Significance
Aberrant activation of the homeostatic Wnt signaling pathway is a hallmark of various types of cancer. In many cases, it is unclear how elevated Wnt levels are maintained in the absence of activating pathway mutations. Here we find that the uncharacterized transcription factor FOXB2, whose expression is usually restricted to the developing brain, is induced in aggressive prostate cancer. FOXB2 strongly activates Wnt signaling via the induction of multiple pathway agonists, particularly the neurogenic ligand WNT7B. Accordingly, our analyses suggest that FOXB2 imposes a neuronal differentiation program on prostate cancer cells, which is associated with treatment failure and poor prognosis. Thus, our work identifies FOXB2 as a tissue-specific Wnt activator that may play a role in prostate cancer progression.
Keywords: FOXB2, Wnt signaling, forkhead, prostate cancer
Abstract
The Wnt signaling pathway is of paramount importance for development and disease. However, the tissue-specific regulation of Wnt pathway activity remains incompletely understood. Here we identify FOXB2, an uncharacterized forkhead box family transcription factor, as a potent activator of Wnt signaling in normal and cancer cells. Mechanistically, FOXB2 induces multiple Wnt ligands, including WNT7B, which increases TCF/LEF-dependent transcription without activating Wnt coreceptor LRP6 or β-catenin. Proximity ligation and functional complementation assays identified several transcription regulators, including YY1, JUN, and DDX5, as cofactors required for FOXB2-dependent pathway activation. Although FOXB2 expression is limited in adults, it is induced in select cancers, particularly advanced prostate cancer. RNA-seq data analysis suggests that FOXB2/WNT7B expression in prostate cancer is associated with a transcriptional program that favors neuronal differentiation and decreases recurrence-free survival. Consistently, FOXB2 controls Wnt signaling and neuroendocrine differentiation of prostate cancer cell lines. Our results suggest that FOXB2 is a tissue-specific Wnt activator that promotes the malignant transformation of prostate cancer.
The Wnt pathway is a major homeostatic signaling cascade in development and stem cell homeostasis (1–3). In the canonical or β-catenin–dependent signaling branch, secreted Wnt ligands engage a transmembrane receptor system consisting of Frizzled family core and LRP5/6 coreceptors to inhibit a multiprotein β-catenin destruction complex. Consequently, cytosolic β-catenin is relieved from constitutive proteasomal degradation and induces the transcription of target genes through association with TCF/LEF family transcription factors.
Specificity of the Wnt signaling output is achieved primarily via the differential expression of a wide range of Wnt ligands and receptors, which exert overlapping but nonredundant functions (2). Interestingly, several Wnt molecules act downstream of β-catenin by synergizing with canonical Wnt ligands. In particular, WNT7B elicits limited pathway activation on its own, as evidenced by its inability to induce LRP6 phosphorylation and β-catenin stabilization to any substantial degree (4). In contrast, WNT7B strongly cooperates with other ligands, primarily WNT1, in driving TCF/LEF-dependent gene transcription. The mechanism of WNT7B-dependent pathway activation is unclear, but it requires additional coreceptors, namely RECK and GPR124 (4–6). The expression of WNT7B and its coreceptors is largely restricted to specific tissues, especially the developing brain, where they contribute to blood–brain barrier formation and maintenance through activation of Wnt/β-catenin signaling (7, 8). Additionally, increased expression of WNT7B and subsequent Wnt pathway activation have been observed in several cancers, including prostate cancer (9, 10). However, as with most Wnt ligands, it remains largely unresolved how WNT7B expression is regulated.
Earlier studies reported that mouse Wnt7b is induced by Ttf-1, Gata-6, Foxa2, Pax6, and p53 in a tissue-specific manner (11–13). Moreover, WNT7B is a transcriptional target of the androgen receptor, and, accordingly, prostate cancer cells exhibit high basal levels of this ligand (10, 14). Here, we identify the uncharacterized forkhead box (FOX) transcription factor FOXB2 as a potent activator of Wnt signaling that drives the expression of various Wnt ligands, primarily WNT7B. Although FOXB2 is predominantly expressed in the developing brain (15), we find that it is induced in advanced prostate cancer. Most prostate tumors initially progress slowly, but clonal evolution of cancer cells may result in androgen resistance and neuroendocrine differentiation, which is associated with treatment failure and exceptionally poor prognosis (16). Chronic Wnt pathway activation is a key driver of malignant prostate cancer progression (17). However, in contrast to, e.g., colorectal cancer, activating pathway mutations are relatively infrequent in prostate cancer, and sustained Wnt signaling is thought to be maintained via tissue-specific pathway activators, including WNT7B (10, 17). Thus, our identification of FOXB2 as major WNT7B regulator may have important implications for developmental and cancer biology.
Results
FOXB2 Activates Wnt/TCF Signaling.
