Significance
Cells interact with their environment using filopodia, thin membrane protrusions supported by actin filaments. Filopodia formation in evolutionarily divergent organisms requires force generation by a MyTH4-FERM (MF) myosin that binds membrane proteins and is designed to promote cytoskeletal reorganization and filopodial transport. The amoeboid filopodial MF myosin is found to be a resilient motor in filopodia formation. Its activity and regulation appear to have emerged by multidomain coevolution for optimized control of filopodia formation and stability. Its properties reflect those of the ancestral MF myosin that gave rise to the metazoan MF myosins. These findings have implications for understanding the fundamental principles of how filopodia form and how MF myosins function in phylogenetically distant organisms.
Keywords: myosin, filopodia, actin, MyTH4-FERM
Abstract
Filopodia are actin-filled protrusions employed by cells to interact with their environment. Filopodia formation in Amoebozoa and Metazoa requires the phylogenetically diverse MyTH4-FERM (MF) myosins DdMyo7 and Myo10, respectively. While Myo10 is known to form antiparallel dimers, DdMyo7 lacks a coiled-coil domain in its proximal tail region, raising the question of how such divergent motors perform the same function. Here, it is shown that the DdMyo7 lever arm plays a role in both autoinhibition and function while the proximal tail region can mediate weak dimerization, and is proposed to be working in cooperation with the C-terminal MF domain to promote partner-mediated dimerization. Additionally, a forced dimer of the DdMyo7 motor is found to weakly rescue filopodia formation, further highlighting the importance of the C-terminal MF domain. Thus, weak dimerization activity of the DdMyo7 proximal tail allows for sensitive regulation of myosin activity to prevent inappropriate activation of filopodia formation. The results reveal that the principles of MF myosin-based filopodia formation are conserved via divergent mechanisms for dimerization.
Filopodia are specialized cellular projections that detect extracellular cues, facilitate adhesion, and aid in directed cell migration (1). They are important for processes ranging from neuronal pathfinding to cancer cell metastasis (2, 3). Parallel bundles of actin support these thin membrane projections that extend from the dendritic actin network at the cell cortex. The structure and function of filopodia has been established, but the molecular mechanism of filopodia initiation is not fully understood. Specifically, it is not yet clear how the cortical actin network at the membrane is locally reorganized into the parallel actin bundles that form nascent filopodia. A core set of conserved proteins including actin bundling proteins and an unconventional myosin are required to make filopodia (4). A favored model is that an activated small GTPase recruits key actin regulators such as formins and VASP at a filopodia initiation site. Actin filaments then converge into a parallel array, and actin polymerization drives filopodia elongation (5). MyTH4-FERM myosins (myosin tail homology–band 4.1, ezrin, radixin, moesin; MF) are molecular motors found at filopodia tips and are necessary for filopodia formation across cell types (6, 7), but the action of MF myosins in the process of filopodia initiation remains poorly understood.
The MF myosins are a group of unconventional myosins with roles in the formation and function of parallel actin-based structures such as filopodia, stereocilia, and microvilli (7–11). They are characterized by the presence of a MF domain(s) in the C-terminal tail. These domains mediate interactions with binding partners and microtubules (e.g., refs. 12–15). The amoeboid MF myosin DdMyo7 and the phylogenetically distant metazoan MF myosin Myo10 are essential for the initiation of filopodia (6, 7). Myo10 is proposed to organize the actin network at initiation sites and/or transport the actin regulator VASP toward the tip during filopodia formation (16–18). These actions of bundling and transport require a dimeric myosin. The metazoan Myo10 monomer is autoinhibited by head–tail interaction, as is typical for many myosins (19, 20). Dimerization is induced by PI(3,4,5)P3-mediated membrane binding that promotes formation of an antiparallel dimer via a region in the proximal tail (20–24). This arrangement confers increased distance between motor heads, gives the molecule flexible reach, and facilitates walking on bundled actin filaments in filopodia cores (23).
Filopodia are found in a diverse array of organisms, and many proteins in the “filopodia toolkit” are paneukaryotic (4). MF myosins are ancient—they arose prior to the Amoebozoa/Holozoa split and are present in unicellular and multicellular Holozoa and Fungi, but Myo10 is notably only found in Holozoa (4, 25). The amoebozoan MF myosin, DdMyo7, and the metazoan Myo10 have evolved independently across ∼600 Ma, yet both have essential roles in filopodia formation, whereas other MF myosins evolved to have roles in the formation and maintenance of microvilli and stereocilia (11, 26). It is not known whether the 2 distinct filopodial myosins employ unique mechanisms to generate filopodia. An understanding of the features employed by the divergent amoeboid MF myosin to form filopodia can provide insight into the conservation or divergence of the mechanism of MF myosin-based filopodia formation. In particular, while DdMyo7 appears to be regulated by head–tail autoinhibition, it lacks a predicted coiled-coil dimerization region in the proximal tail, thus raising the question: Did proximal dimerization evolve to tune the specialized function of metazoan Myo10, or is dimerization an ancient feature of filopodia motors? A functional dissection of the amoebozoan DdMyo7 filopodia myosin was undertaken to address this issue.
Results and Discussion
Autoinhibition Regulates DdMyo7 Activity.
DdMyo7 is essential for the formation of filopodia in Dictyostelium discoideum (7). The first step in this process is the recruitment of the MF myosin to the cell membrane or underlying actin cortex. Myo10 is recruited and dimerized following PH domain-mediated interaction with PI(3,4,5)P3 that promotes both activation, by releasing motor/tail autoinhibition, and dimerization, which is required for activity (20, 22). DdMyo7 lacks both a clear lipid-binding motif and identifiable coiled-coil domain that could mediate dimerization, raising the question of how this myosin is targeted and activated.
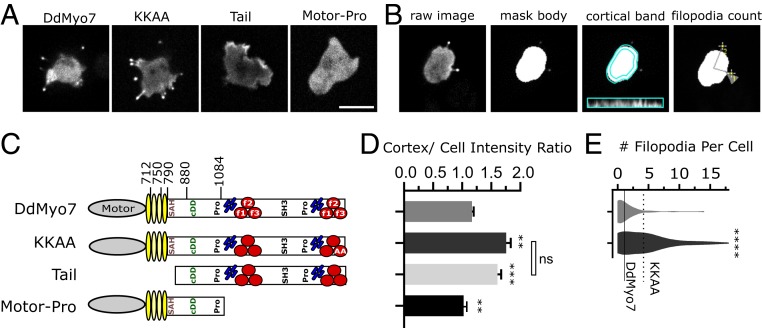
DdMyo7 switches between an autoinhibited state and an active state that is controlled via a basic patch in the C-terminal tail that allosterically inhibits motor activity (26). This motif is found among widely divergent Myo7s (26, 27), and autoinhibition provides spatial and temporal control of motor activity. Domain deletion of DdMyo7 could disrupt the conformation necessary for head–tail autoinhibition, and mutant motors were assessed for both their ability to rescue filopodia formation and extent of efficient targeting (that would increase in case of lack of autoinhibition). GFP-tagged full-length DdMyo7 and mutants were expressed in DdMyo7 null cells (myo7−) (SI Appendix, Fig. S1), and fluorescence intensity at the cortex was measured using automated image analysis (Fig. 1 B and C and ref. 26). GFP-DdMyo7 is enriched at the cortex over the cytoplasm by a ratio of ∼1.2. The motor-Pro fragment is not targeted (Fig. 1 A and D), while the DdMyo7 tail fragment has higher cortical intensity than the wild-type DdMyo7 (a ratio of 1.61 ± 0.06 vs. 1.17 ± 0.03), consistent with a lack of the head–tail interaction leading to increased cortical recruitment (Fig. 1C). The link between autoinhibition and cortical targeting is supported by the finding that mutating key residues for autoinhibition (KKAA; K2333A, K2336A) results in increased cortical targeting (cortical intensity, 1.76 ± 0.08; Fig. 1D). The autoinhibition mutant KKAA also shows a larger percentage of cells making at least 1 filopodia (DdMyo7, 58.3%; KKAA, 78.8%) and more filopodia per cell (Table 1, Fig. 1E, and ref. 26) consistent with autoinhibition via tail/motor interaction controlling targeting and release of autoinhibition leading to increased targeted MF myosin and more filopodia formation.
