Significance
Identification of genomic biomarkers that predict response to anticancer agents is the central research problem of cancer precision medicine. While the vast majority of the human genome encodes long noncoding RNAs (lncRNAs) as compared to protein-coding genes, thus far the characterization of lncRNAs as potential biomarkers has proved challenging. Here, we leverage data from large-scale cancer cell line screens to model response to hundreds of drugs as a function of lncRNA expression or somatic alteration. By carefully accounting for the confounding effects of tissue type, neighboring genes, and established biomarkers, the lncRNA models predict response to most drugs better than existing biomarkers. Thus, our framework can be applied for the discovery of lncRNAs as pharmacogenomic biomarkers in cancer research.
Keywords: long noncoding RNA, pharmacogenomics, drug response prediction, machine learning
Abstract
Large-scale cancer cell line screens have identified thousands of protein-coding genes (PCGs) as biomarkers of anticancer drug response. However, systematic evaluation of long noncoding RNAs (lncRNAs) as pharmacogenomic biomarkers has so far proven challenging. Here, we study the contribution of lncRNAs as drug response predictors beyond spurious associations driven by correlations with proximal PCGs, tissue lineage, or established biomarkers. We show that, as a whole, the lncRNA transcriptome is equally potent as the PCG transcriptome at predicting response to hundreds of anticancer drugs. Analysis of individual lncRNAs transcripts associated with drug response reveals nearly half of the significant associations are in fact attributable to proximal cis-PCGs. However, adjusting for effects of cis-PCGs revealed significant lncRNAs that augment drug response predictions for most drugs, including those with well-established clinical biomarkers. In addition, we identify lncRNA-specific somatic alterations associated with drug response by adopting a statistical approach to determine lncRNAs carrying somatic mutations that undergo positive selection in cancer cells. Lastly, we experimentally demonstrate that 2 lncRNAs, EGFR-AS1 and MIR205HG, are functionally relevant predictors of anti-epidermal growth factor receptor (EGFR) drug response.
Long noncoding RNAs (lncRNAs) are transcripts greater than 200 nucleotides in length that do not contain protein-coding sequences. They act as key regulators of gene expression (1), controlling a diverse set of transcriptional and posttranscriptional processes, including chromatin remodeling, RNA splicing and transport, and protein synthesis (2, 3). Although, less than 1% of lncRNAs have been functionally characterized (4), a comprehensive characterization of lncRNA across thousands of tumors suggests pervasive dysregulation of the lncRNA transcriptome at rates similar to protein-coding genes (PCGs) (5, 6). In addition, some lncRNAs function as either oncogenes or tumor suppressor genes in human cancers (7, 8).
Several large-scale cancer cell line screens systematically investigated the response to hundreds of drugs to identify genomic and transcriptomic biomarkers of cancer drug response (9–13). These studies expanded the repertoire of somatic alterations and gene expression biomarkers linked with drug response but focused exclusively on PCGs. Considering less than 2% of the genome codes for PCGs (14) with nearly 70% of the genome transcribed into noncoding RNAs (15), it seems that the mechanisms of anticancer drug response cannot be explained by PCGs alone (16). Subsequently, a recent study reported lncRNA models are better predictors of drug response compared to PCGs for several drugs (17). However, lncRNAs are expressed with a high degree of tissue specificity and the expression of genic lncRNAs tends to be strongly correlated with the expression of PCGs on complementary strands (15). Therefore, the identification of novel lncRNA biomarkers associated with anticancer drug response requires careful consideration of the potential confounding influence of the tissue lineage along PCGs proximal to the lncRNAs.
Here we report the results of a systematic investigation of the lncRNA transcriptome and genome of cancer cell lines and large-scale drug screens to establish a pharmacogenomic landscape of lncRNAs. We use regularized regression models to predict drug response using lncRNA transcriptome to demonstrate its potency compared to PCGs. To guide the discovery of individual lncRNA biomarkers, we delineate the effects of cis-PCGs on drug–lncRNA associations in regression models. In addition, we identify lncRNA-specific somatic mutations undergoing positive selection in cancer cells and determine their associations with drug response. We further investigate the contribution of lncRNAs in predicting the response for drugs with clinically actionable PCG biomarkers. Based on our analysis, we highlight the role of EGFR-AS1 and MIR205HG as predictors of anti-epidermal growth factor receptor (EGFR) therapeutic response independent from EGFR somatic mutations and experimentally confirm their potential as erlotinib-response biomarkers in lung cancer cells.
