Significance
Aggregatibacter actinomycetemcomitans, a microorganism associated with aggressive periodontitis, can result in tooth loss and systemic complications. The hyperleukotoxin-producing A. actinomycetemcomitans strain demonstrates up-regulation of attachment genes, permitting it to attach better than the wild-type strain in competition with commensal bacteria for space on tooth surfaces. Further, the availability of lactate provided by streptococci and other members of the indigenous plaque microbiota consorting with A. actinomycetemcomitans reduces its competition for glucose as a main carbon source and assists already-attached strains in growth and persistence in the developing plaque biofilm. Data derived from in vivo and in vitro studies illustrate functional strategies that allow A. actinomycetemcomitans to colonize and survive in the competitive and stressful environment of the oral cavity.
Keywords: Aggregatibacter actinomycetemcomitans, leukotoxin, rhesus monkey, l-lactate, microbiome
Abstract
Aggregatibacter actinomycetemcomitans is associated with aggressive periodontitis resulting in premature tooth loss in adolescents. Tooth adherence and biofilm persistence are prerequisites for survival in the oral domain. Here, using a rhesus monkey model, 16S rRNA sequencing, and weighted network analysis, we assessed colonization of A. actinomycetemcomitans variants and ascertained microbial interactions in biofilm communities. Variants in A. actinomycetemcomitans leukotoxin (ltx) were created, labeled, inoculated, and compared with their progenitor strain for in vivo colonization. Samples of tooth-related plaque were assessed for colonization at baseline and after debridement and inoculation of labeled strains. Null, minimal, and hyper-Ltx–producing strains were created and assessed for hydroxyapatite binding and biofilm formation in vitro. Ltx-hyperproducing strains colonized with greater prevalence and at higher levels than wild type or ltx mutants (P = 0.05). Indigenous and inoculated A. actinomycetemcomitans strains that attached were associated with lactate-producing species (i.e., Leptotrichia, Abiotrophia, and Streptoccocci). A. actinomycetemcomitans was found at 0.13% of the total flora at baseline and at 0.05% 4 wk after inoculation. In vivo data were supported by in vitro results. We conclude that hyper-Ltx production affords these strains with an attachment advantage providing a foothold for competition with members of the indigenous microbiota. Increased attachment can be linked to ltx gene expression and up-regulation of adherence-associated genes. Growth of attached A. actinomycetemcomitans in vivo was enhanced by lactate availability due to consorting species. These associations provide A. actinomycetemcomitans with the constituents required for its colonization and survival in the complex and competitive oral environment.
Aggregatibacter actinomycetemcomitans is a gram-negative oral pathobiont implicated in localized aggressive periodontitis (LAgP) that occurs primarily in adolescents of African ancestry and can result in premature tooth loss (1, 2). It is well-established that A. actinomycetemcomitans colonization of teeth is a critical step in the infectious process (3). Colonization is a 2-step process that includes initial attachment followed by growth and persistence of attaching strains (4). Colonization is a key component of survival for microorganisms in the hazardous environment of the oral cavity (4).
Genetic and functional studies have demonstrated that A. actinomycetemcomitans possesses a 14-gene operon (the widespread colonization island) containing the tight adherence (tad) genes that show avid biofilm formation and robust binding to enamel-like substrates in vitro (5–8).
Previously, we demonstrated that inoculation of wild-type A. actinomycetemcomitans colonized the oral cavity of rhesus (Rh) monkeys, while A. actinomycetemcomitans with a deletion in the leukotoxin (ltx) structural gene failed to colonize (9). In vitro experiments determined that deletion of ltxA interfered with expression of attachment genes (tad and flp) required for colonization (9, 10). This previously unknown association between leukotoxin production and attachment (2 formerly unrelated functions) led us to speculate that perhaps A. actinomycetemcomitans’s hyperleukotoxin production, found in the most aggressive forms of LAgP (11), could be linked to up-regulation of attachment genes and increased in vivo colonization (11).
This study therefore aimed to determine whether increased A. actinomycetemcomitans ltxA gene expression would result in up-regulation of tad genes. If so, the study posited that introduction of this novel A. actinomycetemcomitans strain, having an attachment advantage, could gain a foothold among the competing indigenous strains of the same species in our Rh primate colonization model (9, 12). Further, it has been postulated that the virulence attributed to the hyper-Ltx–producing strain of A. actinomycetemcomitans can be modulated and reduced when found in conjunction with wild-type minimally leukotoxic A. actinomycetemcomitans; however, this association has not yet been explored in a competitive in vivo colonization model (11).
A. actinomycetemcomitans strains have been reported to thrive and survive by using lactate as their preferred carbon source (13). Early plaque development resulting from A. actinomycetemcomitans’s competition for nutrients with fast-growing, glucose-utilizing, pioneer tooth-adhering species (i.e., streptococci and Actinomyces spp., lactate-producing species) can be lessened by A. actinomycetemcomitans’s preference for lactate as its main carbon source (13–15). This led us to reason that the growth of A. actinomycetemcomitans strains that attached in our Rh model would be enhanced by lactate availability derived from pioneer lactate producer strains contained within the tooth-associated biofilm milieu.