Fig. 1.
Recruitment to the cortex and release of head–tail autoinhibition promotes filopodia formation. (A) Micrographs of D. discoideum cells expressing GFP-tagged DdMyo7 constructs expressed in myo7 null cells. Note the concentration of GFP-DdMyo7 and the KKAA autoinhibition mutant in the filopodia tip and distinct cortical enrichment of the GFP tail. (Scale bar: 10 μm.) (B) Illustration of the quantitative image analysis pipeline (adapted from ref. 26). Cells expressing GFP-DdMyo7 fusions are analyzed by identifying fluorescent cell bodies and filopodia tips then registering those tips to cells to calculate average number of filopodia per cell. The fluorescence intensity of a 0.8-µm band around the cell periphery is measured and compared to the rest of the cell body to measure cortical targeting. Only cells expressing the fluorescent protein are included in analysis by thresholding. (C) Schematic illustration of DdMyo7 and fragments. N-terminal motor domain (gray oval), light chain binding IQ motifs (yellow ovals), stable α-helix (SAH), candidate dimerization domain (cDD) 2 proline-rich regions (Pro), MyTH4 domains (blue rods), FERM domains (red circles), and a src-homology 3 domain (SH3). Amino acid numbers are indicated above. (D) Graph of cortical band intensity. (E) Violin plot of number of filopodia per cell. Cells expressing the tail or motor fragments lack filopodia and are excluded. (C and D) DdMyo7 and KKAA means are represented by horizontal solid and dashed lines, respectively, for comparison. Significance indicators from Tukey’s test are in comparison to DdMyo7 control, unless otherwise indicated on the graph: ns, not significant; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Table 1.
Quantification of filopodia formation by DdMyo7 mutants in myo7 − cells
| Region | Name | % Cells with filopodia | Filopodia per cell ± SEM | Cortex/cell intensity ratio ± SEM | Filopod length ± SEM, μm | Tip/cell intensity ratio ± SEM | Tip accumulation | n (cells) | N (experiments) | Autoinhibition dysregulated | Activity in filopodia formation |
| DdMyo7 | 58 | 2.26 ± 0.32 | 1.17 ± 0.03 | 3.05 ± 0.12 | 1.86 ± 0.16 | No | 84 | 4 | No | WT | |
| KKAA | 79 | 5.32 ± 0.37 | 1.76 ± 0.08 | 2.69 ± 0.08 | 3.05 ± 0.12 | No | 130 | 3 | Yes | More than WT, due to increase in active form | |
| Motor-Pro | 6 | N/A | 1.02 ± 0.01 | N/A | N/A | N/A | 206 | 4 | Yes | None (lacks required domains) | |
| Motor-Pro-CC | 11 | 1.32 ± 0.14 | 1.38 ± 0.02 | 2.14 ± 0.27 | 1.23 ± 0.18 | No | 181 | 7 | Yes | More targeted, but less active; forms shorter filopodia than WT | |
| Proximal tail | ΔcDD | 59 | 2.52 ± 0.27 | 1.49 ± 0.07 | 3.0 ± 0.16 | 3.15 ± 0.19 | No | 94 | 2 | Yes | More in active form, but not efficient to lack of proximal dimerization |
| ΔPro1 | 51 | 2.49 ± 0.25 | 1.23 ± 0.06 | 2.8 ± 0.18 | 1.5 ± 0.17 | — | 68 | 5 | No | WT | |
| ΔSAH-PLA | 40 | 2.29 ± 0.32 | 2.16 ± 0.17 | 3.1 ± 0.24 | 5.08 ± 0.58 | Yes | 78 | 3 | Yes | Active, less efficient (LA and distal oligomerization sufficient for activity) | |
| MF deletions | ΔMF1-SH3 | 58 | 3.20 ± 0.27 | 1.33 ± 0.04 | 2.38 ± 0.11 | 2.03 ± 0.13 | No | 203 | 5 | Yes | Less active due to less targeting |
| ΔSAH-SH3 | 25 | 1.60 ± 0.13 | 1.26 ± 0.05 | 3.1 ± 0.17 | 2.49 ± 0.19 | Yes | 219 | 4 | Yes | Less efficient in filopodia formation (poor targeting + no proximal dimerization) | |
| ΔPro2-MF2 | 35 | 3.18 ± 0.40 | 1.38 ± 0.08 | 2.7 ± 0.23 | 2.99 ± 0.35 | No | 82 | 5 | Yes | Less active due to less targeting | |
| ΔcDD/MF2 | 3 | N/A | 0.98 ± 0.02 | N/A | N/A | N/A | 84 | 4 | Yes | None (need proximal dimerization when targeting/distal oligomerization is less efficient) | |
| ΔSAH-PLA/MF2 | 1 | N/A | 1.08 ± 0.02 | N/A | N/A | N/A | 165 | 6 | Yes | None (need proximal dimerization when targeting/distal oligomerization is less efficient) | |
| Lever arm | Δ2IQ | 68 | 3.18 ± 0.40 | 1.43 ± 0.06 | 3.8 ± 0.18 | 3.31 ± 0.18 | No | 95 | 4 | Yes | More in active form, less efficient due to shorter lever arm |
| Δ3IQ-PLA | 14 | 1.17 ± 0.17 | 1.70 ± 0.14 | N/A | N/A | N/A | 46 | 3 | Yes | More in active form, less efficient due to short LA and proximal dimerization | |
| Δ2IQ/SAH-PLA | 0 | N/A | 1.51 ± 0.03 | N/A | 4.32 ± 0.26* | No* | 179 | 2 | Υes | None (short lever arm and no proximal dimerization) |
WT, wild type.
WT background.
The Proximal Tail Contributes to Optimal DdMyo7 Activity but Is Not Essential for Either Activity or Autoinhibition.