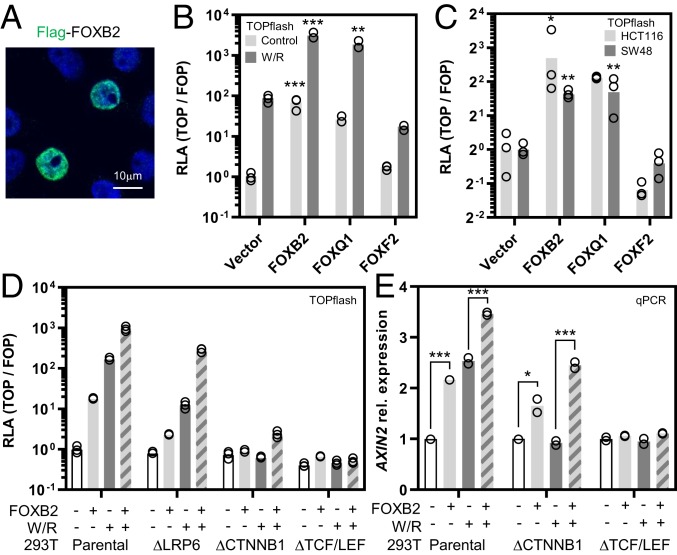
The context-dependent regulation of canonical Wnt signaling is incompletely understood. In an effort to discover pathway regulators, we performed a gain-of-function screen by coexpressing Flag-tagged proteins of interest with a β-catenin/TCF luciferase reporter (TOPflash) in normal and cancer cell lines. Among 17 FOX family proteins tested in this assay, we identified the uncharacterized transcription factor FOXB2 as the most potent candidate Wnt activator (Fig. 1 A and B and SI Appendix, Fig. S1 A and B). We additionally confirmed known positive and negative regulators of Wnt signaling, such as FOXQ1 and FOXF2, respectively (18, 19). FOXB2 strongly promoted TOPflash activity and potently synergized with the pathway agonists Wnt3a/R-spondin 3 (Fig. 1B). Moreover, FOXB2 induced TOPflash activity in HCT116 and SW48 colorectal cancer cells, which harbor activating β-catenin mutations and are thus largely unresponsive to further pathway activation (Fig. 1C). Of note, the most closely related FOX family member, FOXB1, only had a marginal effect on Wnt signaling in these assays (SI Appendix, Fig. S1C).
Fig. 1.
FOXB2 is a Wnt/TCF activator. (A) Over-expression in 293T cells showed nuclear FOXB2 localization. (B) β-Catenin/TCF luciferase reporter assay (TOPflash) in 293T cells. Expression of the indicated Flag-tagged proteins showed that FOXB2 activates Wnt signaling and synergizes with Wnt3a/R-spondin 3 (W/R)-conditioned media in pathway activation. FOXQ1 and FOXF2 were included as positive and negative controls, respectively. RLA, relative luciferase activity. (C) FOXB2 activated TOPflash in HCT116 and SW48 colorectal cancer cells with activating β-catenin mutation. (D) Epistasis assay in normal and gene-edited 293T cells. Loss of LRP6, β-catenin (CTNNB1), or all TCF/LEF transcription factors strongly attenuated FOXB2-dependent reporter activation. (E) Quantitative real-time PCR (qPCR) in 293T cells. FOXB2 induced AXIN2 expression in synergy with Wnt3a/R-spondin 3. This effect was also observed in β-catenin–deficient, but not TCF/LEF-deficient, cells. Note that experiments in B and D were performed using calcium phosphate transfection, which generally produces stronger TOPflash signals compared to lipofection (*P < 0.05, **P < 0.01, and ***P < 0.001 versus control).
FOX transcription factors control Wnt signaling through various mechanisms, such as the regulation of β-catenin nuclear shuttling and stability (18–20). To elucidate the signaling mode of FOXB2, we first performed an epistasis assay in gene-edited 293T cells with genetic deletion of Wnt coreceptor LRP6, β-catenin, or all TCF/LEF transcription factors (21) (Fig. 1D and SI Appendix, Fig. S1 D and E). Loss of LRP6 was sufficient to reduce FOXB2-induced TOPflash activation by ∼90%, while deletion of TCF/LEF essentially blocked reporter activity. In contrast, we observed residual pathway activation by FOXB2 in β-catenin–deficient cells. Some FOX proteins, e.g., FOXQ1 and FOXM1, control β-catenin activity via direct association (20, 22). However, FOXB2 did not bind β-catenin or colocalize with nuclear β-catenin (SI Appendix, Fig. S1 F and G). Since TOPflash is an artificial Wnt reporter, we also tested the regulation of the prototypical Wnt target gene AXIN2 by qPCR (Fig. 1E). Consistent with results from the TOPflash assays, FOXB2 induced AXIN2 expression alone as well as in synergy with Wnt3a/R-spondin 3. As before, partial AXIN2 induction by FOXB2 was also observed in β-catenin–deficient cells, and Wnt3a/R-spondin 3 synergy was retained despite no effect of these ligands by themselves. Collectively, these data identify FOXB2 as a β-catenin–independent Wnt pathway activator.
FOXB2 Induces WNT7B to Activate TCF/LEF.
The most likely explanation for the aforementioned observations is that FOXB2 activates Wnt signaling by inducing one or more canonical Wnt ligands. To test this hypothesis, we first treated FOXB2-transfected 293T cells with porcupine inhibitor LGK974, which blocks the release of endogenous Wnts (Fig. 2A and SI Appendix, Fig. S2 A and B). LGK974 strongly attenuated FOXB2-induced Wnt signaling even at low nanomolar concentration. This effect was more pronounced in cells with additional exogenous R-spondin 3 compared to cells treated with Wnt3a conditioned media, suggesting that FOXB2 induces Wnt ligands rather than R-spondins. Consistently, FOXB2 synergized with recombinant R-spondin 3 protein, but not recombinant WNT3A (SI Appendix, Fig. S2C). Moreover, iCRT14, a TCF and Notch pathway inhibitor that disrupts β-catenin/TCF/DNA interaction, also reduced FOXB2-dependent TOPflash activation, and this effect was less pronounced in the presence of exogenous Wnt3a (SI Appendix, Fig. S2D).
Fig. 2.