Results
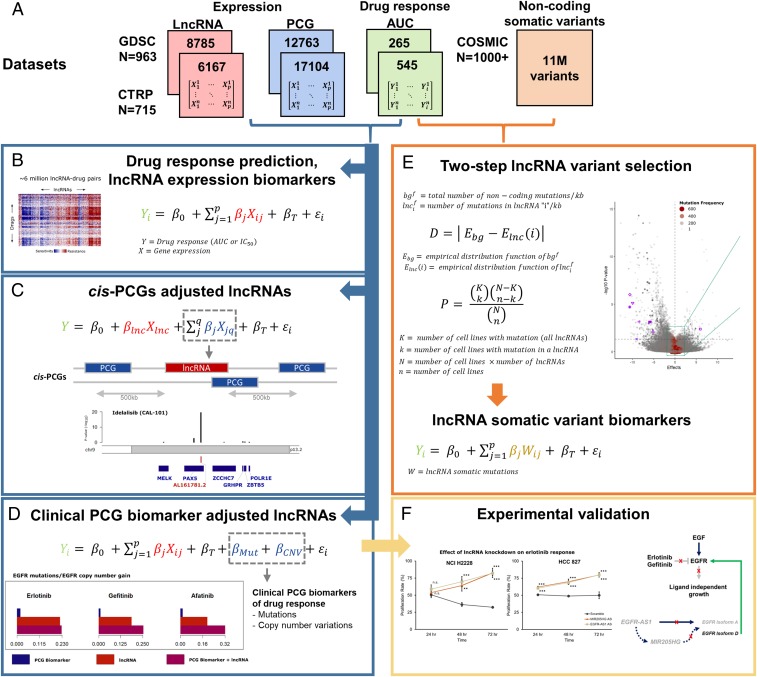
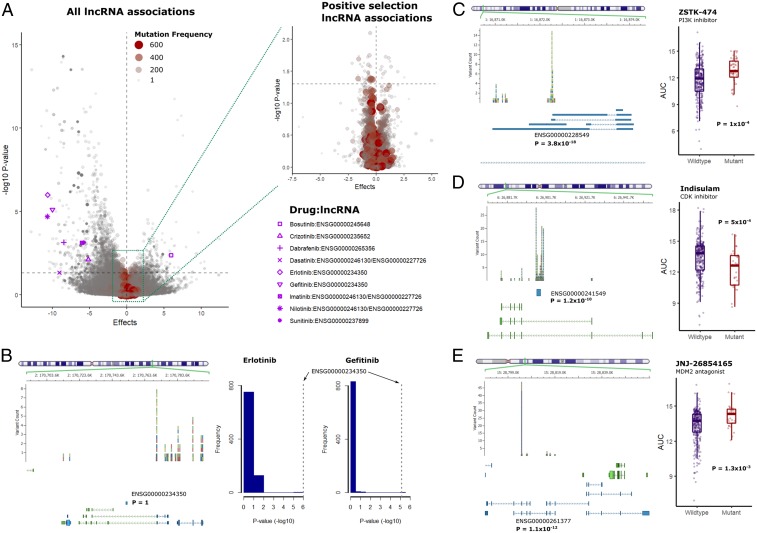
Despite the tremendous success of cancer cell line screens in discovering novel PCG biomarkers of drug response, the contribution of lncRNAs in cancer pharmacogenomics is poorly established. To systematically determine the relevance of lncRNAs as anticancer drug response biomarkers, we propose the following framework to delineate their contribution as response predictors while accounting for the effects of tissue type, proximal cis-PCGs (within ±500 Kb), and known biomarkers (Fig. 1 A–D and SI Appendix, Fig. S1 A and B). In addition, we determine the contribution of somatic alterations specific to lncRNAs in drug response using a statistical approach to determine positively selected mutations (Fig. 1E). Finally, we experimentally validate the functional role of 2 lncRNA predictors of anti-EGFR drug response (Fig. 1F).
Fig. 1.
Framework for lncRNA biomarker discovery. (A) Datasets used in the study, including gene expression (PCG, lncRNA) and drug response profiles corresponding to Genomics of Drug Sensitivity in Cancer (GDSC) and Cancer Therapeutics Response Portal (CTRP) cell lines, and noncoding somatic variants from COSMIC. RNAseq data for PCG and lncRNA transcriptome corresponding to the CTRP cell lines were obtained from the Cancer Cell Line Encyclopedia. For the GDSC cell lines, the lncRNA transcriptome was imputed using the gCSI RNAseq dataset (see Methods for details). “N” indicates the number of cell lines in each dataset, while the number of lncRNA, PCGs, or drugs with the area under the curve (AUC) of drug response in each dataset are indicated inside the colored boxes. (B) Linear model for predicting drug response (AUC) using the PCG or lncRNA transcriptome. (C) Determining significance drug:lncRNA associations after adjusting for the expression levels of neighboring cis-PCGs within a ±500-Kb window. (D) Identification of significant drug:lncRNA associations after adjusting for the mutation or copy number variation status of clinically established PCG biomarkers of drug response. (E) A two-step statistical approach to determine lncRNAs with somatic mutations that undergo positive selection in cancer cell lines. (F) Experimental validation of candidate lncRNAs that augment clinical biomarkers of drug response.
Determining the Contribution of lncRNA Transcriptome as a Predictor of Anticancer Drug Response.
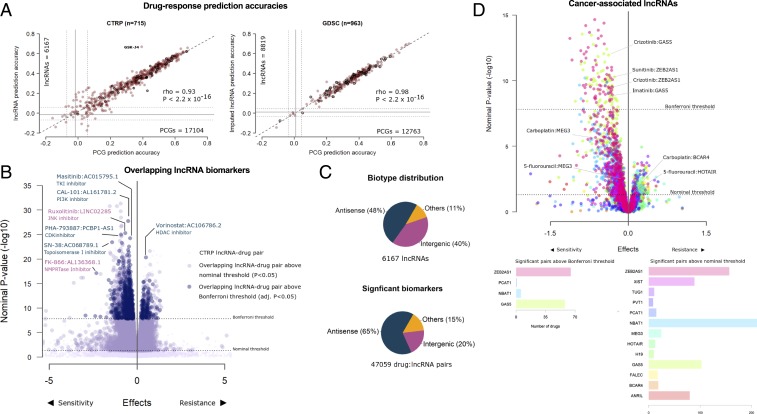
An important finding reported by the cancer cell line screens was the ability to implement machine-learning algorithms that accurately predicted drug response using the baseline PCG transcriptome of cancer cells. Thus, we first compared the ability of lncRNA transcriptome to predict response to 265 and 545 compounds from the Genomics of Drug Sensitivity in Cancer (GDSC) and Cancer Therapeutics Response Portal (CTRP) screens, respectively. In both screens, the lncRNA transcriptome was equally potent at predicting response to individual drugs as PCGs (CTRP Spearman’s ρ = 0.93; GDSC Spearman’s ρ = 0.98) (Fig. 2A), with no difference in median prediction accuracies across all drugs (CTRP P = 0.17; GDSC P = 0.32) (SI Appendix, Fig. S1C). The drug GSK-J4, a potent and highly selective inhibitor of H3K27 histone demethylases JMJD3 and UTX, was an exception that was predicted with better accuracy using lncRNA transcriptome. For the set of drugs that were common to both screens, consistent prediction accuracies were achieved using both lncRNA (P = 0.24) and PCG models (P = 0.07) (SI Appendix, Fig. S1 D and E). Moreover, no differences were observed in GDSC drug response prediction accuracies (P = 0.78) when using either measured Genentech Cell Line Screening Initiative (gCSI) RNAseq or imputed lncRNA profiles from the subset of cell lines that were common between GDSC and gCSI (SI Appendix, Fig. S1F).
Fig. 2.