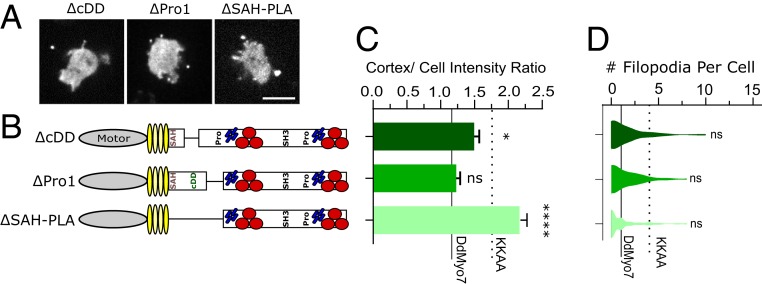
The DdMyo7 proximal tail region, i.e., post lever arm (PLA) (residues 830 to 1117), follows the catalytic motor domain, the lever arm (LA), and a single α-helix (SAH 798 to 830) that extends the LA (without mediating dimerization) (28–30) and encompasses the region up to the MyTH-FERM1 domain. The DdMyo7 PLA consists of a region possibly adopting a folded structure (N-PLA) followed by a proline-rich domain (Pro1, residues 1020 to 1084). Secondary structure prediction tools indeed suggested the PLA was unlikely to form a coiled-coil (31), but its N-terminal sequence (residues 845 to 1000) has a hydrophobic and charged amino acid content compatible with forming a folded structural domain (32), which could possibly permit dimerization or self-association. Sequence alignment of amoeba Myo7s revealed such a conserved candidate dimerization domain (cDD) (residues 884 to 952). The proline-rich (Pro1) region, on the other hand, is a candidate for adaptor binding (33, 34). GFP-DdMyo7 rescues filopodia formation in the myo7 null cells and the myosin is enriched at filopodia tips (26). Deletion of the candidate dimerization domain (ΔcDD) or proline-rich region (ΔPro1) did not affect filopodia formation (meaning fraction of cells making filopodia and filopodia/cell), but the cDD has increased cortical targeting (P < 0.03) (Fig. 2 and Table 1).
Fig. 2.
The proximal tail regulates DdMyo7 activity. (A) Micrographs of D. discoideum cells expressing GFP-tagged deletion constructs in myo7 null cells. (Scale bar: 10 μm.) (B) Schematic illustration of constructs. (C) Cortical band intensity of proximal tail deletion mutants. (D) Violin plot of the distribution of filopodia per cell. (C and D) DdMyo7 and KKAA means are represented by horizontal solid and dashed lines, respectively, for comparison. Significance indicators are in comparison to DdMyo7 control by Dunnett’s multiple-comparison test: ns, not significant; *P < 0.05; ****P < 0.0001.
Interestingly, a DdMyo7 mutant with part of the LA deleted in addition to the proximal tail (ΔSAH-PLA) exhibits increased cortical localization, exceeding that seen for the autoinhibition (KKAA) mutant (Fig. 2C). Mutants are typically compared to control DdMyo7, but it is informative to also compare them to the autoinhibition mutant to evaluate the protein function relative to another uninhibited myosin. Despite this significant cortical enrichment, the ∆SAH-PLA cells do not produce an excess of filopodia suggesting that the overall activity of the ΔSAH-PLA motor to promote filopodia formation is diminished (Fig. 2 C and D; dashed line shows KKAA value). This reveals the role of the SAH-PLA region in optimization of filopodia formation in addition to stabilizing the autoinhibited conformation of DdMyo7. It also suggests that proximal dimerization or targeting via this domain is not absolutely required for filopodia initiation, as it has been proposed to be the case for Myo10 (20): The motor either functions as a monomer to initiate filopodia or other regions of the DdMyo7 tail collaborate to mediate dimerization.
The Proximal Tail Has a Dominant-Negative Effect on Filopodia and Promotes Weak Dimerization.
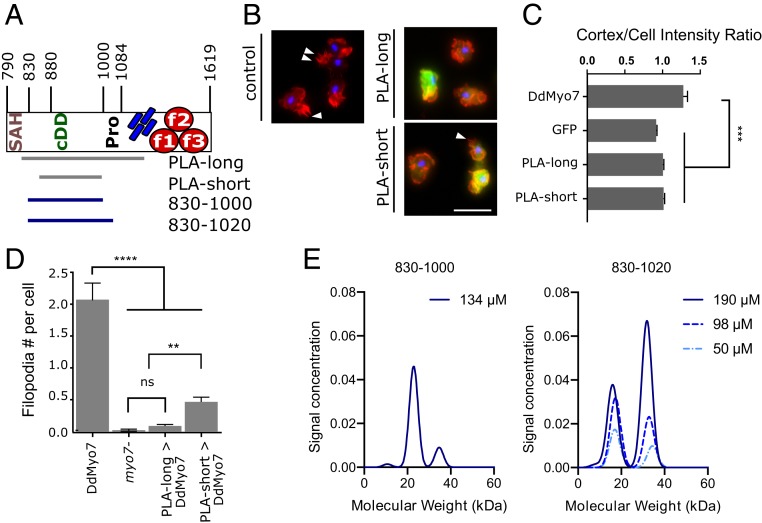
If the proximal tail region promotes dimerization or self-association, then filopodia formation could be inhibited by overexpression of this fragment, as has been shown for Myo10 (9). GFP fusions of a long or short fragment of the PLA (amino acids [aa] 809 to 1154 and aa 848 to 1000, respectively; Fig. 3A) were expressed in wild-type cells. Both the long and short PLA fragments are excluded from filopodia tips, and do not localize to the cell cortex in wild-type cells and therefore do not promote targeting of the myosin (Fig. 3 B and C). Expression of either fragment disrupted filopodia formation (actin projections, >1 μm) of wild-type cells (Fig. 3D). These results cannot distinguish between whether the PLA associates directly with endogenous DdMyo7 in vivo to poison a functional dimeric motor, or whether it plays an inhibitory role by preventing binding and sequestering an (unknown) partner.
Fig. 3.
The proximal tail domain has a dominant-negative effect on filopodia and weakly dimerizes in solution. (A) Schematic of proximal tail region. Numbers represent amino acid number in the full protein. (B) Representative micrographs of AX2 (control) D. discoideum cells fixed and stained with phalloidin (actin; red) and DAPI (nuclei; blue). (Left) Control untransformed cells. The white arrowheads point to filopodia. (Right) GFP-PLAlong fragment or GFP-PLAshort expression in wild-type cells. (Scale bar: 10 µm.) (C) Cortical band intensity of cells expressing either GFP-DdMyo7, GFP alone, or 1 of 2 different GFP-PLA fragments. (D) Graph of phalloidin-stained filopodia per cell. (E) Graph of molecular-weight distribution of PLA830-1000 (single-concentration run) or PLA830-1000 (3 increasing concentrations) by sedimentation velocity analytical centrifugation. Significance indicators: ns, not significant; **P < 0.01; ***P < 0.001.
Sedimentation velocity analytical ultracentrifugation (AUC) was performed on purified fragments of the PLA to test their ability to dimerize directly. Two N-PLA fragments were tested, PLA830-1000 and PLA830-1020. Interestingly, the shorter fragment is primarily monomeric; however, the longer fragment exhibits concentration-dependent dimerization (Fig. 3E), with a significant fraction of PLA830-1020 shifting into a higher molecular-weight species at 100 to 200 µM. The failure of the shorter fragment to dimerize could suggest that the additional C-terminal sequences are required to stabilize the structural domain or the dimer. These results indicate that the PLA can self-associate, consistent with proximal dimerization of DdMyo7 that is weak and context dependent.
The Distal Tail of DdMyo7 Is Critical to Promote Filopodia Length and Number.