FOXB2 activates TCF signaling via WNT7B. (A) Treatment with Wnt secretion inhibitor LGK974 (10 µM) strongly attenuated FOXB2-dependent TOPflash activation in 293T cells, particularly in the presence of exogenous R-spondin 3 (Rspo3). (B) Immunoblot of 293T whole-cell lysates (WCL) and corresponding cytosolic extracts (Cyt). FOXB2 did not activate LRP6 or stabilize β-catenin. Where indicated, cells were treated with Wnt3a-conditioned media for 6 h. (C) qPCR of all 19 Wnt ligands in 293T cells. FOXB2 strongly induced multiple Wnts. Data are displayed as fold change compared to empty vector control. (D) Depletion of WNT7B or its coreceptors RECK and GPR124 by RNA interference attenuated FOXB2-dependent TOPflash activation in 293T cells. Note that these siRNAs had essentially no effect on basal Wnt signaling. (E) Luciferase-based WNT7B promoter/enhancer assay in 293T cells. FOXB2, but not FOXB1, activated the WNT7B reporters (*P < 0.05, **P < 0.01, and ***P < 0.001 versus control).
LRP6 phosphorylation and β-catenin stabilization are early hallmarks of Wnt pathway activation by canonical ligands. However, FOXB2 did not increase phospho-LRP6 and β-catenin levels (Fig. 2B), even though it strongly induced TCF-dependent transcription under comparable experimental conditions (Fig. 2A). To explore these observations further, we examined the regulation of all 19 Wnt ligands by FOXB2 (Fig. 2C). Expression of FOXB2 in 293T cells strongly induced multiple Wnts, particularly WNT1, WNT6, WNT9B, and especially WNT7B. Select Wnt ligands, such as WNT7B, can activate TCF/LEF-dependent gene transcription independently of β-catenin (4). Thus, to determine if WNT7B is the major mediator of FOXB2-induced Wnt signaling, we interrupted this pathway by RNA interference (Fig. 2D and SI Appendix, Fig. S2 E–G). Indeed, depletion of WNT7B or its coreceptors RECK and GPR124 attenuated FOXB2-induced TOPflash activation by ∼55%, 75%, and 35%, respectively. Moreover, knock-down of WNT7B or RECK reduced FOXB2-dependent AXIN2 expression in β-catenin–deficient cells (SI Appendix, Fig. S2E). Finally, to test whether FOXB2 directly regulates WNT7B expression, we generated luciferase reporter constructs containing the 1-kb 5′ promoter region of WNT7B, as well as an intronic WNT7B gene enhancer. FOXB2, but not FOXB1, significantly activated these reporters (Fig. 2E). We conclude that FOXB2 activates Wnt/TCF independently of β-catenin, by engaging a WNT7B/RECK/GPR124 signaling module.
Multiple Structural Features Shape FOXB2-Dependent Wnt Signaling.
The DNA-binding forkhead box is highly conserved across FOX family proteins (23). Nonetheless, despite high sequence similarity between, e.g., the FOXB1 and FOXB2 forkhead domain (SI Appendix, Fig. S3A), only FOXB2 promotes Wnt signaling to any substantial degree. Thus, the control of specific target genes likely requires interaction of FOXB2 with additional transcription regulators, which is consistent with the mode of action of other FOX proteins (24). To explore the structural requirements for FOXB2-induced Wnt signaling, we first generated a series of truncations and point mutants (SI Appendix, Fig. S3B). Importantly, all truncation constructs exhibited strongly reduced activity in the TOPflash assay, including deletion mutants missing the unique central and C-terminal regions (Fig. 3A). Moreover, a FOXB2 point mutant that is unable to bind DNA (P14A/P15A [23]) was completely inert in this assay. In contrast, mutation of a putative engrailed homology (EH1) motif (G277A) had no effect on TOPflash activation. Based on these results, we tested the ability of select FOXB2 mutants to induce WNT7B expression (Fig. 3B). Indeed, WNT7B induction was essentially blocked in FOXB2 constructs lacking either the N-terminal or C-terminal domain. Additionally, we replaced the unique, highly unstructured central domain of FOXB2 (109–336) with the corresponding region of FOXB1 (SI Appendix, Fig. S3 C and D). The resulting chimeric protein phenocopied FOXB1, i.e., it had a minimal impact on TOPflash activity (Fig. 3C). We conclude that regulation of Wnt signaling by FOXB2 requires multiple structural domains in addition to DNA binding.
Fig. 3.
FOXB2-dependent Wnt signaling requires multiple protein domains. (A) FOXB2 mutation or truncation decreased its TOPflash activity in 293T cells, except for a G277A mutation. Numbers indicate amino acid positions. Major protein features are shown in the inset cartoon. FKH, forkhead domain; NLS, nuclear localization sequence; EH1, engrailed homology 1 motif. (B) qPCR analysis in 293T cells showed that FOXB2 N- or C-terminal truncation blocked WNT7B induction. (C) A FOXB2/B1 chimera with the central domain of FOXB1 was largely inert in TOPflash activation, similar to FOXB1. Protein expression levels in A–C were generally comparable for all constructs (SI Appendix, Fig. S3 B and C; *P < 0.05, **P < 0.01, and ***P < 0.001 versus control).
FOXB2-Dependent Wnt Signaling Is Controlled by Transcriptional Coregulators.