LncRNAs as predictors and biomarkers of anticancer drug response. (A) Scatterplot of 545 CTRP or 265 GDSC prediction accuracies using PCG transcriptome (X axis) or imputed lncRNA transcriptome (y axis) as predictors. Each point on the scatter plot shows the accuracy of predicting response to a drug by using models generated using PCG or lncRNA transcriptome. The bolded points are drugs common to both GDSC and CTRP. Gray lines indicate prediction accuracies with confidence intervals from a null model. (B) Volcano plots of drug–lncRNA associations in the cohort of drug–lncRNA pairs common to CTRP and GDSC screens displaying effect sizes (X axis) and P values (y axis) of the regression analyses. The light-blue enlarged circles indicate significant (FWER-adjusted) CTRP drug–lncRNA pairs also significant in GDSC at the nominal threshold, while dark-blue enlarged circles are significant in GDSC at the FWER threshold. The dark blue labels indicate examples of drugs paired with antisense lncRNAs, and pink labels show intergenic lncRNAs. (C) Distribution of lncRNA biotypes across all cell lines and within the subset of significant drug–lncRNA pairs (FWER-adjusted) identified from linear regression analysis of the CTRP screen adjusted for tissue type. (D) Volcano plot displaying effect sizes (X axis) and P values (y axis) of oncogenic and tumor suppressor lncRNAs associated with drug response after adjusting for cis-PCGs. The bar plots below indicate the frequency of drugs associated with the lncRNAs at the nominal threshold and FWER-adjusted threshold.
Next, we modeled drug response as a function of individual lncRNA transcripts to determine significant biomarkers. Across the set of drug–lncRNA pairs common to both CTRP and GDSC screens, 68% of the significant [familywise error rate (FWER) < 0.05] CTRP drug–lncRNA associations were also significant in GDSC at the nominal threshold (P < 0.05), while about 28% were significant at the FWER threshold (Fig. 2B). Both antisense and intergenic transcripts were represented in the cohort of top significant overlapping drug pairs; for example, vorinostat resistance was associated with the antisense lncRNA AC106786.2 or ruxolitinib sensitivity with the intergenic lncRNA LINC02285 (Fig. 2B). The biotypes of lncRNAs were nearly equally distributed between genic (including antisense) and intergenic RNAs, while antisense transcripts were overrepresented in the cohort of significant (FWER < 0.05) drug–lncRNA pairs as compared to intergenic transcripts (Tukey’s P < 10−11) (Fig. 2C and Dataset S1).
Next, we evaluated the pharmacogenomic relevance of the well-characterized lncRNAs that have been implicated as oncogenes or tumors suppressors in human cancers (18) (Fig. 2D and SI Appendix, Fig. S1G). For example, the expression of putative tumor suppressor lncRNA MEG3 was associated with sensitivity to carboplatin and 5-fluorouracil, while the putative oncogenes BCAR4 and HOTAIR were associated with resistance to carboplatin and 5-fluorouracil, respectively (Dataset S2). These observations are in line with previous in vitro studies that show elevated MEG3 expression to be associated with sensitivity (19, 20) while high BCAR4 (21) and HOTAIR (22) expression were linked with resistance to cytotoxic anticancer agents. Within in the cohort of significant cancer-associated lncRNAs, we observed the expression of GAS5 and ZEB2AS1 was associated with the sensitivity of more than 50 drugs (Fig. 2D), suggesting these lncRNAs could be candidates for further evaluation as multidrug response predictors. These results suggest the potential of known cancer-associated lncRNAs to serve as determinants of anticancer drug response.
Characterizing the Impact of Proximal cis-PCGs on Drug–lncRNA Associations.
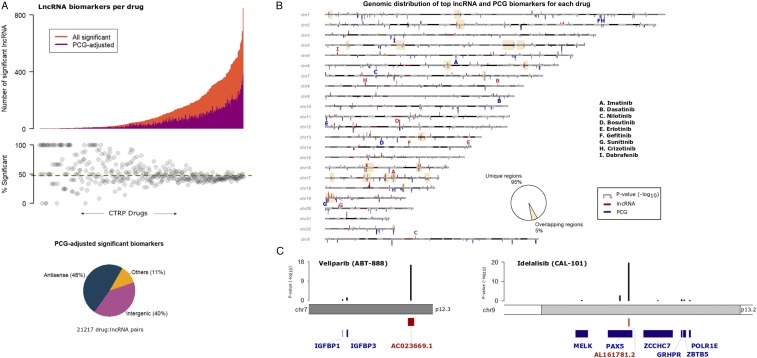
The strong correlation between the expression levels of antisense lncRNAs and overlapping PCGs could potentially result in spurious associations with drug response. To deconvolute the contribution of lncRNAs from the effects of proximal PCGs, we next analyzed drug response as a function of lncRNA expression while adjusting for the effects of proximal cis-PCGs (any PCG located in the −500-Kb region upstream of transcription start site to +500 Kb downstream of transcript end). Nearly 50% of the remaining significant drug–lncRNA associations were affected by the expression levels of the cis-PCGs (Fig. 3A). As expected, adjusting for cis-PCGs mostly affected the proportion of significant antisense lncRNAs over intergenic transcripts (Fig. 3A).
Fig. 3.
Relevance of cis-PCG adjusted lncRNA biomarkers. (A) Distribution of significant lncRNAs associated with each CTRP drug (X axis). The orange bars indicate a number of significant lncRNAs for a drug while the magenta bars indicate the number of lncRNAs that are statistically significant after adjusting for the expression levels of every cis-PCG (within 1 Mb) of the lncRNA transcript. The piechart below shows biotype distribution of the lncRNAs in the cohort of significant drug–lncRNA pairs adjusted for cis-PCGs. (B) Ideogram of the human chromosomes displaying the top lncRNA and PCG associated with each CTRP drug. P values of the drug–gene associations are indicated as red bars for lncRNAs above the chromosomes and as blue bars for PCGs below the chromosomes at their respective locus. The yellow highlighted boxes indicate the cytobands with overlapping top lncRNA and PCG loci associated with a drug. Bolded letters (red = lncRNA, blue = PCG) indicate signals associated with the set of drugs with clinically actionable biomarkers. (C) Examples of top lncRNA associated with veliparib and idelalisib response, with black bars indicating adjusted P values for lncRNA and cis-PCGs.