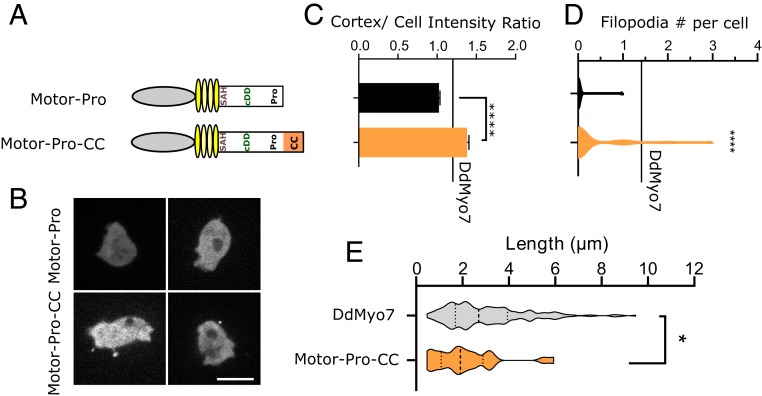
Models have been proposed for how dimeric and monomeric motors such as Myo10 and Myo1b would contribute to filopodia initiation and stabilization. For a dimeric motor with proximal dimerization, the mechanism underlying filopodia initiation could be the reorganization of the actin network by the motor reaching out to different actin filaments (18, 23). The monomeric plus-end motor Myo1b has been proposed to function by stabilizing parallel actin bundles along the filopodia membrane (35). Whether dimerization of the DdMyo7 would be sufficient for function was tested by the addition of an exogenous dimerization region to the C terminus of the DdMyo7 motor-Pro fragment (Fig. 1 C–E). The coiled-coil sequence from mouse Myo5a followed by the leucine zipper GCN4 were added to generate a stable dimeric DdMyo7 motor (motor-Pro-CC; Fig. 4A). The chimera is stably expressed and highly enriched at the cortex, exceeding the full-length DdMyo7 (Fig. 4 B and C and SI Appendix, Fig. S1D), and it restores filopodia formation, unlike motor-Pro (Fig. 4D), showing that DdMyo7 can function as a dimer. It should be noted that, while significant, the rescue is relatively weak with a smaller fraction of cells (11% vs. 60% for DdMyo7) making a reduced number of filopodia (1.32 ± 0.14 per cell vs. 2.26 ± 0.32 per cell for DdMyo7). Interestingly, unlike any of the other DdMyo7 deletion mutants, filopodia generated by motor-Pro-CC are shorter than wild-type filopodium (Fig. 4E and Table 1; 2.14 ± 0.27 vs. 3.05 ± 0.13 µm).
Fig. 4.
Filopodia formation promoted by a forced dimer of the motor. (A) Schematic illustration of the motor-Pro1 region or forced dimer (CC). (B) Localization of the motor-Pro1 and motor-Pro1-CC motors in myo7 null cells. (Scale bar: 10 µm.) (C) Cortical intensity of expressed motor fragments. Motor-Pro-CC also enriched relative to DdMyo7 control (***, horizontal line). (D) Violin plot of filopodia per cell. The horizontal line represents the mean of DdMyo7 control. (E) Measurement of filopodia length of motor-Pro1-CC compared to the control DdMyo7. Significance indicators from Tukey’s test are in comparison to DdMyo7 control, unless otherwise noted: ns, not significant; *P < 0.05; ****P < 0.0001.
The ability of the motor-Pro-CC forced dimer to promote filopodia formation is consistent with previous studies showing that initiation of filopodia occurs when a dimeric tailless Myo10 motor presumably brings actin filament barbed ends close together adjacent the membrane (18). In the case of Myo10, it is not known how effectively the forced dimer promotes filopodia formation compared to full-length Myo10. The weak rescue by motor-Pro1-CC shown here demonstrates that the full-length DdMyo7 requires the distal tail to efficiently initiate and fully extend filopodia.
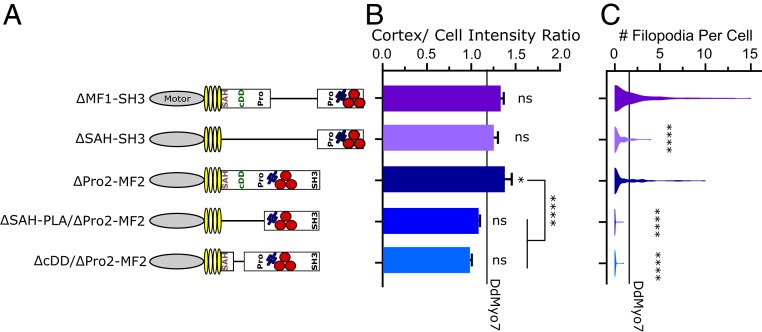
Deletion of both N-PLA and MF2 Domains Abolishes Activity.
The MF domains and proline-rich regions of the DdMyo7 distal tail are likely cargo or partner binding sites (34, 36, 37). DdMyo7 lacking either the MF1 (ΔMF1-SH3) or the MF2 region (ΔPro2-MF2) in the distal tail are able to cortically target and rescue filopodia formation to wild-type levels (Fig. 5 and ref. 26). However, depletion of Pro2-MF2 in combination with the proximal tail (ΔSAH-PLA/Pro2-MF2) completely abolished targeting and filopodia formation (Fig. 5 B and C and SI Appendix, Fig. S2A). In fact, removal of the candidate dimerization region in combination with Pro2-MF2 (ΔcDD/Pro2-MF2) is sufficient to abolish cortical targeting and filopodia formation, indicating either that the proximal dimerization region acts (partially) redundantly with the MF2 domain or that the 2 regions cooperate to promote filopodia formation. In contrast, a mutant lacking the entire proximal tail and MF1 region (ΔSAH-SH3) is targeted to wild-type levels and is capable of rescuing filopodia formation, albeit less efficiently than full-length DdMyo7, the proximal tail mutant (ΔSAH-PLA) or MF1 mutant (ΔMF1-SH3; Fig. 5 B and C, SI Appendix, Fig. S2A, and Table 1). It should be noted that the lack of cortical enrichment by ΔcDD/Pro2-MF2 indicates that the Pro1 and MF1 are not sufficient for targeting the motor to the cortex.
Fig. 5.
Functional cooperation of the proximal tail and MF2 regions. (A) Schematic illustration of proximal tail and MyTH4-FERM double domain mutants. (B) Cortical intensity of tail deletion mutants; significant values are compared to DdMyo7 control (horizontal line). (C) Violin plot of filopodia per cell. The horizontal line represents the mean of DdMyo7 control. Significance indicators from Tukey’s test are in comparison to DdMyo7 control, unless otherwise noted: ns, not significant; *P < 0.05; ****P < 0.0001.
The ability of ∆SAH-SH3 to promote filopodia formation indicates that the presence of the Pro2-MF2 domain is sufficient for function and supports a model whereby binding of a partner to the MF2 permits function in the absence of the PLA. This could be by promoting indirect dimerization or clustering of the motor at the cortex. These data also show that the MF2 domain but not the MF1 acts redundantly in concert with the proximal tail. The PLA region could contribute to activity by either increasing targeting via the recognition of an important partner, or by stabilizing motor dimerization. The lack of targeting by PLA-long (Fig. 3B) and weak dimerization by the PLA830-1020 fragment (Fig. 3E) would suggest that this region has a role in promoting dimer formation. This does not entirely rule out an alternative model that this region binds to partners critical for function, and further studies are needed to address this possibility.
Note also that the ΔSAH-PLA/ΔPro2-MF2 or ΔcDD/ΔPro2-MF2 mutants do not localize to the tips of filopodia in wild-type cells in contrast to ΔPro2-MF2 (SI Appendix, Fig. 2B), potentially because they fail to dimerize with endogenous DdMyo7. However, it is also possible that they are unable to bind an adaptor protein that would facilitate tip targeting. Thus, although the MF1 domain contributes to cortex enrichment, it is not sufficient to allow recruitment of the motor at the tip of filopodia without the presence of either the N-PLA or MF2 domains.