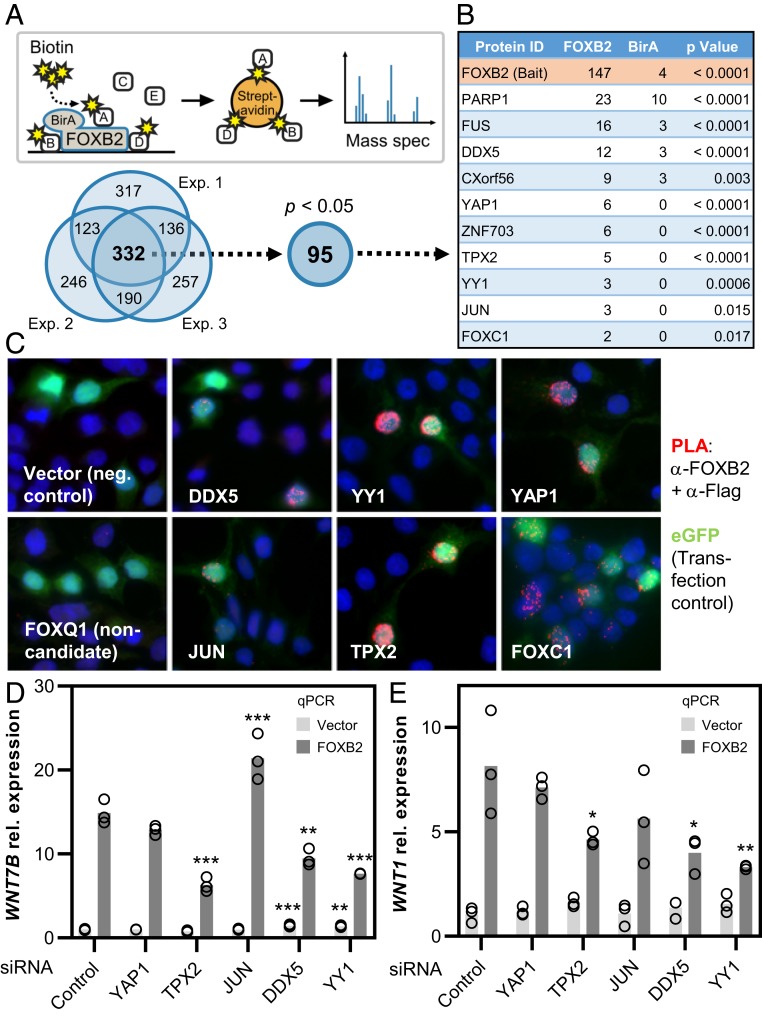
Our data so far suggested that FOXB2 interacts with other regulatory proteins. In order to identify possible FOXB2 interactors, we generated an N-terminal BirA–FOXB2 fusion construct for proximity labeling (25), which retained full activity in TOPflash (SI Appendix, Fig. S4 A–C). Mass spectrometry following streptavidin pull-down of pulse-labeled proteins revealed numerous candidates that were enriched compared to free BirA control, with high consistency across multiple experiments (Fig. 4A). Statistical analysis narrowed the initial list down to 95 high-confidence hits. These included several candidates that have previously been linked to Wnt signaling, such as YAP1 and JUN (Fig. 4B and Dataset S1). As expected, Gene Ontology analysis of high-confidence FOXB2 interactors showed that these proteins are primarily involved in the regulation of gene transcription and mRNA splicing (SI Appendix, Fig. S4D). In contrast, and in agreement with our earlier observations, we did not pull down β-catenin in any experiment.
Fig. 4.
Transcription coregulators contribute to FOXB2-dependent Wnt signaling. (A) Schematic summary of the BioID assay used to identify FOXB2 interactors. The Venn diagram below indicates the number of proteins identified in 3 independent BioID experiments performed in 293T cells after subtraction of BirA background. High-confidence FOXB2 interactors were analyzed further. (B) Partial list of candidate interactors (see also Dataset S1). Numbers indicate the average score from 3 experiments. (C) Validation of candidate interactors by in situ proximity ligation (PLA). Association of Flag-tagged proteins of interest with FOXB2 is indicated by red dots. (D and E) Expression of WNT7B (D) and WNT1 (E) in 293T cells following depletion of candidate FOXB2 interactors (*P < 0.05, **P < 0.01, and ***P < 0.001 versus control).
We first validated the BioID data by in situ proximity ligation (Fig. 4C and SI Appendix, Fig. S4E). Indeed, with the possible exception of JUN, all tested candidates strongly interacted with FOXB2 in this assay. Of note, the promoter regions of WNT7B and WNT1, as well as the WNT7B enhancer investigated here, contain numerous putative binding sites for candidate FOXB2 interactors, including YAP1, JUN, and YY1, and many of these sites overlap with predicted TCF/LEF motifs (SI Appendix, Fig. S5 A and B). We thus tested the functional role of candidate interactors in FOXB2-dependent Wnt signaling by RNA interference and coexpression (Fig. 4 D and E and SI Appendix, Fig. S6 A–E). Indeed, all tested candidates exerted broad, discrete effects primarily on FOXB2-dependent WNT1 and WNT7B expression, as well as WNT7B reporter activation and TOPflash activity. We conclude that FOXB2 interacts with a multiprotein transcriptional complex to promote Wnt gene transcription and TCF-dependent Wnt pathway activity.
FOXB2 Is Induced in Aggressive Prostate Cancer.