Given the obvious influence of proximal PCGs on drug–lncRNA associations and to better understand the genomewide distribution of lncRNA biomarkers relative to PCGs, we mapped the significant lncRNA and PCG biomarker for all drugs analyzed in the CTRP screen (SI Appendix, Fig. S2 A and B). For individual drug, we also mapped the top predictive lncRNA or PCG biomarkers based on their genomic loci (Fig. 3B). Interestingly, we observed that for most drugs, the top lncRNAs and PCG biomarkers were located on distinct loci. In fact, the top lncRNA and PCG loci overlapped for only 5% of the drugs analyzed. As a case in point, we highlight the set of drugs with established clinically actionable biomarkers (23). For example, the top PCG markers for gefitinib and crizotinib response were located on chromosomes 1 while the lncRNAs markers were located on 14 and 8, respectively. Based on these observations, we propose that despite the obvious impact of proximal cis-PCGs, it is possible to discover strong drug–lncRNA associations by carefully accounting for the effects of cis-PCGs.
As additional examples, we zoom-in to highlight the top lncRNA associated with veliparib and idelalisib response along with the proximal cis-PCGs. The intergenic lncRNA AC023669.1 was associated with veliparib response (P = 3.9 × 10−17) after adjusting for the neighboring PCGs IGFBP1 (P = 0.7) and IGFBP3 (P = 0.08) (Fig. 3C). The antisense lncRNA AL161781.2 was associated with idelalisib sensitivity (P = 2.9 × 10−20) while the expression levels of proximal cis-PCGs showed considerably weaker association with the drug (e.g., PAX5 P = 0.002) (Fig. 3C).
lncRNAs Augment Drug Response Predictions from Known PCGs Biomarkers.
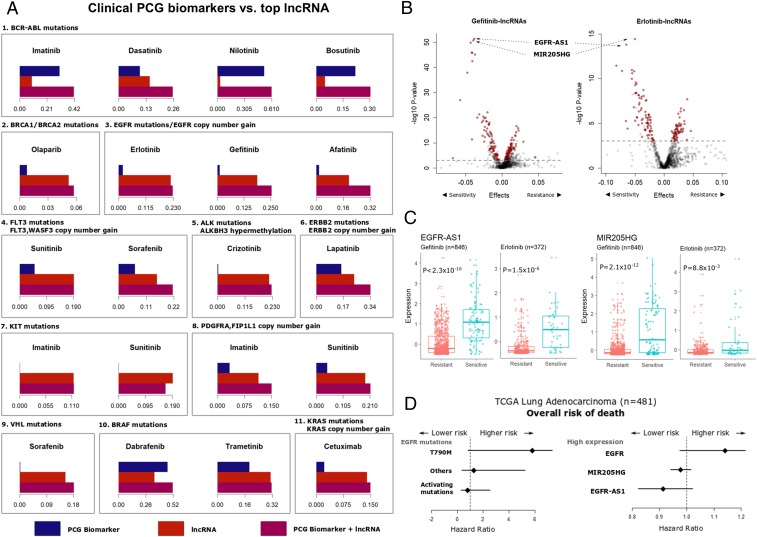
Currently, a small number of PCG mutations and copy number variations (CNVs) are being used in the clinic as biomarkers to guide treatment decisions (23). We evaluated if the top lncRNA biomarkers for such drugs provide any additional benefit over the clinical biomarkers. We modeled drug response as a function of lncRNA expression and known PCG biomarkers and compare the individual and combined contribution of each predictor at explaining the variability in drug response (Fig. 4A). In the case of BCR-ABL targeting tyrosine kinase inhibitors (TKIs), like imatinib and nilotinib, the BCR-ABL1 fusion event was a stronger predictor compared to the top lncRNA. However, the response to dasatinib, a TKI with several other targets besides BCR-ABL, was better explained by the lncRNA. Similarly, BRAF mutations were strong predictors of response to dabrafenib and trametinib, and ERBB2 mutations/CNVs for lapatinib sensitivity. In each of these cases, the addition of the lncRNA biomarker improved the proportion of variance explained by the model compared to the PCG biomarker alone. Other mutations and CNVs in genes like KIT, PDGFR, KRAS, ALK, and VHL actually explained a very small proportion of the variance of the respective drugs and were supplemented by the addition of lncRNA biomarkers. Interestingly, EGFR mutations and CNVs, one of the most prominent examples of cancer pharmacogenomic biomarkers for the EGFR targeting TKIs, were also augmented by the inclusion of expression of the top lncRNA biomarker. These results provide strong evidence to support the utility of lncRNA expression as biomarkers for anticancer drugs beyond PCGs.
Fig. 4.
LncRNAs augment clinical drug response biomarkers. (A) Barplots showing the proportion of variance in drug response (R2 of the regression model adjusted for tissue type) explained by the known PCG biomarker (blue bar), top lncRNA biomarker (red bar), or model combining the two (violet bar). The PCG biomarkers are listed above each subpanel. (B) Volcano plots showing lncRNAs associated with gefitinib or erlotinib response in the GDSC dataset adjusting for mutations and copy number variations in the EGFR gene. Each point on the plot represents an lncRNA–drug pair, with red points indicating lncRNAs common to both erlotinib and gefitinib above the nominal significance threshold of false discovery rate (FDR) < 0.05 (dashed gray line). (C) Distribution of EGFR-AS1 and MIR205HG expression in gefitinib or erlotinib resistant (AUC > 0.9) or sensitive (AUC < 0.9) cell lines. (D) Hazard ratios obtained from Cox-proportional hazard models for overall survival of TCGA LUAD patients based on EGFR mutation status or elevated expression levels of the EGFR, EGFR-AS1, or MIR205HG genes.