This study was thus designed to examine ltxA overexpression in relationship to A. actinomycetemcomitans colonization in the Rh primate model. We coupled in vivo colonization with in vitro assessment of Ltx production, expression of adherence genes, and in vitro adherence to tooth homologs. Using the primate model, we also estimated the association of a variety of lactate-producing pioneer species in relation to the growth and persistence of A. actinomycetemcomitans in vivo. The clear importance of A. actinomycetemcomitans’s ability to colonize, grow, and persist in the competitive oral environment led us to hypothesize that 1) Ltx-hyperproducing strains would promote A. actinomycetemcomitans attachment in vivo in spite of competition with native strains, and 2) increased availability of lactate in the biofilm environment would enhance the growth and persistence of both native and inoculated A. actinomycetemcomitans strains that were already attached.
Results
Confirmation of Leukotoxin Activity of A. actinomycetemcomitans Variant Strains.
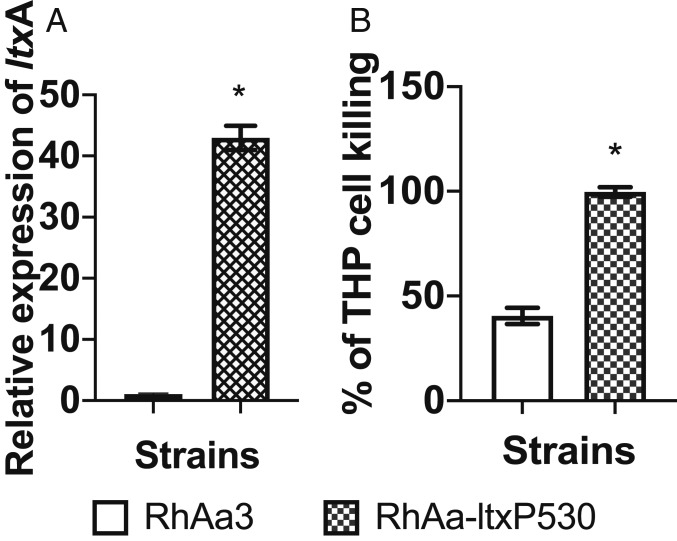
Western blotting showed elevated levels of leukotoxin for RhAa-ltxP530 (the hyper-Ltx–producing strain) when compared with RhAa3 (the wild-type minimal Ltx-producing strain) and RhAa-VS2 (the null-Ltx–producing strain) (SI Appendix, Fig. S1) (16). RhAa-ltxP530 exhibited significantly higher levels of expression of ltxA as compared with the wild-type RhAa3 strain (P < 0.01; Fig. 1A). The culture supernatants from RhAa-ltxP530 exhibited significantly higher LtxA activity as compared with the RhAa3 strain (Fig. 1B). RhAa-VS2 was used as a negative control.
Fig. 1.
Leukotoxin expression levels in A. actinomycetemcomitans strains. (A) The expression of ltxA was significantly higher in the RhAa-ltxP530 strain as compared with the RhAa3 strain and was analyzed by qRT-PCR to measure gene transcripts. (B) Functional leukotoxin production as determined by THP killing was significantly higher in the RhAa-ltxP530 strain as compared with the RhAa3 strain. All of the experiments were performed in biological triplicate and the data shown are the standard error of the mean (±SEM). The differences between the groups were calculated by the Student’s t test (*P < 0.05).
In vivo studies.
Colonization efficiencies of inoculated and labeled A. actinomycetemcomitans strains.
Baseline sampling of indigenous tooth-related plaque was performed prior to debridement of the oral cavity. Oral samples obtained by scalers were placed in tubes for plating and identification (Materials and Methods). After sampling, oral prophylaxis was performed using dental scalers followed by rubber cup polishing after which oral surfaces were subjected to Listerine antiseptic to reduce contamination. A confluent suspension of 1 of the 3 spectinomycin-resistant (SpecR) strains of A. actinomycetemcomitans (RhAa-ltxP530; RhAa3; RhAa-VS2) was applied independently to teeth in each of the 5 monkeys in each inoculation group (SI Appendix, Fig. S1). The control group received no inoculation of foreign A. actinomycetemcomitans.
We examined tooth-related biofilms at baseline prior to cleaning and 1, 2, and 4 wk after the final inoculation of SpecR strains of A. actinomycetemcomitans. Supragingival colonization and persistence of inoculated A. actinomycetemcomitans variants were compared and assessed in the context of the overall tooth-related microbiome derived from our Rh model (SI Appendix, Table S1).
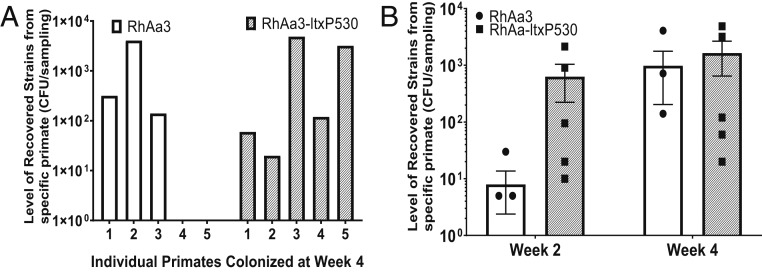
Indigenous A. actinomycetemcomitans colonized routinely in all monkeys. A. actinomycetemcomitans was always found at relatively low abundance (∼0.13% of the total flora at baseline, and 0.05% at the 4-wk assessment; SI Appendix, Table S2) when assessed against the background of the total biofilm microbiome. Four weeks post inoculation, 3 of 5 monkeys in the RhAa3 group colonized, while all 5 monkeys infected with the RhAa-ltxP530 group colonized and persisted (Fig. 2A). None of the monkeys in the RhAa-VS2 group-labeled strains (given the ltx-null strain) were detected in any of the 5 monkeys over the 4-wk period (SI Appendix, Fig. S1). In addition to increased prevalence, the RhAa-ltxP530 group, inoculated with the hyper-LtxA–producing strain, showed an increased level of colonization and persistence as compared with the RhAa3 group at all time periods. At week 2, RhAa-ltxP530 was significantly elevated (P = 0.03; Fig. 2B). The recovered foreign A. actinomycetemcomitans (SpecR) was found at about 7.4 to 16.5% of the total indigenous A. actinomycetemcomitans obtained from the Rh primates (SI Appendix, Table S2).