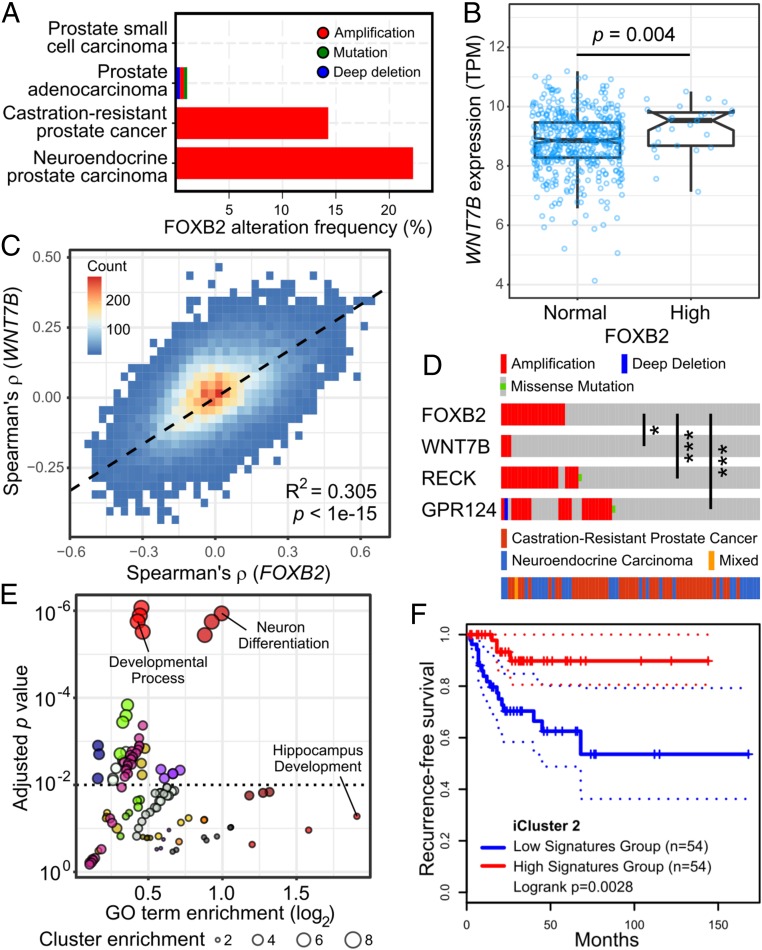
The only major expression site of mammalian FOXB2 is the developing brain, particularly the thalamus and hypothalamus (15). In adult mice, limited Foxb2 expression has been observed in some tissues, specifically the brain, thymus, ovary, and testis (26). Although analysis of public gene expression databases showed that FOXB2 levels are exceedingly low in normal tissues, we found that it is induced in some cancers, including thymomas, ovarian cancer, and testicular germ cell cancer (SI Appendix, Fig. S7A). In particular, we observed that FOXB2 transcript levels were frequently increased in prostate cancer. Here, FOXB2 amplification was detected mainly in aggressive, castration-resistant and neuroendocrine tumors (Fig. 5A). On the molecular level, highest FOXB2 expression was observed in the iCluster 2 prostate cancer subtype (27), which is predominantly characterized by ERG fusions, as well as PTEN and TP53 mutation (SI Appendix, Fig. S7B). Of note, these genetic lesions are individually associated with prostate cancer progression and poor prognosis, further suggesting that FOXB2 is specifically induced in aggressive cancers.
Fig. 5.
FOXB2 is induced in advanced prostate cancer. (A) FOXB2 genomic alterations across 14 prostate cancer studies curated in the cBioPortal for Cancer Genomics. FOXB2 is amplified specifically in advanced, castration-resistant and neuroendocrine cancers. (B) RNA-seq expression analysis in prostate adenocarcinoma (TCGA dataset PRAD) showed increased WNT7B levels in tumors with FOXB2 up-regulation. (C) Genome-wide correlation density plot in dataset PRAD. Data analysis showed a high degree of correlation between the WNT7B and FOXB2-associated transcriptome. (D) OncoPrint analysis of aggressive prostate cancers (39) revealed significant coamplification of FOXB2 and the WNT7B coreceptors RECK and GPR124, as well as WNT7B itself (*P < 0.05 and ***P < 0.001). (E) Gene Ontology (GO) analysis of the top 500 positively FOXB2/WNT7B-associated genes in prostate cancer (see also Dataset S2). GO terms were clustered based on functional relation, indicated by different colors. Individual GO terms of interest are highlighted. Only terms with a cluster enrichment score >2 are shown. (F) Kaplan–Meier survival plot in aggressive iCluster 2-type prostate cancer (27) based on the top 50 FOXB2/WNT7B negatively correlated genes (see also SI Appendix, Fig. S8 D and E). Dashed lines indicate the 95% CI.
Subsequent data analysis revealed that FOXB2 up-regulation in prostate cancer was associated with increased WNT7B levels (Fig. 5B), and that WNT7B transcript levels were highest in iCluster 2 tumors (SI Appendix, Fig. S7C). Importantly, genome-wide analysis of prostate cancer RNA-seq data showed a high degree of correlation between the FOXB2- and WNT7B-associated transcriptome, suggesting that they act in the same pathway in cancer (Fig. 5C). Consistent with this notion, the WNT7B receptors RECK and GPR124, and, to a lesser extent, WNT7B itself, are significantly coamplified with FOXB2 in castration-resistant and neuroendocrine prostate cancers (Fig. 5D). In contrast, FOXB1 and FOXA2 were not associated with increased WNT7B expression (SI Appendix, Fig. S7D).
Gene Ontology analysis of the most strongly FOXB2/WNT7B cocorrelated genes revealed that positively correlated genes are primarily involved in neurogenesis and cell migration (Fig. 5E and Dataset S2), which is consistent with the anticipated physiological role of Foxb2 in brain development (15). Conversely, negatively correlated genes mainly control small molecule metabolism and metal ion transport, required for normal prostate function (28) (SI Appendix, Fig. S7E and Dataset S2). Taken together, these data suggest that FOXB2, presumably via WNT7B, may regulate a transcriptional program involved in the neuronal differentiation of prostate cancer cells, a feature that is linked to cancer progression and poor prognosis (16).