We further evaluated the top lncRNA predictors of EGFR-targeting TKI response, as the inclusion of these lncRNAs in the model resulted in substantial improvement in the proportion of variability in drug response explained by EGFR mutations or CNVs alone. Somatic mutations in the EGFR tyrosine kinase domain, including in-frame deletions in exon 19, single-nucleotide variations in exon 21 and amplification improve sensitivity, while an exon 20 (T790M) secondary mutation causes resistance to the anti-EGFR drugs gefitinib and erlotinib (24–27). However, these well-defined biomarkers can only explain a small proportion of the variance in drug response in (Fig. 4A). This observation is consistent with data from nonsmall cell lung cancer patients, where the response to anti-EGFR therapy is determined by EGFR-activating mutations and CNVs in about 10 to 30% of patients (28). However, about 1 in 4 patients that respond to gefitinib or erlotinib do not carry these activating alterations (29). We found 2 lncRNAs, the EGFR antisense RNA 1 (EGFR-AS1; ENSG00000224057) and the MIR205 host gene (MIR205HG; ENSG00000230937), as the top 2 candidates biomarkers of anti-EGFR drug response independent of known PCG biomarkers (Fig. 4B). The addition of EGFR-AS1 and MIR205HG expression substantially improved the proportion of variance explained by the drug response models to 12 to 18% for erlotinib and 25 to 30% for gefitinib when combined with the EGFR functional events across all cell lines (SI Appendix, Fig. S3A). Without considering EGFR functional events, both EGFR-AS1 and MIR205HG expression levels were higher in the cells sensitive to gefitinib or erlotinib (Fig. 4C and SI Appendix, Fig. S3B). Moreover, the drug–lncRNA associations for EGFR-AS1 (erlotinib P = 1.37 × 10−10; gefitinib P = 2.2 × 10−16) and MIR205HG (erlotinib P = 2.04 × 10−4; gefitinib P = 2.2 × 10−16) were significant after adjusting for EGFR mutations and CNVs (SI Appendix, Fig. S3C).
Building on these results, we validated the correlation between EGFR-AS1 and MIR205HG expression with imputed erlotinib response in lung adenocarcinoma (LUAD) patients from the cancer genome atlas (TCGA) project (30). The analysis of imputed drug response showed higher expression of both EGFR-AS1 and MIR205HG were associated with sensitivity to erlotinib (SI Appendix, Fig. S4 A–D). The significance of correlation between the lncRNAs and erlotinib response was within the same order of magnitude as EGFR mutation status (SI Appendix, Fig. S4 E and F).
We next evaluated the impact of the candidate lncRNA biomarkers on patient survival outcomes, in comparison with EGFR mutation status. As expected in the TCGA LUAD cohort, the presence of EGFR secondary resistance mutation (T790M) or high EGFR expression were both associated with worse overall prognosis (Fig. 4D). In contrast, the elevated expression of MIR205HG and EGFR-AS1 were associated with reduced risk of lung cancer death, a trend similar to the presence of EGFR-activating mutations (Fig. 4D). These results are consistent in the directionality suggested by our analysis, that is, the expression of both EGFR-AS1 and MIR205HG are indicative of improved sensitivity and better prognosis. Thus, further inquiry into the prognostic value of MIR205HG and EGFR-AS1 for anti-EGFR therapeutics will be crucial.
Determining lncRNA-Specific Genomic Alterations Associated with Drug Response.
The majority of the mutation profiles utilized in existing cancer pharmacogenomic studies were generated using exome sequencing. As a result, the impact of somatic variants that specifically affect the lncRNA genome has largely remained unexplored. The study of lncRNA variants is complicated by the lack of a clear definition of “passenger” and “driver” noncoding mutations. Two recent efforts attempted to identify driver lncRNA mutations that were positively selected in human cancers (31, 32). In some form, each method identified noncoding loci or noncoding genes that were mutated at a frequency significantly greater than the background rate across all noncoding loci or across samples, respectively. We adopted a similar framework to define positively selected lncRNA-specific somatic variants using whole-genome sequencing data from about 1,000 catalogs of somatic mutations in cancer (COSMIC) cell lines (11). We excluded all genic noncoding variants (intron, promoter, or untranslated regions of PCGs) and identified lncRNA genes with mutation frequencies greater than the length-adjusted background noncoding mutation frequency.
We analyzed drug response as a function of the mutation status of each candidate lncRNA (Fig. 5A and SI Appendix, Fig. S5 A and B). In contrast with lncRNA expression, only a small set of lncRNA mutations undergoing positive selection were associated with drug response (Fig. 5 A, Inset and SI Appendix, Fig. S5 C and D). In the case of drugs with actionable PCG biomarkers, we found drugs with a similar mechanism of actions tend to share the top lncRNA mutation predictors (Fig. 5A). For example, AC007405.1 (ENSG00000234350) was the top lncRNA associated with sensitivity to both erlotinib and gefitinib (Fig. 5B), suggesting a possible functional link between these lncRNA loci and the mechanism of action of the drugs.
Fig. 5.
lncRNA-specific somatic variations and association with drug response. (A) Volcano plot of drug–lncRNA associations based on somatic mutations in lncRNA genes, with the size of points scaled according to the frequency of mutations across all cell lines. Nominal P value thresholds are indicated by the horizontal dashed gray lines. (Inset) Volcano plot shows lncRNAs satisfying the statistical threshold for positive selection in the COSMIC cell lines. Significant associations above the nominal threshold for drugs with clinically actionable PCG biomarkers are indicated with pink markers. (B, Left) Chromosomal locus of the candidate lncRNA and frequency of noncoding variants identified in the COSMIC cancer cell lines. The exon structure of the proximal PCGs (green) and lncRNAs (blue) are displayed below the variants. (Right) The P-value distribution of the lncRNAs associated with erlotinib and gefitinib sensitivity. (C–E) Examples of lncRNAs with mutation frequencies above the statistical threshold for positive selection in the COSMIC cell lines, with the left showing frequency of noncoding variants and exon structure of proximal genes, and the right showing the comparison of drug AUC in wild-type or mutant cell lines.
We next determined associations for the lncRNAs with somatic mutation frequencies above the statistical threshold for positive selection (Fig. 5 A, Inset). As an example, the intergenic lncRNA BX284668.2 (ENSG00000228549) (P = 3.8 × 10−18) was associated with sensitivity to the PI3K inhibitor ZSTK-474 (P = 1 × 10−4) (Fig. 5C). Similarly, mutations in the glucuronidase beta pseudogene 2 (ENSG00000241549) (P = 1.8 × 10−10) were linked with resistance to the CDK inhibitor indisulam (P = 5 × 10−4) (Fig. 5D). Additionally, mutations in the programmed cell death 6 interacting protein pseudogene 2 (ENSG00000261377) were determined as positively selected in the COSMIC cell lines (P = 1.1 × 10−12) and predicted sensitivity to the MDM2 antagonist JNJ-26854165 (P = 0.001) (Fig. 5E). While the biological function of these lncRNAs is virtually unknown, these examples focusing on lncRNAs undergoing positive selection in cancer cells hint at the existence of PCG-independent associations between somatic alterations in the lncRNA genome and drug sensitivity. At the very least, it is clear that further studies are warranted to characterize the function of somatic lncRNAs variants and study their associations with anticancer drug response.