Fig. 2.
Recovery of labeled A. actinomycetemcomitans strains in plaque. (A) All 5 primates in the RhAa-ltxP530 group showed colonization, while only 3 in the RhAa3 group colonized at week 4. (B) The level of recovery was significantly higher in the RhAa-ltxP530 group vs. RhAa3 at week 2 (P = 0.03) but not in week 4, illustrating the difference between early attachment and growth and persistence. No strains from the RhAa-VS2 strain were recovered in any time period (SI Appendix, Fig. S1, colonization at 1, 2, and 4 wk). CFU, colony-forming unit.
Pattern of associations of oral microbes with A. actinomycetemcomitans in mature and developing plaque.
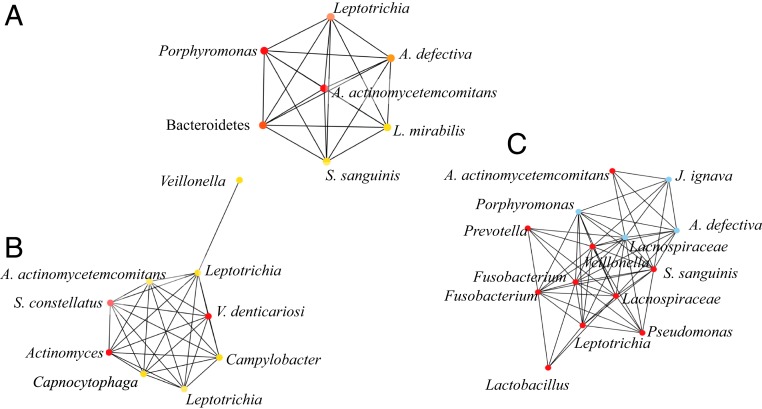
Network analysis (17, 18) was performed using total A. actinomycetemcomitans (indigenous and SpecR-inoculated) and the other top 50 most abundant microbes assessed by 16S rRNA next-generation sequencing. At baseline, in mature plaque, A. actinomycetemcomitans was highly associated with Leptotrichia, Abiotrophia, and streptococci, all lactate-producing species (Fig. 3 and SI Appendix). In 4-wk plaque, once again, A. actinomycetemcomitans was associated with Leptotrichia, Lactobacillus, Abiotrophia, and streptococci, confirming cooperative associations between A. actinomycetemcomitans and these unrelated species (Fig. 3). Co-occurrence network analysis (CoNet) of taxa confirmed associations made with the weighted gene correlation network analysis (WGCNA) and further validated the association of A. actinomycetemcomitans with interconnected species (19) (see detailed methods in SI Appendix, Fig. S2).
Fig. 3.
WGCNA showing the relationship of A. actinomycetemcomitans to its colonization partners in all of the inoculation groups. Supragingival plaque samples for WGCNA assessed at baseline (A), week 2 (B), and week 4 (C). At baseline, 3 of 6 of the most closely associated bacteria at the genus level were l-lactate producers: streptococci, Leptotrichia, and Abiotrophia. Analysis included total A. actinomycetemcomitans (both inoculated and indigenous). At week 2, after the final inoculation, Leptotrichia and Streptococcus spp. were prominent while at 4 wk the top co-occurrence l-lactate producers were Leptotrichia, Lactobaccillus, streptococci, and Abiotrophia. Color coding reflects the strength of the association/co-occurrence among the OTUs (red, strong; blue, intermediate; and yellow, weak). A. actinomycetemcomitans in subnetworks was subjected to coding (SI Appendix).
Estimation of lactate availability in inoculation groups over time.
The operational taxonomic units (OTUs) for the top 50 microbial groups were assessed. Prominent lactate producers were found in plaque derived from all inoculation groups. The total OTUs from the most prominent lactate-producing strains included Abiotrophia spp., Leptotrichia spp., Lactobacillus spp., and Streptococcus spp. Data derived from these lactate producers were compared with Veillonella, a species with a known lactate requirement. In our effort to understand the increased in vivo persistence of A. actinomycetemcomitans strains in the hyper-Ltx–producing group (RhAa-ltxP530) and the wild-type A. actinomycetemcomitans group (RhAa3), we assessed community levels (total and labeled A. actinomycetemcomitans) and lactate producers and utilizers in each group. To estimate available lactate, we calculated total OTUs in the most prominent lactate-producing strains, as compared with Veillonella, the most prominent lactate utilizer. Lactate availability was estimated by calculating the ratio of prominent lactate producers as compared with the most prominent lactate utilizer. The RhAa-ltxP530 hyperproducer group exhibited an elevated ratio of lactate availability as compared with the wild-type group consistently over the course of the 4-wk period (Table 1).