The finding that the PLA showed redundancy with MF2 and not MF1 was unexpected as deletion of either of the 2 MF domains alone did not impact targeting or filopodia formation (Fig. 4 and ref. 26). The observation that deletion of only MF2 in combination with the proximal tail dimerization domain results in a complete loss of filopodia formation (Fig. 4D) most likely reveals a specialization of the MF2 domain in targeting and potentially inducing the dimerization or clustering of DdMyo7. In contrast, it appears that the MF1 region promotes cortical localization (likely by contributing to recognition of partners) but not dimerization or self-association of DdMyo7 (required for filopodia initiation). Overall, these results are consistent with a model whereby MF1 and MF2 domains target DdMyo7 to cortical initiation sites. The resulting increase in concentration of active monomers at initiation sites via MF2 binding partner proteins would, in turn, be stabilized by the weak self-association of the dimerization domain in the proximal tail.
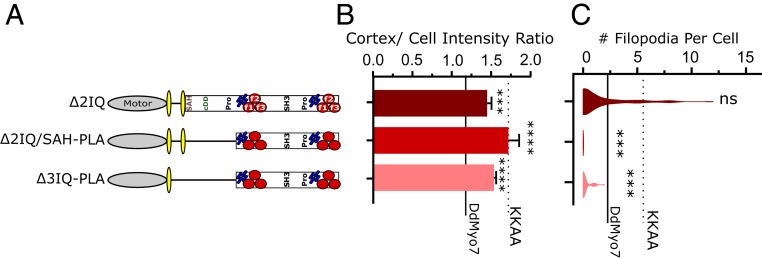
Shortening the LA and PLA Disrupts Filopodia Formation.
Models of filopodia initiation and maintenance suggest that a dimeric MF myosin at the cortex could bind adjacent filaments and coerce the actin into a parallel arrangement and then deliver cargo to the tip as the filopodium grows (18, 23, 38). The LA is important for interhead distance and step size; thus, the LA that sets the distance between catalytic heads of MF myosin dimers could govern the efficiency of both filopodia initiation and maintenance. In the context of a monomeric myosin, the LA can also play a role in tuning the force-sensing activity of myosin as previously shown for mammalian Myo1b (39). The DdMyo7 LA is composed of 4 IQ motifs and the single α-helix (SAH). The structural features of DdMyo7 required to promote filopodia formation were explored by designing myosins with shorter LAs (Fig. 6A). Deletion of IQ domains 2–4 or IQ 2–4-SAH apparently destabilized the myosin based on the inability to obtain stable cell lines for analysis. Deletion of the 2 internal IQs 2–3 (Δ2IQ) resulted in increased cortical localization compared to wild-type DdMyo7 (Fig. 6 A and B and SI Appendix, Fig. S2B). Interestingly, despite increased cortical targeting, Δ2IQ produced wild-type numbers of filopodia. This differs from the increased numbers of filopodia seen for the activated KKAA mutant (Fig. 6C, KKAA: P < 0.0001). On the other hand, the Δ2IQ makes more filopodia compared to the proximal tail mutant ΔSAH-PLA (P < 0.0001) that is, like Δ2IQ, targeted to the cortex more efficiently than DdMyo7 (Table 1). These findings suggest that the Δ2IQ LA mutant has both dysregulated autoinhibition that increases targeting and a shorter power stroke or reach between actin filaments that decreases the efficiency of filopodia formation. These observations highlight the role of the LA and PLA regions to finely regulate the step size and proximal dimerization for DdMyo7 to have optimal activity.
Fig. 6.
Shortening the LA and PLA disrupts filopodia formation. (A) Schematic illustration of deletion mutants. (B) Cortical intensity of LA and PLA region deletion mutants; significant values are compared to DdMyo7 control (horizontal line). (C) Violin plot of filopodia per cell. The solid horizontal line represents mean of DdMyo7 control; the dashed line represents KKAA. Significance indicators from Tukey’s test are in comparison to DdMyo7 control, unless otherwise noted: ns, not significant; ***P < 0.001; ****P < 0.0001.
Shortening of the LA in combination with deleting the PLA is predicted to disrupt motor coordination and function by both reducing the LA of the motor and impairing proximal dimerization. Deletion of 2IQ/SAH-PLA or 3IQ-PLA causes increased cortical localization, and thus appears to disrupt autoinhibition (Fig. 6B and Table 1). Comparison of these activated motors suggests that a proper LA, and subsequent motor function, actually contributes to motor function. Despite the increased cortical targeting, the ∆3IQ-SAH-PLA mutant also showed a significant defect in filopodia formation compared to ∆SAH-PLA (Fig. 6C, Table 1, and SI Appendix, Fig. S2) consistent with filopodia formation requiring proper spacing and force generation between DdMyo7 heads.
The distal tail domains of Δ2IQ/ΔSAH-PLA (notably the MF1 region that now abuts IQ4) are not able to elongate the LA of this mutant, and dimerization via partner binding by the distal tail is not sufficient to promote efficient function (Fig. 6), possibly by loss of coordination (gating) between heads. As stated earlier, the functionality of the ΔSAH-PLA mutants indicates that the full IQ region (4IQ motifs) provides a sufficiently long LA and that proximal dimerization is not an absolute requirement for filopodia initiation as long as the motor can cluster following proper cortical recruitment and oligomerization via the MF2 domain, as is the case with the even more dramatic deletion in ΔSAH-SH3 (Fig. 5). Although filopodia can form despite the lack of proximal dimerization, the function of these activated truncated myosins is lower than for the full-length myosin, as shown most clearly by the comparison with the activated autoinhibition mutant, KKAA (Fig. 1). Thus, filopodia formation by DdMyo7 is resilient to changes and perturbations in the LA (Fig. 6) and proximal and distal tails (Figs. 2 and 4, respectively), yet it is optimized with redundant functions within the tail. The minimum requirement for (near) wild-type filopodia formation includes either a N-PLA or MF2 to cluster monomers, and a LA composed of either 4IQs or 2IQ+SAH.
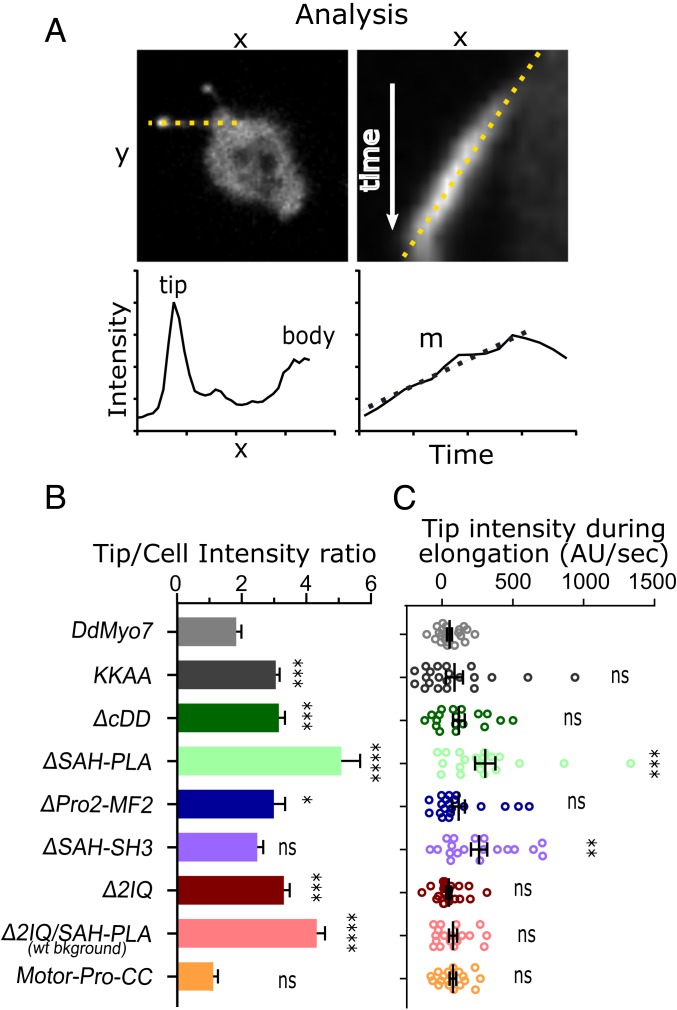
Disruptions in the Proximal Tail Cause DdMyo7 Accumulation in Filopodia Tips.