In support of this hypothesis, we observed that both FOXB2 and WNT7B are associated with worse recurrence-free survival (RFS) in prostate cancer (SI Appendix, Fig. S8 A and B), albeit nonsignificantly in the case of FOXB2. Moreover, numerous FOXB2/WNT7B-correlated genes are individually associated with altered RFS in prostate cancer, and, remarkably, they cluster in a near-binary manner (SI Appendix, Fig. S8C): genes that are positively correlated with FOXB2 preferentially predict worse survival, whereas negatively correlated genes are linked to better survival. Indeed, we observed that a gene signature consisting of the top 50 FOXB2/WNT7B negatively correlated genes was strongly associated with improved RFS in iCluster 2-type prostate cancer (Fig. 5F and SI Appendix, Fig. S8 D and E), where FOXB2 and WNT7B expression peaks. We thus hypothesize that the FOXB2/WNT7B-associated prostate cancer transcriptome promotes disease progression by favoring malignant cell differentiation.
FOXB2 Drives Wnt Signaling and Neuroendocrine Differentiation in Prostate Cancer Cells.
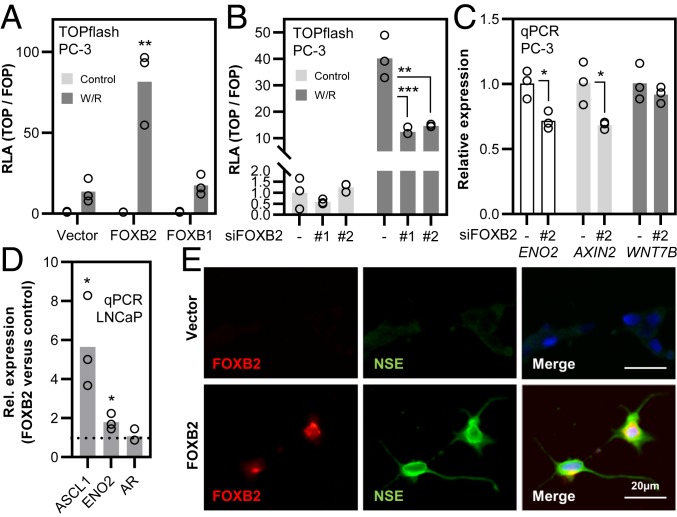
To test experimentally whether FOXB2 activates Wnt signaling in prostate cancer cells as well, we first assessed the regulation of all Wnt ligands in LNCaP and PC-3 cells, which resemble prostate adenocarcinoma and neuroendocrine (NE) prostate cancer, respectively (29). FOXB2 strongly induced the expression of multiple Wnts, and, indeed, the expression profiles largely followed the one observed in 293T cells (SI Appendix, Fig. S9A). Importantly, FOXB2 also significantly increased the expression of WNT7B in both cell lines, despite the very high basal levels of this ligand in LNCaP and PC-3 cells (14) (SI Appendix, Fig. S9B). Consistently, FOXB2 strongly activated TOPflash activity in both cell lines in synergy with Wnt3a/R-spondin 3, whereas FOXB1 had limited or no effect (Fig. 6A and SI Appendix, Fig. S10A). Conversely, depletion of FOXB2 in PC-3 using 2 separate siRNAs significantly inhibited Wnt/R-spondin induced Wnt signaling (Fig. 6B).
Fig. 6.
FOXB2 induces Wnt activity and neuroendocrine differentiation in prostate cancer cells. (A) TOPflash assay in PC-3 cells. FOXB2, but not FOXB1, synergized with Wnt3a/R-spondin 3 (W/R) in pathway activation. (B) TOPflash assay in PC-3 cells following depletion of FOXB2 with 2 independent siRNAs. Scrambled siRNA was used as control. (C) qPCR expression analysis of ENO2 (encoding neuron-specific enolase; NSE), AXIN2, and WNT7B in PC-3 cells following depletion of FOXB2. (D) Expression analysis of androgen receptor (AR) and neuroendocrine differentiation markers ASCL1 and ENO2 in LNCaP cells. FOXB2 was over-expressed transiently, and data were normalized to empty vector control. (E) Immunofluorescence staining of NSE and FOXB2 in stably transfected LNCaP cells following G418 selection for more than 2 wk. FOXB2-overexpressing cells exhibited high levels of NSE protein. Nuclei are shown in blue (*P < 0.05, **P < 0.01, and ***P < 0.001 versus control).
Wnt pathway activation in prostate cancer has been linked to NE differentiation (30, 31), which is associated with exceptionally poor prognosis. FOXB2 knock-down in PC-3 cells reduced AXIN2 expression concomitantly with ENO2, encoding the prototypical NE marker neuron-specific enolase (NSE), whereas the effect on WNT7B levels was marginal (Fig. 6C). Conversely, FOXB2 over-expression in LNCaP cells significantly increased the expression of NE markers ASCL1 and ENO2 (Fig. 6D). Since these results suggested that FOXB2 controls the malignant differentiation of prostate cancer cells, we additionally assessed changes in LNCaP morphology following FOXB2 expression. Transient transfection of FOXB2 for up to 4 d had no apparent effect on the phenotype of LNCaP cells, although we noticed a slight increase in NSE protein levels in FOXB2-positive cells at this time point (SI Appendix, Fig. S10 B and C). In contrast, we observed a striking morphological change in LNCaP cells that stably expressed FOXB2 for several weeks (Fig. 6E and SI Appendix, Fig. S10B). FOXB2-expressing cells adopted a neuron-like morphology with compact body and very long protrusions. Additionally, these cells stained strongly for NSE compared to controls, indicative of bona fide NE differentiation (Fig. 6E).
Discussion
The key conclusions from our study are (i) that FOXB2, an uncharacterized protein, is a potent regulator of Wnt ligand expression and TCF signaling; and (ii) that FOXB2 drives the neuroendocrine differentiation of prostate cancer cells. Our results thus add FOXB2 to the growing list of FOX transcription factors involved in Wnt pathway regulation and pathobiology. FOX proteins constitute an evolutionarily conserved transcription factor family with 43 members in humans, which play critical roles in development and tissue homeostasis (24, 32). Consistently, dysregulation of FOX signaling is a common feature of major human diseases, notably cancer. Despite this, the function of many FOX proteins, including FOXB2, remains poorly understood.