Table 1.
Estimate of l-lactate producers vs. a l-lactate utilizer
| Strains | RhAa3, no. of OTUs | RhAa-ltxP530, no. of OTUs | ||||
| Week 1 | Week 2 | Week 4 | Week 1 | Week 2 | Week 4 | |
| l-lactate producers a* | 6,568 | 3,313 | 3,210 | 4,476 | 3,788 | 4,506 |
| Streptococci b | 4,560 | 22,167 | 16,650 | 7,386 | 12,052 | 10,022 |
| l-lactate producers (P)** | 11,128 | 25,480 | 19,860 | 11,862 | 15,840 | 14,528 |
| Veillonella (U) | 3,722 | 13,690 | 1,037 | 2,402 | 3,388 | 422 |
| P/U ratio | 3.0 | 1.9 | 19.2 | 4.9 | 4.7 | 34.4 |
P, producer; U, utilizer.
l-lactate producers include Abiotrophia, Lactobacillus, and Leptotrichia.
Total lactate producers include a and b.
In vitro studies.
Assessment of A. actinomycetemcomitans binding to salivary-coated hydroxyapatite.
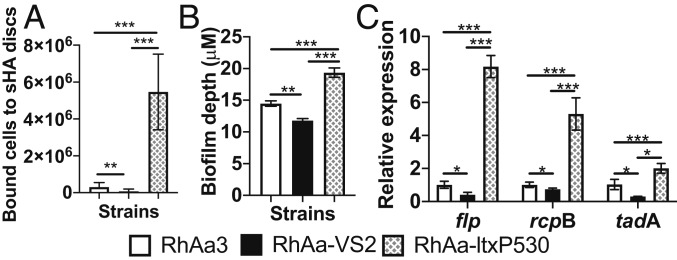
Increased in vivo colonization levels of the hyper-Ltx–producing group could be due to the ability of these strains to attach to tissues with greater efficiency. The RhAa-ltxP530 strain showed significantly increased binding to salivary-coated hydroxyapatite (sHA) discs as compared with the RhAa3 and RhAa-VS2 strains (Fig. 4A). Experiments were performed in biological quadruplicate.
Fig. 4.
sHA binding, biofilm formation, and gene expression in A. actinomycetemcomitans variant strains in vitro. (A) The RhAa-ltxP530 strain showed increased binding to sHA discs as compared with the RhAa-VS2 and RhAa3 strains (P = 0.0001). Experiments were performed in biological quadruplicate. (B) The confocal analysis of biofilm depth was significantly increased in RhAa-ltxP530 vs. RhAa3 and RhAa-VS2. (C) Attachment gene expression was significantly elevated in RhAa-ltxP530 as compared with the 2 other strains (P = 0.0001). *P = 0.1, **P = 0.001, ***P = 0.0001. Experiments in B and C were performed in triplicate. The significant high-level expression was calculated by 1-way analysis of variance using the Tukey’s post hoc test to determine differences between groups (*P < 0.05).
Assessment of biofilm growth.
Confocal microscopic analysis enabled comparison of biofilm formation in the 3 strains. The hyperleukotoxin-producing strain showed a significant increase in biofilm depth when compared with the other strains (Fig. 4B).
Expression of genes involved in attachment and biofilm formation.
In order to better understand the in vivo colonization data, we examined the functional relationship of gene expression of the A. actinomycetemcomitans variants in vitro (16). Among the genes in the tad locus, flp-1, rcpB, and tadA were elevated in RhAa-ltxP530 (Fig. 4C). Genes for aae, apiA, and rcpA were also significantly up-regulated in RhAa-ltxP530 as compared with RhAa-VS2 (SI Appendix, Fig. S3). Experimental results as shown above were analyzed by 1-way ANOVA with Tukey’s post hoc multiple comparison, setting P = 0.05 as the significance level.
Effect of l-lactate on biofilm growth and gene expression.
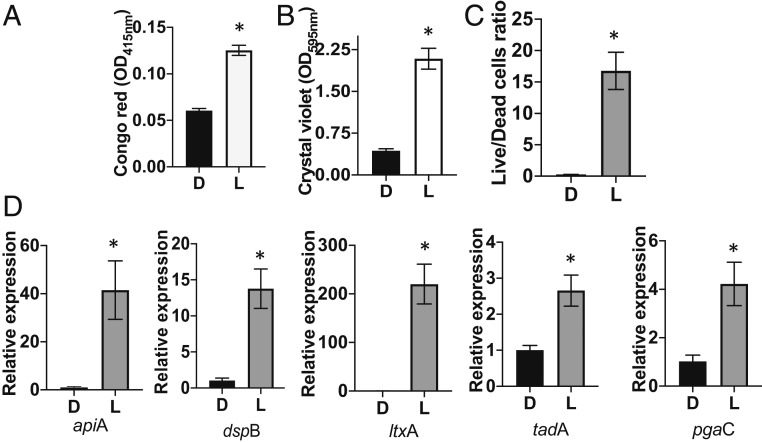
RhAa-ltxP530 cells grown for 72 h under anaerobic conditions in l-lactate–supplemented chemically defined media (CDM) (nitrate added to CDM acted as the terminal electron acceptor under anaerobic conditions) showed a significant increase in biofilm growth and formation (crystal violet staining) and exopolysaccharide production (Congo red staining) (Fig. 5 A and B). More live cells were also seen in CDM supplemented with lactate vs. dextrose at 72 h for the hyper-Ltx strain (see Fig. 5C and SI Appendix, Fig. S4 for confocal images). Similar results were shown for RhAa3 (SI Appendix, Figs. S4 and S5A). We also examined the mRNA expression of genes (Fig. 5D) related to tight adherence (tadA), dispersin B (dspB), leukotoxin (ltxA), apiA, and exopolysaccharide (pgaC) in CDM supplemented with dextrose vs. l-lactate for RhAa-ltxP530. RhAa-ltxP530 showed a significant increase in all genes assessed at a level of P < 0.01 in l-lactate–supplemented media (Fig. 4D). Similar results were observed for RhAa3 (SI Appendix, Fig. S5B). The above experimental data were analyzed by the Student’s t test (P < 0.05).