Experimental Validation of the Influence of EGFR-AS1 and MIR205HG Expression on Erlotinib Response in Lung Cancer Cells.
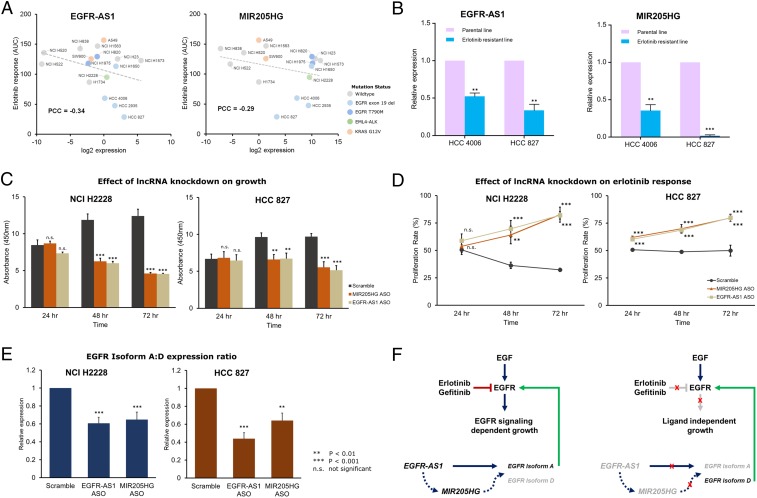
We determined the correlation between EGFR-AS1 and MIR205HG expression with erlotinib response in a cohort of 16 lung cancer cell lines (SI Appendix, Fig. S6A). As expected, the cell lines with EGFR-activating mutations (exon 19 deletions) including HCC 4006, HCC 2935, and HCC 827, were most responsive to erlotinib treatment (Fig. 6A). Our experiments yielded similar results as observed in the cancer cell line screens, with higher expression of both lncRNA transcripts associated with increased sensitivity to erlotinib (EGFR-AS1 PCC = −0.34, MIR205HG PCC = −0.29) (Fig. 6A).
Fig. 6.
In vitro validation of EGFR-AS1 and MIR205HG as determinants of erlotinib response. (A) Scatter plots showing expression levels of EGFR-AS1 and MIR205HG along with erlotinib response (AUC) for 16 lung cancer cell lines labeled on the plot. Cell lines carrying EGFR-activating mutations (L858R, exon 19 del) are encircled in blue, while lines with resistance mutation (T790M) are encircled in red. The dashed gray line indicates a linear fit. (B) Comparison of relative EGFR-AS1 and MIR205HG expression levels in 2 EGFR-responsive cell lines––HCC 4006 and HCC 827. Violet bars indicate expression levels in parental lines, while blue bars indicate expression levels in derived, erlotinib-resistant, lines. (C) Barplots indicating the growth of NCI H2228 and HCC 827 cells upon ASO-mediated k.d. of EGFR-AS1 or MIR205HG over a period of 72 h post-k.d. (D) Effect of ASO-mediated EGFR-AS1 or MIR205HG k.d. on erlotinib response determined as proliferation rate relative to untreated NCI H2228 and HCC 827 cells measured over a period of 72 h. (E) The ratio of relative expression levels of EGFR Isoform A: Isoform D in NCI H2228 and HCC 827 cells with ASO-mediated EGFR-AS1 or MIR205HG k.d. (F) Schematic of the proposed lncRNA-driven pathway that determines response to erlotinib in the presence (Left) or absence (Right) of EGFR-AS1 and MIR205HG-mediated regulation of EGFR isoforms. **P < 0.01, ***P < 0.001; n.s., not significant.
Next, we measured the expression of the 2 lncRNAs in 2 erlotinib-resistant lines generated from erlotinib-sensitive parental lines (HCC 4006 and HCC 827) carrying EGFR-activating mutations (Fig. 6B). The expression levels of EGFR-AS1 were about 50 to 60% lower in the resistant lines as compared to the parental HCC 4006 and HCC 827 cell lines (both P < 0.01). Similarly, MIR205HG expression levels were 60 to 90% lower in the resistant lines (HCC 4006 P < 0.01; HCC 827 P < 0.001). Based on these results, we hypothesized that these 2 lncRNAs have a functional influence on erlotinib response and do not merely serve as predictive biomarkers.
To confirm their functional impact, we performed anti-sense oligonucleotide (ASO) mediated knockdown (k.d.) of EGFR-AS1 and MIR205HG in 2 cell lines with different levels of erlotinib sensitivity – HCC 827 (strong response) and NCI H2228 (weak response) (SI Appendix, Fig. S6 B and C). The k.d. of either transcripts resulted in a reduction in the growth of the 2 cell lines over time, with significantly slower growth observed at 48 and 72 h (Fig. 6C). However, in the presence of erlotinib, the rates of proliferation were elevated in both the EGFR-AS1 and MIR205HG k.d. cell lines as compared to control (Fig. 6D). These results corroborate the outcome of our analyses and hint at a possible functional impact of the 2 lncRNAs on erlotinib response.
To further investigate its functional role, we investigated the relative expression levels of 2 EGFR isoforms that were recently described as regulatory targets of EGFR-AS1 (33). The relative ratio of these isoforms could affect the ligand-dependent activation of the EGFR signaling pathway (34). The product of EGFR transcript 1 (ENST00000275493.6 or NM_005228.5) translates into the full-length Isoform A of the protein while transcript variant 4 (ENST00000344576.6 or NM_201284.1) translates into the truncated Isoform D of the protein. Only the extracellular domain is present in the shorter isoform and lacks the tyrosine kinase domain. Thus, abundant expression Isoform D may act as an antagonist of ligand-dependent EGFR action.