FOX transcription factors affect tumorigenesis in part by controlling Wnt signaling, which is a key oncogenic pathway in various cancers, including prostate cancer (1, 17); however, it is unclear to what extent FOX transcription factors control Wnt activity in these diseases. Our results show that FOXB2 levels are increased in aggressive prostate cancer, and that FOXB2 activates the Wnt pathway via induction of agonistic ligands, primarily WNT7B. WNT7B expression in prostate cancer has been linked to tumor growth and drug resistance (9, 10), and may thus contribute to malignant cancer progression. Interestingly, Zheng et al. (10) reported that WNT7B is a direct transcriptional target of the androgen receptor (AR). AR expression is frequently lost in advanced prostate cancer, particularly neuroendocrine prostate cancer (33). FOXB2 induction in these aggressive tumors might maintain high WNT7B levels in the absence of androgen signaling, and thereby drive cancer cell differentiation and treatment resistance. We note, however, that both canonical and noncanonical Wnt signaling modes have been implicated in prostate cancer neuroendocrine differentiation (30, 31), and FOXB2 may engage both pathways in parallel through induction of multiple Wnt ligands.
A major unresolved question in the field is how the expression of Wnt ligands is controlled at the transcriptional level (34). Our data suggest that FOXB2 is a promiscuous Wnt activator, in that it induces at least 13 of the 19 human Wnt ligands in different epithelial cell lines. How FOXB2 achieves this broad effect is currently unclear, although we have reason to believe that it acts as a molecular hub for various transcription (co)factors that synergistically drive Wnt expression. Indeed, experimental and in silico analyses of, for example, the mouse Wnt7b promoter show that this region is highly enriched for numerous transcription factor binding sites, including forkhead box motifs (11, 35). Thus, we consider it worthwhile to further investigate the DNA and protein binding properties of FOXB2.
With regard to developmental biology, prior studies suggest that there is substantial overlap between Foxb2, Wnt7b, and Axin2 expression domains in the developing brain, particularly in the early thalamus (15, 36, 37). During embryogenesis, WNT7B signaling is primarily involved in angiogenesis and the formation of the blood–brain barrier, which it controls redundantly with WNT7A (38). Our data show that FOXB2 strongly induces WNT7B as well as WNT7A at least in epithelial cells. Thus, FOXB2 may act as a rheostat of Wnt signaling activity and angiogenesis in the developing brain.
In summary, we identify the cryptic forkhead box transcription factor FOXB2 as a potent activator of Wnt/TCF signaling in normal and cancer cells. Given the putative roles of FOXB2 in neurogenesis and the neuroendocrine differentiation of advanced prostate cancer, it is possible that FOXB2 controls both processes through a common, potentially WNT7B-dependent signaling mode, which is aberrantly reactivated in cancer. However, it remains to be formally established whether the malignant cancer cell differentiation induced by FOXB2 is indeed driven by Wnt/WNT7B signaling. Nonetheless, we believe that further exploration of the FOXB2/WNT7B-associated transcriptome may uncover new therapeutic vulnerabilities in rare, aggressive cancers.
Methods Summary
Cell Culture.
Authenticated 293T, HCT116, SW48, LNCaP, PC-3, L, and L/Wnt3a cells were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ), the American Type Culture Collection (ATCC), and the European Collection of Authenticated Cell Cultures (ECACC). 293T ΔLRP6 cells were generated by CRISPR-Cas9–mediated gene editing. 293T ΔCTNNB1 and ΔTCF/LEF cells have been described elsewhere (21). All experiments were performed using low-passage cells from confirmed mycoplasma-free frozen stocks, as determined by analytical qPCR (Eurofins Genomics).
Molecular Cloning.
Expression constructs were generated by restriction cloning of full-length cDNA from in-house epithelial cell libraries into a pCS2+ vector with N-terminal Flag or V5 tag. For stable expression, cDNAs were subcloned into a pcDNA3 vector, and transfectants were isolated by G418 selection. Point and truncation mutants were generated by restriction cloning and PCR-based mutagenesis. All plasmids were validated by partial sequencing (Eurofins Genomics).
BioID and Mass Spectrometry.
The BioID assay was performed essentially as described (25). Briefly, N-terminal BirA-FOXB2 and BirA plasmid were transfected into 293T cells using Lipofectamine 2000 (Thermo Fisher). After transfection, cells were treated with 50 µM biotin. Biotinylated proteins were precipitated with streptavidin beads (GE Healthcare) and digested using spectrometry-grade trypsin (Thermo Fisher). BioID samples were analyzed by mass spectrometry using an Easy nano LC II HPLC interfaced with a nanoEasy spray ion source (Thermo Fisher) connected to an Orbitrap Velos Pro mass spectrometer (Thermo Fisher).
Public Dataset Analyses.
The results in this study are in large part based upon data generated by the TCGA Research Network (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). Dataset analyses were performed primarily in the cBioPortal for Cancer Genomics, GEPIA 2, and DAVID, with additional analyses and visualization done in R 3.6.1 (R Foundation for Statistical Computing).
Reproducibility and Statistical Analyses.
All experimental data represent at least 2 independent experiments with comparable results. Assays were performed with 3 biological replicates. Bar graphs display the group mean with individual data points. Assay-specific controls at identical concentrations were included in all experiments. A detailed report on statistical methods and software packages can be found in the SI Appendix, Table S2. For better readability, only the most relevant statistical results are indicated in the figure panels. All other analyses are shown in Dataset S3.