Fig. 5.
Effect of lactate on growth, gene expression, and survival of the RhAa-ltxP530 strain. (A) Increased biofilm layering (Congo red staining). (B) Elevated growth (crystal violet staining). (C) Increased live/dead cell ratios for the RhAa-ltxP530 strain grown in l-lactate (L)–supplemented media compared with dextrose (D) under anaerobic conditions. RhAa3 showed identical results, which are shown in SI Appendix, Fig. S5. (D) Level of virulent gene expression when RhAa-ltxP530 was grown in media supplemented with l-lactate as compared with dextrose. Growth was significantly higher in cells grown on l-lactate as compared with dextrose. Images were analyzed by calculating the live/dead cell ratio (propidium iodide/live and SYTO9/total) (SI Appendix, Fig. S4). The ratio of live cells was significantly higher in lactate media compared with dextrose. Differences were calculated by the Student’s t test (*P < 0.05).
Discussion
Studies of dental plaque formation were among the first to describe biofilms and microbial ecology in a well-defined human habitat (20, 21). Seminal in vivo studies documented microbial successional events that took place on tooth surfaces after thorough debridement (22). Immediately after debridement, gram-positive, pioneer facultative aerobic glucose-utilizing streptococcal species were shown to attach to salivary-coated enamel surfaces (21–23). These pioneer species developed stacks of parallel arrays of conjoined cocci, forming perpendicular to tooth surfaces, that gave the impression of skyscrapers emerging from the streets of New York City (24). Interspersed in the spaces between these skyscraper-like parallel arrays were capnophilic gram-negative species, such as the lactate-utilizing Veillonella spp. (4, 25, 26). These pioneer colonizers resisted the forces of mastication and salivary flow due to their inherent ability to attach and persist (24). Further growth was dependent on the metabolic adaptability of these already-attached microorganisms (25).
The Rh primate colonization model allowed us to investigate A. actinomycetemcomitans, an important understudied pathobiont, in an ecology and oral anatomy which resembles that observed in humans. The role of A. actinomycetemcomitans in plaque/biofilm formation has been difficult to document because it rarely occurs in the mouths of healthy subjects (26) and it has a low abundance even when isolated from diseased patients (1, 27). However, A. actinomycetemcomitans is found with great regularity in the mouths of periodontally healthy Rh primates (28), suggesting that Rh monkeys could provide an ideal model to study A. actinomycetemcomitans in a natural environment. In fact, a limited number of studies conducted in monkeys and humans support the concept that A. actinomycetemcomitans is found in early plaque formation, which reenforces its in vitro attachment characteristics (8, 23, 29). Previously, the Rh primate model provided data indicating that newly introduced and labeled A. actinomycetemcomitans, a capnophilic/lactate-utilizing species, could be recovered in early-developing plaque, albeit at low abundance (8).
Data from this current study demonstrated that the newly introduced hyper-Ltx–producing A. actinomycetemcomitans strain rarely exceeded 18% of total recoverable A. actinomycetemcomitans, and typically was found in 8% of the total indigenous A. actinomycetemcomitans flora at the end of the experimental period. Further, the overall cumulative A. actinomycetemcomitans percentage of the total flora ranged from 0.13% at baseline to 0.05% over the 4-wk period of plaque accumulation, which followed intensive debridement. Of particular interest is the power of indigenous A. actinomycetemcomitans to reclaim its dominance in its natural habitat, despite thorough mechanical debridement and antiseptic application prior to inoculation of repeated high doses of labeled A. actinomycetemcomitans. The colonization model also permitted us to observe the way in which inoculated/foreign A. actinomycetemcomitans can gain traction in a microbiota dominated by pioneer colonizing species.
The hyperleukotoxic A. actinomycetemcomitans strain found in humans (equivalent to RhAa-ltxP530 created in Rh monkey strains) is thought to be a highly pathogenic A. actinomycetemcomitans strain in aggressive periodontitis seen in adolescents of African and Middle Eastern descent (2, 11). Clinical findings show more disease in individuals who have the hyper-Ltx strains, which is typically explained by the fact that increased levels of Ltx have been shown to kill polymorphonuclear leukocytes, macrophages, and lymphocytes efficiently to overcome host defenses (30). The findings presented in this study suggest that increased Ltx levels can also be associated with up-regulation of adherence genes that can result in increased attachment of A. actinomycetemcomitans to tooth surfaces (the first step in the infection process).