In the EGFR-AS1 and MIR205HG k.d. cell lines, the expression levels of the consensus EGFR sequence were not significantly altered (SI Appendix, Fig. S6D). While the Isoform A levels were lower, the reduction was not statistically significant in either lncRNA k.d. in each cell line (SI Appendix, Fig. S6E). The expression levels of Isoform D were elevated in both EGFR-AS1 and MIR205HG k.d. cell lines, but were significant only in HCC 827 cells (SI Appendix, Fig. S6F). However, upon considering the ratios of Isoform A to Isoform D, we found a significant reduction in the relative abundance of the 2 isoforms in both NCI H2228 (EGFR-AS1 k.d. P = 1 × 10−4; MIR205HG k.d. P = 1.6 × 10−3) and HCC 827 (EGFR-AS1 k.d. P = 1.6 × 10−6; MIR205HG k.d. P = 3.8 × 10−7) cells (Fig. 6E). These results indicate a reduction in ligand-dependent growth of the cells and, consequently, reduced impact of erlotinib on the cell lines with lncRNA k.d. (Fig. 6F). It is interesting to note that the MIR205HG k.d. also resulted in a similar phenotypic impact as EGFR-AS1 k.d. along with reduced Isoform A:D ratio. Moreover, we observed a significant reduction in MIR205HG expression in the EGFR-AS1 k.d. cells, hinting at a possible mediatory role of MIR205HG that calls for further investigation.
Discussion
Although representing over 80% of the human transcriptome, the pharmacogenomic relevance of lncRNAs in drug response is largely unknown. This gap in knowledge motivated us to comprehensively study the lncRNA transcriptome and genome to determine their relevance in cancer pharmacogenomics. Recent large-scale drug screening efforts (9–13) generated invaluable response data for hundreds of drugs measured across over 1,000 cell lines, along with exome sequencing and PCG expression data. We leveraged these datasets to perform an in-depth analysis of the relationship among the lncRNA transcriptome, genome, and response to drugs. Previously, expression and somatic alterations of PCGs were successfully utilized to predict drug response using various machine-learning approaches (35). From the early drug screens and subsequent prediction efforts, it is clear that the tissue lineage of cancer cell lines has a strong confounding effect on the drug response prediction models (10). Considering the tissue-specific expression patterns of lncRNAs (36), we emphasized the inclusion of tissue type of the cell lines as a covariate in all of our analyses. Moreover, we addressed the unique challenge of identifying drug-response-related lncRNAs that are independent of the effects of proximal cis-PCGs and well-established PCG biomarkers. Together with confounding effects of tissue type, adjusting for effects of cis-PCGs or known biomarkers is of critical importance in determining potential lncRNA biomarkers. These factors were not taken into account in previous attempts at predicting drug response using lncRNAs (17).
In this study, we first analyzed over 3.3 million drug–lncRNA expression associations in the CTRP screen and about 2.3 million associations in the GDSC screen. We determined a large proportion of significant drug–lncRNA associations overlapped in the 2 independent screens. Furthermore, we hypothesized and validated that the lncRNA transcriptome could effectively predict drug response with equal efficacy as PCGs in both screens.
The physical proximity of a portion of lncRNAs to PCGs and redundant expression patterns raise concerns whether lncRNAs actually provide any additional information. We addressed this important concern by carefully accounting for the possible effects of neighboring PCGs. The choice of ±500-Kb boundary that defined cis-PCGs in our study was based on the definition of cis-eQTLs from the genotype tissue expression project (37). Based on this conditional analysis, we determined about half of the initially observed drug–lncRNA associations were redundant with cis-PCG expression. However, for most drugs, the genomic loci for the top lncRNA biomarker do not overlap with the PCGs, including for the drugs with established clinical biomarkers.
Among the known oncogenic and tumor suppressor lncRNAs, we found intriguing associations between GAS5 and ZEB2AS1 expression with >50 drugs in both GDSC and CTRP. The GAS5 (growth arrest-specific 5) lncRNA acts as a tumor suppressor with various proposed mechanisms of action, including, cell cycle control (38–40), proliferation (41, 42), and regulation of epithelial to mesenchymal transition (EMT) program (43). Considering the multifaceted mechanisms by which GAS5 functions as a tumor suppressor, it is plausible that its expression may be predictive of response to multiple drugs. In contrast, from what we know so far about ZEB2AS1, it appears this transcript up-regulates ZEB2 expression to induce EMT program in cancer cells (43, 44). One possible mechanism could be the expression of this transcript is indicative of aggressive cancer cells that generally show a better response to drugs in vitro. Nevertheless, both lncRNAs are candidates for experimental validation as multidrug response prediction biomarkers.
In addition to the lncRNA transcriptome, we study the associations between somatic alterations in the lncRNA region and drug response. Currently, there are no gold standards to distinguish driver vs. passenger noncoding somatic variants. Unlike PCGs, noncoding variants cannot be classified as synonymous or nonsynonymous, which complicates identification of relevant variants. We attempted to identify lncRNA variants that did not overlap with any PCGs and were positively selected in the cell lines based on the total background noncoding mutation frequency and lncRNA mutation frequency across all cell lines. While preliminary, the emergence of significant somatic lncRNA associations with drug-response warrants future studies focusing on elucidating the biological role of such variants.