Data Availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (40) partner repository with the dataset identifier PXD015725 and 10.6019/PXD015725. Public dataset analyses can be found in supplemental Dataset S2.
Supplementary Material
Acknowledgments
The authors thank Drs. Christof Niehrs, Claudio Cantù, and Lennart Svensson for cell lines and reagents. We also thank all investigators who have made materials and data available through public repositories. Technical support from the microscopy and mass spectrometry core facilities at Linköping University is gratefully acknowledged. S.K. is a Wallenberg Molecular Medicine fellow and receives financial support from the Knut and Alice Wallenberg Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906484116/-/DCSupplemental.
References
- 1.Nusse R., Clevers H., Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 2.MacDonald B. T., Tamai K., He X., Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 17, 9–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch S., Extrinsic control of Wnt signaling in the intestine. Differentiation 97, 1–8 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Alok A., et al. , Wnt proteins synergize to activate β-catenin signaling. J. Cell Sci. 130, 1532–1544 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Vallon M., et al. , A RECK-WNT7 receptor-ligand interaction enables isoform-specific regulation of wnt bioavailability. Cell Rep. 25, 339–349.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanhollebeke B., et al. , Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/β-catenin pathway during brain angiogenesis. eLife 4, e06489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y., Nathans J., Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Dev. Cell 31, 248–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhnert F., et al. , Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science 330, 985–989 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto D. T., et al. , RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng D., et al. , Role of WNT7B-induced noncanonical pathway in advanced prostate cancer. Mol. Cancer Res. 11, 482–493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weidenfeld J., Shu W., Zhang L., Millar S. E., Morrisey E. E., The WNT7b promoter is regulated by TTF-1, GATA6, and Foxa2 in lung epithelium. J. Biol. Chem. 277, 21061–21070 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Brynczka C., Merrick B. A., The p53 transcriptional target gene wnt7b contributes to NGF-inducible neurite outgrowth in neuronal PC12 cells. Differentiation 76, 795–808 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi M., Sato K., Nomura T., Osumi N., Manipulating gene expressions by electroporation in the developing brain of mammalian embryos. Differentiation 70, 155–162 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Zhu H., et al. , Analysis of Wnt gene expression in prostate cancer: Mutual inhibition by WNT11 and the androgen receptor. Cancer Res. 64, 7918–7926 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Kaestner K. H., Schütz G., Monaghan A. P., Expression of the winged helix genes fkh-4 and fkh-5 defines domains in the central nervous system. Mech. Dev. 55, 221–230 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Parimi V., Goyal R., Poropatich K., Yang X. J., Neuroendocrine differentiation of prostate cancer: A review. Am. J. Clin. Exp. Urol. 2, 273–285 (2014). [PMC free article] [PubMed] [Google Scholar]
- 17.Murillo-Garzón V., Kypta R., WNT signalling in prostate cancer. Nat. Rev. Urol. 14, 683–696 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Higashimori A., et al. , Forkhead box F2 suppresses gastric cancer through a novel FOXF2-IRF2BPL-β-catenin signaling axis. Cancer Res. 78, 1643–1656 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Peng X., et al. , FOXQ1 mediates the crosstalk between TGF-β and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol. Ther. 16, 1099–1109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang N., et al. , FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell 20, 427–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doumpas N., et al. , TCF/LEF dependent and independent transcriptional regulation of Wnt/β-catenin target genes. EMBO J. 38, e98873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagati A., et al. , Melanoma suppressor functions of the carcinoma oncogene FOXQ1. Cell Rep. 20, 2820–2832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C., Tucker P. W., DNA-binding properties and secondary structural model of the hepatocyte nuclear factor 3/fork head domain. Proc. Natl. Acad. Sci. U.S.A. 90, 11583–11587 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myatt S. S., Lam E. W., The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 7, 847–859 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaestner K. H., et al. , Six members of the mouse forkhead gene family are developmentally regulated. Proc. Natl. Acad. Sci. U.S.A. 90, 7628–7631 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network , The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costello L. C., Franklin R. B., The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: Connecting the dots. Mol. Cancer 5, 17 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai S., et al. , PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate 71, 1668–1679 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uysal-Onganer P., et al. , Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol. Cancer 9, 55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X., et al. , A human- and male-specific protocadherin that acts through the wnt signaling pathway to induce neuroendocrine transdifferentiation of prostate cancer cells. Cancer Res. 65, 5263–5271 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Golson M. L., Kaestner K. H., Fox transcription factors: From development to disease. Development 143, 4558–4570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonkhoff H., Neuroendocrine differentiation in human prostate cancer. Morphogenesis, proliferation and androgen receptor status. Ann. Oncol. 12 (suppl. 2), S141–S144 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Tian Q., Jin H., Cui Y., Guo C., Lu X., Regulation of Wnt gene expression. Dev. Growth Differ. 47, 273–281 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Kandyba E., Kobielak K., Wnt7b is an important intrinsic regulator of hair follicle stem cell homeostasis and hair follicle cycling. Stem Cells 32, 886–901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimogori T., et al. , A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767–775 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bluske K. K., Kawakami Y., Koyano-Nakagawa N., Nakagawa Y., Differential activity of Wnt/beta-catenin signaling in the embryonic mouse thalamus. Dev. Dyn. 238, 3297–3309 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenman J. M., et al. , Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science 322, 1247–1250 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Beltran H., et al. , Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moparthi L., et al. , Mass spectrometry analysis of FOXB2 interactors using BioID. ProteomeXchange. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD015725. Deposited 6 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (40) partner repository with the dataset identifier PXD015725 and 10.6019/PXD015725. Public dataset analyses can be found in supplemental Dataset S2.