Individuals who carry hyper-Ltx–producing strains have been shown to ascribe a significant increase in relative risk when compared with individuals who carry the wild-type strain in longitudinal studies of LAgP (31). Despite the decrease in relative risk shown when wild-type A. actinomycetemcomitans strains are seen in conjunction with the hyper-Ltx–producing strains, this combination of A. actinomycetemcomitans variants has not been studied and could be relevant to disease (11). Results obtained in our primate model provide an opportunity to study these 2 variants in the same ecosystem (11).
This study has demonstrated that high levels of Ltx production in vitro caused elevated levels of expression of flp-1, apiA, and tad locus genes (attachment-related genes), which results in increased attachment to hydroxyapatite and increased biofilm depth in vitro (32, 33). This previously undocumented association between leukotoxin and adherence genes (10) has been supported in an erythromycin stress model, which showed that increased ltxB expression was associated with increased expression of flp-1 and pga, both related to colonization and biofilm formation (15).
With respect to persistence and survival, recent evidence indicates cooperative associations between streptococci, the preeminent early plaque formers, and A. actinomycetemcomitans (34). Due to its slow growth rate and limited carbon catabolic activity, A. actinomycetemcomitans shows a predilection for lactate produced by streptococci as its favored carbon source (34, 35). This preferential selection permits A. actinomycetemcomitans to avoid competition for glucose with the rapidly growing, highly catabolic strains of streptococci seen in abundance in early-stage supragingival plaque. Moreover, the association of A. actinomycetemcomitans with streptococci, well-established early pioneer colonizers, that are known to be the initial colonizers of tooth surfaces immediately after debridement (21), provides a biologically plausible rationale for the presence of A. actinomycetemcomitans in the neighborhood of lactate-rich early-forming dental plaque (13).
A. actinomycetemcomitans interactions with streptococci have previously been illustrated both in in vitro and in vivo models (15). The mutually beneficial/cooperative interactions between streptococci and A. actinomycetemcomitans provide a window into the complex associations in human climax communities. Our results indicate that A. actinomycetemcomitans is highly associated with Streptococcus spp., Leptotrichia spp., Lactobacillus spp., and Abiotrophia, all lactate-producing species present in plaque found on tooth surfaces.
To test this concept, we adapted WGCNA, originally used to examine gene expression in cancer biology, to assess community associations between dominant plaque microorganisms and A. actinomycetemcomitans (17, 18, 36). WGCNA allowed us to examine lactate-producing species in both fully formed plaque, as seen at baseline, and in plaque developing de novo over the period from 2 to 4 wk in our Rh monkey model (18, 36). Reanalysis of the same data using CoNet combined with Cytoscape yielded similar associations (37, 38). These associations were consistent with results found in a human longitudinal model of healthy subjects who developed aggressive periodontitis in which Streptococcus parasanguinis, a lactate producer, was found to be associated with A. actinomycetemcomitans in the anaerobic subgingival habitat in periodontal pockets in subjects prior to developing aggressive periodontitis (26). The estimate of increased l-lactate availability in vivo illustrated in this study suggests that this metabolic interaction is present in a competitive stochastically determined environment.
In conclusion, this study reports that deletion of 530 bp in the promoter region of A. actinomycetemcomitans resulting in increased expression of Ltx is associated with increased expression of the tad and apiA genes, increased attachment to sHA, and increased attachment to teeth in an in vivo model that resembles that seen in humans. In addition, this study shows that streptococci as well as other lactate-producing members of the indigenous plaque flora that are found in the vicinity of A. actinomycetemcomitans can contribute to its persistence in both mature and developing plaque biofilm. Moreover, combining both in vitro and in vivo data in this study supports the concept that A. actinomycetemcomitans colonization and persistence in a competitive and complex supragingival domain are fostered by the combination of increased expression of attachment genes and the association of attached A. actinomycetemcomitans with a variety of lactate-producing plaque microbes.
Materials and Methods
Analysis of Leukotoxic Activity of Rhesus A. actinomycetemcomitans Strains.
Cell-free culture supernatants were equalized to 45 μg of protein derived from all A. actinomycetemcomitans strains and evaluated in vitro by Western blot and for THP-1 cell viability (SI Appendix, Fig. S6) (39). The viability of THP-1 cells exposed to normalized supernatants was used to assess Ltx activity as evaluated by the amount of ATP produced by live THP-1 cells exposed to Ltx and measured by luminescence using the Infinite M200 PRO microplate reader (Tecan). In all experiments, uncultured liquid media and purified leukotoxin standards were used as negative and positive controls, respectively. Experiments were conducted in biological triplicate. Relative gene expression of ltxA was assessed by RT-PCR (16).
Bacterial Strains and Growth Conditions.
Strains used in this study are listed in SI Appendix, Table S1. A. actinomycetemcomitans strains were routinely grown on brain heart infusion agar (BHI; Becton Dickinson) containing 0.6% yeast extract supplemented with 0.8% dextrose and 0.4% sodium bicarbonate. Spontaneous SpecR RhAa3, RhAa-VS2, and RhAa-ltxP530 strains were developed to discriminate between inoculated and indigenous A. actinomycetemcomitans strains (9). Total A. actinomycetemcomitans was enumerated using BHI supplemented with bacitracin (75 μg/mL) and vancomycin (5 μg/mL) (BHIBV plates). Inoculated SpecR A. actinomycetemcomitans strains were enumerated using BHIBV plates supplemented with spectinomycin (50 μg/mL) (BHIBVSPEC). Plates were incubated for 24 to 48 h at 37 °C in a 10% CO2 environment.