The filopodia tip complex is a hub of activity that directs incorporation of actin monomers to drive filopodia growth and contains receptors that sense the environment and adhesion receptors for binding to substrates (1). MF myosins are strikingly localized in filopodia tips, a result of either active transport by walking along actin, via restricted diffusion, or assembling in an initiation complex and “surfing” out on actin as the filopodium elongates (6, 18, 40). In fact, there is evidence that Myo10 can undergo intrafilopodia motility, accumulating in filopodia tips over time, suggesting Myo10 utilizes active transport to localize to tips (6, 38, 41).
The intensity of DdMyo7 in filopodia tips is consistently 2-fold higher compared to the cell body (Fig. 7B). The filopodia tip intensity is uniform during filopodia elongation, with little change in intensity as elongation proceeds (Fig. 7 A and C and ref. 26). Intrafilopodial transport and accumulation of DdMyo7 in growing filopodia tips was not observed for control DdMyo7 despite multiple efforts to visualize this process. These data are consistent with DdMyo7 motors assembling into an initiation complex at the cortex that is pushed out as the filopod extends. While the tip intensity was the same for wild type and many of the mutants, one unexpected observation was the striking enrichment of tip intensity signal from several DdMyo7 mutants, including ΔSAH-PLA and Δ2IQ (Fig. 7B and SI Appendix, Fig. S2).
Fig. 7.
Deletion of the proximal tail region causes DdMyo7 accumulation in filopodia tips. (A) Method for analysis of tip intensity during filopodia extension. Shown is a myo7 null expressing ∆SAH-Pro1. (Left Top) Sample micrograph dotted line drawn from the filopodia tip to cell body. Axis labels “x” and “y” are distance units (microns). (Left Bottom) Sample graph representing the fluorescence intensity of GFP-fusion protein along filopodia. The x axis is distance along the length of a filopodia starting and the tip, and y axis represents GFP intensity in arbitrary units. Note the 2 intensity maxima at the filopodia tip and cell body/cortex. (Right Top) Sample kymograph of an elongating filopodium (generated from the line on the Left). The line is drawn along the extending filopodia tip. The x axis is distance (microns), and the y axis is time (seconds). (Right Bottom) Sample graph representing the intensity measurement of the filopodia tip over time from the above kymograph. The x axis is time (seconds), and the y axis is GFP intensity (arbitrary units). (B) Graph of the tip/cell body intensity ratios of GFP-DdMyo7 and mutants. All constructs are expressed in myo7 null cells except for the 2IQ/SAH-PLA mutant that does not rescue filopodia formation and was expressed in the AX2 wild-type strain (noted on graph). (C) Graph of filopodia tip intensity accumulation rates (change in tip intensity during elongation) of GFP-DdMyo7 and mutants. Note that a negative rate indicates the tip gets dimmer. Comparisons are to DdMyo7 control: ns, not significant; *P < 0.05, **P < 0.01; ***P < 0.001, and ****P < 0.0001.
Quantitative analysis revealed that ΔSAH-PLA had increased signal in filopodia tips (Fig. 7 A and B). The autoinhibition mutant “KKAA” and smaller proximal tail deletion “ΔcDD” also showed brighter filopodia tips to a lesser extent (Fig. 7B and Table 1). Kymographs were generated from time-lapse images and filopodia tip intensity was measured during elongation. The change in tip intensity over time (slope, m) for each filopodia tip was determined (Fig. 7A). In contrast to the control, ΔSAH-PLA shows a significant increase in accumulation in the tips of elongating filopodia over time (Fig. 7C). Despite their overall higher filopodia tip intensity, KKAA and ΔcDD did not show significant changes in tip intensity during extension (Fig. 7C). Despite the observed accumulation, the signal in the filopodia shaft is diffuse and no moving DdMyo7ΔSAH-PLA puncta were observed. These data do not distinguish active transport of the motor along the actin bundles from dimensionally restricted diffusion of the molecule into filopodia tips, but they do highlight the role of DdMyo7 autoinhibition in regulation of the filopodia tip complex.
The ΔSAH-PLA mutant has an MF2 domain that could play a critical role in targeting the mutants to the tip complex and a large change in the LA length that significantly impairs autoinhibition. These mutants could accumulate due to dysregulation of DdMyo7 recycling in and out of the tip complex of the filopodia, allowing their enrichment at the tip complex over time. Alternatively, the proximal tail could bind a partner that facilitates the off-state or tip recycling, and deleting the domain abolishes that interaction and reduces recycling.
In both cases, control of autoinhibition may facilitate the rearward transport from the tip to the cell body by a compact DdMyo7, as the geometry of the mutant could be unlikely to favor the off-state.
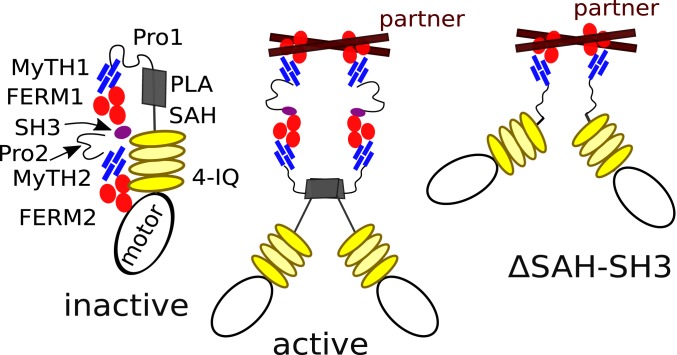
Summary and Conclusion.