In clinical practice, a handful of somatic variants are routinely profiled to guide treatment decisions in cancer patients (23). Among these, somatic mutations that activate EGFR activity and their impact on anti-EGFR therapeutics like erlotinib and gefitinib are some of the first and extensively studied clinically actionable biomarkers (45). An important consideration in identifying novel lncRNA biomarkers is accounting for such well-known PCG somatic alterations to prevent redundancy. In this direction, we identified and characterized the impact of 2 lncRNAs, EGFR-AS1 and MIR205HG, which are strong predictors of response to erlotinib and gefitinib independent of EGFR somatic mutation status. A recent study proposed the theory that EGFR-AS1 stabilizes the EGFR signaling pathway by influencing the transcript ratio of the full-length EGFR Isoform A to the truncated Isoform D (33). Our results lend support to this idea, demonstrating the effect of EGFR-AS1 k.d. on the ratio of the 2 isoforms. Moreover, we reported a shift in both the growth pattern and erlotinib sensitivity of cell lines upon EGFR-AS1 k.d. confirming the functional impact of this lncRNA on addiction to ligand-dependent EGFR signaling. The MIR205HG lncRNA undergoes posttranscriptional processing leading to the synthesis of miRNA-205. Several studies have focused on the functions of this miRNA; however, its mechanism of action remains conflicting, with both oncogenic and tumor suppressor activities proposed (46, 47). Similar to EGFR-AS1, the k.d. of MIR205HG also resulted in a change in EGFR isoform ratio and phenotypic outcome. Additionally, MIR205HG expression levels were affected by EGFR-AS1 k.d., suggesting this gene could be an intermediator in EGFR isoform regulation. While miRNA-205 does not bind and regulate EGFR expression directly, it could modulate secondary transcriptional repressors that regulate the expression of the transcripts (48) (Fig. 6F). Besides an intriguing functional link with EGFR signaling, the expression of these 2 lncRNAs could predict response in patients who do not carry known activating EGFR mutations and thus are strong candidates for clinical evaluation.
We are still in the early stages of understanding the biology of lncRNAs, although sufficient evidence points toward the direction of altered lncRNA’s involvement in human cancers. By comprehensively analyzing the associations between lncRNA transcriptome, genome, and drug response, we have shown that lncRNAs are indeed biomarkers of drug response. Furthermore, we have provided compelling evidence that these associations are not just dependent on the correlative structure with PCGs. Future studies focusing on the mechanism of lncRNA action would be invaluable in improving our understanding of cancer progression and drug response.
Materials and Methods
Materials and methods are detailed in SI Appendix.
Supplementary Material
Acknowledgments
This study was supported by an NIH/National Cancer Institute (NCI) Grant 1R01CA204856-01A1. R.S.H. also receives support from a research grant from the Avon Foundation for Women.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 21963.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909998116/-/DCSupplemental.
References
- 1.Mattick J. S., Rinn J. L., Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5–7 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Kopp F., Mendell J. T., Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang K. C., Chang H. Y., Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quek X. C., et al. , lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 43, D168–D173 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan X., et al. , Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28, 529–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer M. K., et al. , The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huarte M., The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Schmitt A. M., Chang H. Y., Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnett M. J., et al. , Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barretina J., et al. , The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iorio F., et al. , A landscape of pharmacogenomic interactions in cancer. Cell 166, 740–754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu A., et al. , An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seashore-Ludlow B., et al. , Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 5, 1210–1223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djebali S., et al. , Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derrien T., et al. , The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malek E., Jagannathan S., Driscoll J. J., Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget 5, 8027–8038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., et al. , Systematic identification of non-coding pharmacogenomic landscape in cancer. Nat. Commun. 9, 3192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutschner T., Diederichs S., The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 9, 703–719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., et al. , The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS One 10, e0114586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L., et al. , MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin-induced cell apoptosis. Oncol. Rep. 38, 1383–1392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godinho M. F. E., et al. , Relevance of BCAR4 in tamoxifen resistance and tumour aggressiveness of human breast cancer. Br. J. Cancer 103, 1284–1291 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z., et al. , The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregulation of p21(WAF1/CIP1) expression. PLoS One 8, e77293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Relling M. V., Evans W. E., Pharmacogenomics in the clinic. Nature 526, 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pao W., et al. , Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pao W., et al. , EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U.S.A. 101, 13306–13311 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez J. G., et al. , EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Moroni M., et al. , Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: A cohort study. Lancet Oncol. 6, 279–286 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Gazdar A. F., Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 28 (suppl. 1), S24–S31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma S. V., Bell D. W., Settleman J., Haber D. A., Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Geeleher P., et al. , Discovering novel pharmacogenomic biomarkers by imputing drug response in cancer patients from large genomics studies. Genome Res. 27, 1743–1751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanzós A., et al. , Discovery of cancer driver long noncoding RNAs across 1112 tumour genomes: New candidates and distinguishing features. Sci. Rep. 7, 41544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juul M., et al. , Non-coding cancer driver candidates identified with a sample- and position-specific model of the somatic mutation rate. eLife 6, e21778 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan D. S. W., et al. , Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat. Med. 23, 1167–1175 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Reiter J. L., et al. , Comparative genomic sequence analysis and isolation of human and mouse alternative EGFR transcripts encoding truncated receptor isoforms. Genomics 71, 1–20 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Costello J. C., et al. , A community effort to assess and improve drug sensitivity prediction algorithms. Nat. Biotechnol. 32, 1202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang C., et al. , Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 7, 7120–7133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonsdale J., et al. ; GTEx Consortium , The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., et al. , lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci. Rep. 5, 10159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazar J., Rosado A., Shelley J., Marchica J., Westmoreland T. J., The long non-coding RNA GAS5 differentially regulates cell cycle arrest and apoptosis through activation of BRCA1 and p53 in human neuroblastoma. Oncotarget 8, 6589–6607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo G., et al. , LncRNA GAS5 inhibits cellular proliferation by targeting P27Kip1. Mol. Cancer Res. 15, 789–799 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P., Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3, ra8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., et al. , Long non-coding RNA growth arrest specific transcript 5 acts as a tumour suppressor in colorectal cancer by inhibiting interleukin-10 and vascular endothelial growth factor expression. Oncotarget 8, 13690–13702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuang J., et al. , TGFβ1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci. Rep. 5, 11924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beltran M., et al. , A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 22, 756–769 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jimeno A., Hidalgo M., Pharmacogenomics of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors. Biochim. Biophys. Acta 1766, 217–229 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Greene S. B., Herschkowitz J. I., Rosen J. M., The ups and downs of miR-205: Identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol. 7, 300–304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu K., Shen W., Zhang Y., Zhao Y., Lu Y., MiR-205 promotes motility of ovarian cancer cells via targeting ZEB1. Gene 574, 330–336 (2015). [DOI] [PubMed] [Google Scholar]
- 48.De Cola A., et al. , miR-205-5p-mediated downregulation of ErbB/HER receptors in breast cancer stem cells results in targeted therapy resistance. Cell Death Dis. 6, e1823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.