In Vivo Rh Monkey Colonization Experimental Design.
All experimental procedures were performed in accordance with the Institutional Animal Care and Use Committees (IACUCs) of Rutgers University and the New England Primate Research Center. Overall, 20 male Rh monkeys (Macaca mulatta, 2 to 5 y old) were studied. Monkeys were monitored daily as per IACUC recommendations (SI Appendix, Fig. S7, flow diagram).
Sample Acquisition and Oral Inoculation.
Monkeys were anesthetized with 10 to 20 mg/kg of ketamine and medetomidine intramuscularly prior to oral procedures. After sampling and oral prophylaxis, Listerine was applied to reduce contamination of oral surfaces. Sterile water was applied to remove all residual Listerine. Oral plaque samples collected in tubes containing 1 mL of PBS were plated on BHIBV or BHIBVSPEC plates (9, 12) and colonies were examined and enumerated (Olympus SZ61 microscope) for their star-shaped morphology and then independently confirmed as A. actinomycetemcomitans by PCR.
A second inoculation identical to the procedure previously described (Results) was performed on day 7. Sampling was done on weeks 1, 2, and 4 after inoculation. Samples were evaluated for 1) total bacterial counts using 16S sequencing OTUs derived from next-generation DNA sequencing, 2) total OTUs found, and 3) recovery of the 3 types of inoculated/SpecR A. actinomycetemcomitans strains via plate counting.
Supragingival plaque was collected from different quadrants on weeks 1, 2, and 4. Collected plaque was split into 2 sample vials; one vial was used for DNA analysis of bacterial strains (40, 41), and the second for recovery of inoculated A. actinomycetemcomitans by culture methods.
Human Oral Microbiome Next-Generation Sequencing.
For DNA analysis, 10 to 50 ng of total DNA obtained from plaque samples was PCR-amplified using V3 to V4 primers and HotMasterMix (5 Prime). PCR samples were purified (AMPure beads; EpiGentek). Each library was pooled, gel-purified, and quantified using qPCR. Detection levels were reported as levels at or above 105 OTUs. MiSeq was used (Illumina; see SI Appendix for more details).
Estimation of Patterns of Association of A. actinomycetemcomitans and Other Members of the Rh Primate Oral Microbiota Using WGCNA and CoNet.
We obtained OTUs from human oral microbiome next-generation sequencing (HOMINGS) analysis and subjected those identified to the genus level to WGCNA in order to estimate the microbes most associated with A. actinomycetemcomitans found in the plaque sampled (36). Over 700 taxa were identified. The relationship between A. actinomycetemcomitans and other members of the flora was restricted to the top 50 microbes by the percentage of the total calculated. Network analysis performed by WGCNA was applied to complex biofilm communities (18). A correlation matrix containing all pairwise Pearson correlations between all probe sets across all monkeys was performed. Outlier samples were identified using an absolute hierarchical cluster analysis and the top 50 microbes were used to construct a weighted network by choosing a beta thresholding power such that co-occurrence similarity could be calculated (17, 36). Briefly, this method was used to identify groups of highly correlated OTUs that co-occur across different monkey samples. WGCNA uses a weighted OTU co-occurrence adjacency matrix to include the actual strength of the association between OTUs and their abundance. In this analysis, the power (soft thresholding value) and R2 (scale of independence) were between 8 and 10, and 0.75 and 0.84, respectively, for the 3 sets of samples. Baseline samples were assessed, as were samples obtained after the final inoculation. For our analysis of A. actinomycetemcomitans, 1 of the top 50 microbes in the analysis was selected to determine the most closely associated microbes. Network centralities were assessed using Cytoscape (18, 38). The patterns of microbial co-occurrence were also assessed by CoNet, a second analysis system designed to examine significant mutual coinhabitants (37) (see SI Appendix for added information).
Estimation of Lactate Availability in Primate Plaque Communities.
Total OTUs for all species were determined by HOMINGS and then the top 50 species were calculated using the WGCNA methodology. As such, Abiotrophia, Leptotrichia, Lactobacillus, and Streptococcus species emerged as the most prominent lactate producers in all 3 groups for baseline and over the 4-wk period after the final inoculation. In addition, since our intent was to determine lactate availability as it related to persistence in vivo, we also calculated OTUs for the Veillonella species, the most prominent lactate utilizer as determined by WGCNA. These calculations were done for all 3 groups for the network centralities via Cytoscape. However, since only RhAa3 (wild type) and RhAa-ltxP530 (hyper-Ltx producer) colonized in the in vivo model, we limited our comparison to these 2 groups. Therefore, we calculated the total OTUs for each species in the 2 groups over 4 wk and computed a ratio for differences in specific lactate producers as compared with the most prominent l-lactate utilizer for each group. These computed ratios represented the estimated lactate availability over time when the 2 groups were compared.
In Vitro Analysis of Attachment to Salivary-Coated Hydroxyapatite Discs.
A modification of the sHA binding assay was performed as previously described by our laboratory (10). The statistical significance in binding between strains was calculated by 1-way ANOVA using Tukey’s post hoc multiple-comparison test (SI Appendix).
In Vitro Analysis of Biofilm Growth and Viability and Gene Expression.