Filopodia formation requires the action of a MF myosin in Metazoa and Amoebozoa, providing the opportunity to address questions of fundamental mechanisms of action by phylogenetically distinct MF myosins. This study and others support a model whereby filopodia formation relies on a dimeric MF myosin. In the case of DdMyo7, the proximal and distal tail cooperate to dimerize (or cluster) the motor (Fig. 5). The MF2 domain of DdMyo7 has a specialized role in filopodia formation likely by efficient partner binding and an ability to oligomerize DdMyo7 motors. The ability of the Myo10 MF to functionally substitute for the DdMyo7 MF2 domain (26) highlights the conservation of key attributes of filopodial MF myosins. Interestingly, during the evolution of filopodia myosins, the internal MF present in the Dictyostelium DdMyo7 tail (and likely present in an ancestral MF myosin; ref. 25) has been replaced in Myo10 by a membrane interaction domain. This reveals that filopodia formation does not require a precise tail design (2 MF domains or PH domains) as long as it serves to target the motor to the membrane and promote self-association. The data presented here are consistent with a model of a weak, context-dependent self-association domain of DdMyo7 being present in the proximal tail region and an MF2 domain serving to interact with a binding partner that promotes dimerization or clustering of the motor (Fig. 8). Partner binding would lead to activation of the motor by preventing internal autoinhibitory interactions and likely also lead to sufficiently high local concentrations of the motor to favor proximal dimerization. The SAH-PLA mutant indicates that this region is required to promote autoinhibition and is thus key to regulate the time the motor stays associated in an active form. This design is likely to favor both optimized function and precise regulation of the molecule.
Fig. 8.
Model of DdMyo7 Inhibition and activation states of DdMyo7. The major domains are shown as in Fig. 1. (Left) The inactive state where MF2 interacts with the motor domain to autoinhibit activity. (Middle) Active DdMyo7 that is dimerized via partner binding and dimerization mediated by a dimerization region (gray rectangle) of weak affinity. (Right) Proposed active state for the ΔSAH-SH3 mutant that lacks the central region of the DdMyo7 encompassing the SAH, PLA, MF1, and SH3 domains. Shown here is a pair of myosin heavy chains dimerized by partner binding to the MF2 and extended motor domains that can bind 2 actin filaments.
Myosins such as the MF myosins required for filopodia formation are regulated, sophisticated enzymes and likely resulted from multidomain coevolution. The large tail domain of DdMyo7 (and others) is required to not only bind partners that regulate its activity, but also stabilize the off-state of a myosin with a long LA. Head–tail interaction and formation of the off-state controls both cortical targeting and filopodia formation (Fig. 1 and ref. 26). The proximal tail of DdMyo7 is implicated in regulating the inactivation of DdMyo7 (Fig. 2). Disruptions in the LA and PLA that impair the myosin’s ability to form filopodia (Fig. 6) could result from compromising the ability of the motors to correctly orient actin filaments at the plasma membrane or to traffic actin regulators along the growing filopodium. The shortened LA mutants are sufficient for filopodial formation (Fig. 6); however, more structural and reconstitution studies are required to precisely define the design of MF myosin required for reorientation of actin filaments at the membrane.
The SAH-PLA region regulates the levels of DdMyo7 in the filopodia tip complex as evidenced by dysregulation of tip turnover seen for ∆SAH-PLA and ∆SAH-SH3 deletion mutants (Fig. 7). The increasing intensity of both mutants as the filopodium extends is consistent with this region contributing to the ability of DdMyo7 to be turned off and recycled back to the cytosol and reveals a previously unappreciated requirement for filopodia tip recycling. MF myosin recycling in filopodia may be required for discontinuous growth of filopodia as the elongation pauses and the tip interacts with the extracellular matrix, possibly when partners such as talin are incorporated into the filopodia tip complex via integrin binding, before resuming elongation (41, 42). Identification of DdMyo7 binding partners at the cortex and in the tip will provide needed information on how this myosin is incorporated and maintained in the initiation sites and the tip complex.
The amoebozoan filopodial myosin DdMyo7 and metazoan MF myosins are proposed to have evolved from a common ancestor that had 2 MF domains, with an overall domain structure similar to DdMyo7, metazoan Myo7, Myo22, and Myo15 (25). As with DdMyo7, the majority of MF myosin family members either lack a clear region of coiled-coil sequence that could promote dimerization or they only have a short stretch of coiled coil. Thus, the partner-mediated dimerization mechanism employed by DdMyo7 likely reflects the properties of its ancestral MF myosin. In the case of Myo7A, it has a small coiled-coil sequence that appears insufficient for dimer formation, although binding to its partner MyRIP activates transport, consistent with partner-mediated dimerization (43). Myo7B lacks any identifiable coiled-coil sequence, similar to DdMyo7, and it also appears to be assembled into either a transport or anchoring complex through binding to partners (12, 14). A similar mode of partner-mediated dimerization has been shown for Myo6 (44), indicating that control of myosin dimerization through partner interaction is a widely used and ancient strategy for activating and modulating myosin function.
DdMyo7 and Myo10 can both promote filopodia formation when the motor domains alone are stably dimerized (Fig. 4 and ref. 18). The mechanism of dimerization is clearly distinct for DdMyo7 and Myo10. The metazoan filopodial MF myosin Myo10 has evolved a specialized mode for forming antiparallel coiled-coil dimers via a short self-association region of rather high affinity (21, 23). Further studies of Myo10 are required to demonstrate whether the recycling or detachment of the motor from the tip complex controls its enrichment at the tip. In the case of DdMyo7, the results here suggest that it may function as a dimer via partner binding. Evolution has solved the interesting and complex problem of modulating motor function through coevolution of head, LA, and tail to maintain stable, yet activatable off-state, and a mechanism for precisely recruiting and activating the motor when and where necessary for cellular function. The findings here not only provide a better understanding of the formation and regulation of these widely used cell sensors but also provide insight into how other metazoan MF myosins that evolved from a common ancestor (such as those making stereocilia and microvilli) may use an array of optimized regulatory methods including partner-mediated dimerization and PLA-regulated autoinhibition to carry out their specialized cellular functions.
Materials and Methods
Cells Lines and Microscopy.
Dictyostelium control/wild-type (AX2) or myo7 null (HTD17-1) (7) were cultured in HL5 media. Cell lines were generated and screened as described (26, 45, 46). All expression plasmids were based on the cloned myoi gene (dictyBase:DDB_G0274455; ref. 47). The complete list of plasmids and oligos is provided in SI Appendix, Table S1. Live-cell imaging was done as previously described (26). Briefly, cells are adhered to cover glass and starved for 45 to 75 min in nutrient-free buffer (SB, 16.8 mM phosphate, pH 6.4), and then imaged at 1 to 4 Hz on a spinning disk confocal with a 1.4 numerical aperture, 63× objective (3i Marianas or Zeiss AxioObserver Z.1).
Data Analysis.
Images were quantified using a custom FIJI plugin “Seven” (26, 48). Cells not expressing transgenic proteins were excluded from the analysis. Statistical analysis was performed in Prism (GraphPad). One-way ANOVA analysis with post hoc Tukey test or Dunnett’s multiple comparison to wild-type control was used to compare groups; Student’s t test was used when only comparing 2 datasets. Automated analyses data points deemed definite outliers (0.1%) by Rout method were excluded. Error bars are SEM, unless noted. P values are represented on graphs as follows: ns, P ≥ 0.05; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Significant differences are in comparison to control (DdMyo7), unless noted. Filopodia formation in cells expressing PLA fragments were manually scored for the presence of actin-rich protrusions. Filopodia tip accumulation was analyzed by creating kymographs of initiating filopodia in FIJI, and then drawing a line along the filopodia shaft and calculating the intensity of the line.
