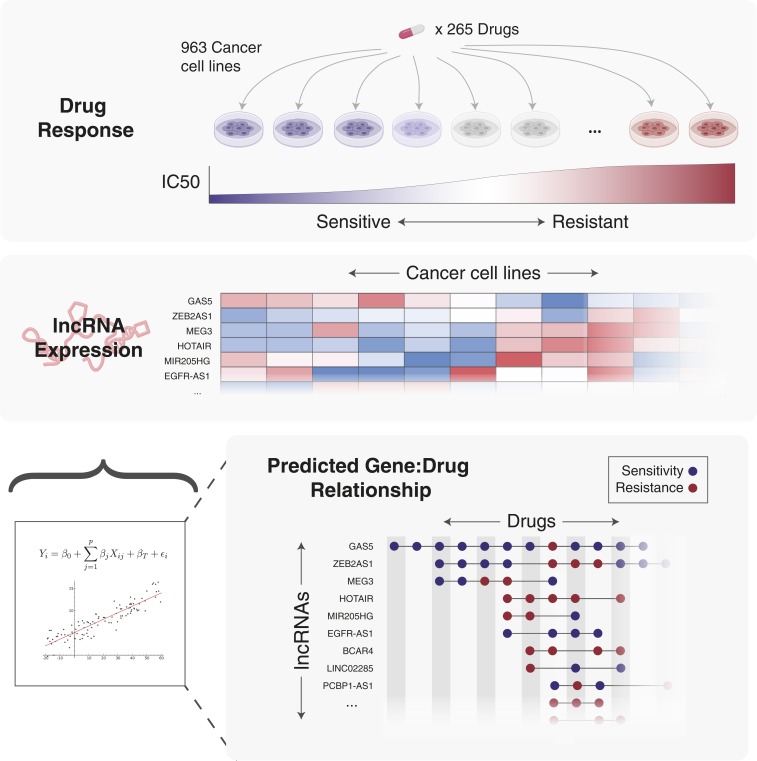
In PNAS Nath et al. (1) take a major step forward by demonstrating that both PCG and lncRNA expression patterns are predictive of response to anticancer drugs. The study is a tour de force of gene-expression analysis. Using the baseline gene-expression data and corresponding drug response data from 2 massive cancer cell line screens (Genomics of Drug Sensitivity in Cancer, 265 compounds × 963 cancer cell lines; and Cancer Therapeutics Response Portal, 545 compounds × 715 cancer cell lines), Nath et al. (1) find that indeed many individual lncRNAs can be considered biomarkers for anticancer drug resistance or sensitivity (Fig. 1).
Fig. 1.
Baseline lncRNA expression predicts drug sensitivity in cancer cell lines. Thousands of cancer cell lines have been tested for cell viability in response to the addition of hundreds of compounds. The same cell lines have been extensively characterized, including the measurement of transcriptome-wide RNA levels. Using the expression of lncRNAs as predictor variables in regularized linear regression models, individual lncRNAs that are strongly predictive of sensitivity to a drug across cancer cell lines can be uncovered. Thus, these drug–lncRNA relationships have potential to be used as biomarkers to inform patient care.
Since the transcription rate of a gene can affect its genomic neighborhood, Nath et al. (1) were very careful to take into account “cis” (or local) changes in expression in lncRNAs nearby PCGs. This local cis effect accounted for 50% of the lncRNA–drug pairs that were considered predictive. Outside these local effects Nath et al. (1) identified many lncRNAs that were independently informative outside their associations with PCGs. For example, the GAS5 lncRNA alone could predict sensitivity to more than 50 anticancer drugs. Moreover, simply adding EGFR-AS1 and MIR205HG lncRNAs to existing models of erlotinib resistance could explain 25% to 30% more of the variance in drug response than using PCG expression levels by themselves. Thus, lncRNAs represent distinct and independent signals of how cancer cells become resistant to cancer therapies.
Even if the vast majority of lncRNAs are not functional, their expression is driven by the same processes as those of PCGs and hence they provide additional information in predicting the cell’s response to a specific drug. In fact, due to the tissue- and context-dependent expression patterns of lncRNAs, they could be an even more information-rich source to define tissue/disease subtypes and poised states for drug sensitivity. This is consistent with numerous previous studies that have demonstrated that lncRNAs have at the very least equal discriminative power to PCGs in determining cell state or “cell specificity” (2–8). In some cases, lncRNA levels can distinguish cortical neurons from other cell types better than PCG levels (3). Of note, lncRNAs, on average, have much lower expression than PCGs. It is critical to account for the lower abundance of lncRNAs when determining their cell and condition specificity; many of the statistics to determine cell specificity are highly sensitive to distribution shifts (3). This was extensively examined (and code documented) in ref. 8. Thus, collectively Nath et al. (1) extend if not translate these findings toward more personalized diagnostic anticancer therapies.
In the future the gene–drug response associations found by Nath et al. (1) could be used in personalized medicine. By simply measuring the patient’s tumor transcriptome, one could find the best predicted drug–patient match. However, the use of transcriptomics to inform individual medicine does not end there. It is now possible to create “living laboratories” personalized to an individual. Patient tumor biopsies can be transplanted onto mice (patient-derived xenographs [PDX]) and patient cells can be induced to a pluripotent state (induced pluripotent stem cells [iPSCs]) and cultured in the laboratory. These “avatar” cell lines or tumors can be screened for tailored drug treatments and combination therapies. In this context, genome-wide transcriptome profiling can guide diagnoses, drug/drug combinations to screen, and future experimental strategies to target additional drug resistance genes in cancer and beyond (9).
A great recent example of how genome-wide RNA profiling has advanced personalized medicine is the case of a patient with pancreatic cancer (9). This patient was monitored for 5 y using numerous “omic profiling” as well as avatar PDX models to guide cancer treatments through time and metastases. When an unexpected drug response occurred in the patient, that drug was administered to the PDX models and followed by RNA-seq. This uncovered mechanisms of action that were driving that particular drug response. Moreover, these “omic” profiles revealed that many aspects of the noncoding genome changed during tumor evolution (9).
Nath et al. (1) demonstrate that the noncoding transcriptome allows us a more complete view into cellular state, but perhaps there is even more to gain from looking at the genome’s dark matter. It is likely that at least some of the lncRNAs associated with drug response and disease subtype may have a functional role in mediating that response or disease. Nath et al. (1) make this observation and show evidence for functionality for 2 lncRNA genes associated with increased sensitivity to erlotinib in lung cancer cell lines. Upon knockdown of each lncRNA, the cell lines showed increased resistance to the drug. Overall, Nath et al. (1) open up avenues for personalized profiling that are universally applicable to our ever-growing precision medicine efforts.
Footnotes
The authors declare no competing interest.
See companion article on page 22020.
References
- 1.Nath A., et al. , Discovering long noncoding RNA predictors of anticancer drug sensitivity beyond protein-coding genes. Proc. Natl. Acad. Sci. U.S.A. 116, 22020–22029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabili M. N., et al. , Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molyneaux B. J., et al. , DeCoN: Genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauli A., et al. , Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 22, 577–591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulitsky I., Bartel D. P., lincRNAs: Genomics, evolution, and mechanisms. Cell 154, 26–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien T., et al. , The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Z., et al. , Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 20, 908–913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goff L. A., On the tissue specificity of lncRNAs. Github. https://github.com/Loyale/lncRNASpec. Accessed 29 September 2019.
- 9.Wolff R. A., et al. , Dynamic changes during the treatment of pancreatic cancer. Oncotarget 9, 14764–14790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]