Additional information about methodology, including details of expression plasmid generation and AUC, is in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Dr. Borries Demeler, Director of the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio, for providing initial AUC data. Thanks also to Annika Schroder and Casey Eddington for help with data analysis; Dr. Holly Goodson (Notre Dame) for helpful discussions and comments on the manuscript; Dr. Karl Petersen and Himanshu Jain for development of the “Seven” FIJI script; Dr. Gant Luxton [University of Minnesota (UMN)] for use of the spinning disc confocal microscope; Dr. Ron Rock (University of Chicago) for kindly providing the Myo5 coiled-coil clone; and University Imaging Centers, UMN, for additional imaging support. The work was supported by the CNRS, ANR-17-CE11-0029-01, and Ligue Contre le Cancer RS16 grants (A.H.). The A.H. team is part of the LabexCelTisPhyBio:11-LBX-0038, which is part of the Initiatives of Excellence of Université Paris Sciences et Lettres (ANR-10-IDEX-0001-02PSL). Support was also provided by UMN’s Undergraduate Research Opportunities Program (L.D.S.) and grants from the UMN Medical Foundation, UMN Graduate School, the American Heart Association (to M.A.T.), and NIH National Institute of General Medical Sciences (F31GM128325 to A.L.A. and R01GM122917 to M.A.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901527116/-/DCSupplemental.
References
- 1.Heckman C. A., Plummer H. K. 3rd, Filopodia as sensors. Cell. Signal. 25, 2298–2311 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Davenport R. W., Dou P., Rehder V., Kater S. B., A sensory role for neuronal growth cone filopodia. Nature 361, 721–724 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Jacquemet G., Hamidi H., Ivaska J., Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr. Opin. Cell Biol. 36, 23–31 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Sebé-Pedrós A., et al. , Insights into the origin of metazoan filopodia and microvilli. Mol. Biol. Evol. 30, 2013–2023 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svitkina T. M., et al. , Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409–421 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg J. S., Cheney R. E., Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 4, 246–250 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Tuxworth R. I., et al. , A role for myosin VII in dynamic cell adhesion. Curr. Biol. 11, 318–329 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Belyantseva I. A., et al. , Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat. Cell Biol. 7, 148–156 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Bohil A. B., Robertson B. W., Cheney R. E., Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl. Acad. Sci. U.S.A. 103, 12411–12416 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z. Y., et al. , Myosin-VIIb, a novel unconventional myosin, is a constituent of microvilli in transporting epithelia. Genomics 72, 285–296 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Weck M. L., Grega-Larson N. E., Tyska M. J., MyTH4-FERM myosins in the assembly and maintenance of actin-based protrusions. Curr. Opin. Cell Biol. 44, 68–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., et al. , Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins. Proc. Natl. Acad. Sci. U.S.A. 114, E3776–E3785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber K. L., Sokac A. M., Berg J. S., Cheney R. E., Bement W. M., A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature 431, 325–329 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Yu I. M., et al. , Myosin 7 and its adaptors link cadherins to actin. Nat. Commun. 8, 15864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H., et al. , Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 6, 523–531 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Kerber M. L., Cheney R. E., Myosin-X: A MyTH-FERM myosin at the tips of filopodia. J. Cell Sci. 124, 3733–3741 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokuo H., Ikebe M., Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 319, 214–220 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Tokuo H., Mabuchi K., Ikebe M., The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J. Cell Biol. 179, 229–238 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heissler S. M., Sellers J. R., Various themes of myosin regulation. J. Mol. Biol. 428, 1927–1946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umeki N., et al. , Phospholipid-dependent regulation of the motor activity of myosin X. Nat. Struct. Mol. Biol. 18, 783–788 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Lu Q., Ye F., Wei Z., Wen Z., Zhang M., Antiparallel coiled-coil-mediated dimerization of myosin X. Proc. Natl. Acad. Sci. U.S.A. 109, 17388–17393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plantard L., et al. , PtdIns(3,4,5)P3 is a regulator of myosin-X localization and filopodia formation. J. Cell Sci. 123, 3525–3534 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Ropars V., et al. , The myosin X motor is optimized for movement on actin bundles. Nat. Commun. 7, 12456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vavra K. C., Xia Y., Rock R. S., Competition between coiled-coil structures and the impact on Myosin-10 bundle selection. Biophys. J. 110, 2517–2527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kollmar M., Mühlhausen S., Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol. Biol. 17, 211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen K. J., et al. , MyTH4-FERM myosins have an ancient and conserved role in filopod formation. Proc. Natl. Acad. Sci. U.S.A. 113, E8059–E8068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y., et al. , A FERM domain autoregulates Drosophila myosin 7a activity. Proc. Natl. Acad. Sci. U.S.A. 106, 4189–4194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baboolal T. G., et al. , The SAH domain extends the functional length of the myosin lever. Proc. Natl. Acad. Sci. U.S.A. 106, 22193–22198 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight P. J., et al. , The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 280, 34702–34708 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Peckham M., Knight P. J., When a predicted coiled coil is really a single α-helix, in myosins and other proteins. Soft Matter 5, 2493–2503 (2009). [Google Scholar]
- 31.Lupas A., Van Dyke M., Stock J., Predicting coiled coils from protein sequences. Science 252, 1162–1164 (1991). [DOI] [PubMed] [Google Scholar]
- 32.Callebaut I., et al. , Deciphering protein sequence information through hydrophobic cluster analysis (HCA): Current status and perspectives. Cell. Mol. Life Sci. 53, 621–645 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rath A., Davidson A. R., Deber C. M., The structure of “unstructured” regions in peptides and proteins: Role of the polyproline II helix in protein folding and recognition. Biopolymers 80, 179–185 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Williamson M. P., The structure and function of proline-rich regions in proteins. Biochem. J. 297, 249–260 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prospéri M. T., et al. , Myosin 1b functions as an effector of EphB signaling to control cell repulsion. J. Cell Biol. 210, 347–361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moen R. J., Johnsrud D. O., Thomas D. D., Titus M. A., Characterization of a myosin VII MyTH/FERM domain. J. Mol. Biol. 413, 17–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Planelles-Herrero V. J., et al. , Myosin MyTH4-FERM structures highlight important principles of convergent evolution. Proc. Natl. Acad. Sci. U.S.A. 113, E2906–E2915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerber M. L., et al. , A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr. Biol. 19, 967–973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laakso J. M., Lewis J. H., Shuman H., Ostap E. M., Control of myosin-I force sensing by alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 107, 698–702 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baboolal T. G., Mashanov G. I., Nenasheva T. A., Peckham M., Molloy J. E., A combination of diffusion and active translocation localizes Myosin 10 to the filopodial tip. J. Biol. Chem. 291, 22373–22385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He K., Sakai T., Tsukasaki Y., Watanabe T. M., Ikebe M., Myosin X is recruited to nascent focal adhesions at the leading edge and induces multi-cycle filopodial elongation. Sci. Rep. 7, 13685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacquemet G., et al. , L-type calcium channels regulate filopodia stability and cancer cell invasion downstream of integrin signalling. Nat. Commun. 7, 13297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai T., Umeki N., Ikebe R., Ikebe M., Cargo binding activates myosin VIIA motor function in cells. Proc. Natl. Acad. Sci. U.S.A. 108, 7028–7033 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sweeney H. L., Houdusse A., Structural and functional insights into the myosin motor mechanism. Annu. Rev. Biophys. 39, 539–557 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Galdeen S. A., Stephens S., Thomas D. D., Titus M. A., Talin influences the dynamics of the myosin VII-membrane interaction. Mol. Biol. Cell 18, 4074–4084 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaudet P., Pilcher K. E., Fey P., Chisholm R. L., Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2, 1317–1324 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Titus M. A., A class VII unconventional myosin is required for phagocytosis. Curr. Biol. 9, 1297–1303 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.