A. actinomycetemcomitans strains RhAa3, RhAa-VS2, and RhAa-ltxP530 were grown on BHI agar plates overnight. Colonies were scraped from plates and resuspended (10). Aggregates were subjected to a handheld motorized pestle to break up clumping cells to obtain a uniform seed inoculum (Kimble Chase). The optical density of the cells was adjusted to OD600 nm = 0.2. Sixty microliters of A. actinomycetemcomitans cells was seeded onto 25 mL of BHI media and incubated for 16 h in an atmosphere of 10% CO2. To measure the biofilm depth, the cells were seeded onto 35-mm glass-bottom microwell culture dishes and after film tracer biofilm LIVE/DEAD staining images were acquired with an A1R-A1 confocal microscope (Nikon Instruments) using the objective lens Plan Apo VC 60× WI DIC N2 (16). The depth of the biofilm was measured in 9 different spots of 3 independent dishes and the averaged data were analyzed by 1-way ANOVA with Tukey’s post hoc multiple-comparison test. P ≤ 0.05 was considered significant. For qRT-PCR analysis of all gene transcripts, the cells were seeded onto 100 × 15-mm Petri dishes. After 16-h growth the cells were collected and the total RNA was extracted. To test the effect of the carbon source on biofilm growth and gene expression analysis, a similar concentration of seed culture was inoculated into CDM supplemented with 40 mM dextrose or l-lactate and the cells were grown for 48 to 72 h. All experiments were conducted in biological triplicate and the significant difference was calculated by the Student’s t test.
Statistical Analysis.
Data analysis is provided in the appropriate section of the paper and in SI Appendix.
Supplementary Material
Acknowledgments
We thank the animal care staff at the New England Primate Research Center. We also would like to acknowledge funding derived from Grant R21 DE 021172 (to D.H.F.) from the National Institute of Dental and Craniofacial Research for partial support for the work performed in this study. In addition, D.H.F. would like to thank Dean Cecile Feldman, Rutgers School of Dental Medicine, for added support for these studies. We would also like to thank Dr. S. Horvath for help with the Weighted Coexpression Gene Network Analysis System.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905238116/-/DCSupplemental.
References
- 1.Zambon J. J., Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12, 1–20 (1985). [DOI] [PubMed] [Google Scholar]
- 2.Haubek D., Ennibi O. K., Poulsen K., Benzarti N., Baelum V., The highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans and progression of periodontal attachment loss. J. Dent. Res. 83, 767–770 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Fine D. H., Kaplan J. B., Kachlany S. C., Schreiner H. C., How we got attached to Actinobacillus actinomycetemcomitans: A model for infectious diseases. Periodontol. 2000 42, 114–157 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Marsh P. D., Zaura E., Dental biofilm: Ecological interactions in health and disease. J. Clin. Periodontol. 44 (suppl. 18), S12–S22 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Fine D. H., et al. , Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: Implications for virulence. Microbiology 145, 1335–1347 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Haase E. M., Zmuda J. L., Scannapieco F. A., Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 67, 2901–2908 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kachlany S. C., et al. , Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182, 6169–6176 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Planet P. J., Kachlany S. C., Fine D. H., DeSalle R., Figurski D. H., The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34, 193–198 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Velusamy S. K., Sampathkumar V., Godboley D., Fine D. H., Survival of an Aggregatibacter actinomycetemcomitans quorum sensing luxS mutant in the mouths of rhesus monkeys: Insights into ecological adaptation. Mol. Oral Microbiol. 32, 432–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velusamy S. K., Sampathkumar V., Godboley D., Fine D. H., Profound effects of Aggregatibacter actinomycetemcomitans leukotoxin mutation on adherence properties are clarified in in vitro experiments. PLoS One 11, e0151361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haubek D., et al. , Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A prospective longitudinal cohort study. Lancet 371, 237–242 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Fine D. H., et al. , Colonization and persistence of labeled and “foreign” strains of Aggregatibacter actinomycetemcomitans inoculated into the mouths of rhesus monkeys. J. Oral Biol. (Northborough) 2, 10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsey M. M., Rumbaugh K. P., Whiteley M., Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 7, e1002012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh P. D., Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8, 263–271 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Narayanan A. M., Ramsey M. M., Stacy A., Whiteley M., Defining genetic fitness determinants and creating genomic resources for an oral pathogen. Appl. Environ. Microbiol. 83, e00797-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sampathkumar V., Velusamy S. K., Godboley D., Fine D. H., Increased leukotoxin production: Characterization of 100 base pairs within the 530 base pair leukotoxin promoter region of Aggregatibacter actinomycetemcomitans. Sci. Rep. 7, 1887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath S., et al. , Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc. Natl. Acad. Sci. U.S.A. 103, 17402–17407 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duran-Pinedo A. E., Paster B., Teles R., Frias-Lopez J., Correlation network analysis applied to complex biofilm communities. PLoS One 6, e28438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loe H., Theilade E., Jensen S. B., Experimental gingivitis in man. J. Periodontol. 36, 177–187 (1965). [DOI] [PubMed] [Google Scholar]
- 21.Ritz H. L., Microbial population shifts in developing human dental plaque. Arch. Oral Biol. 12, 1561–1568 (1967). [DOI] [PubMed] [Google Scholar]
- 22.Moore W. E., Microbiology of periodontal disease. J. Periodontal Res. 22, 335–341 (1987). [DOI] [PubMed] [Google Scholar]
- 23.Li J., et al. , Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97, 1311–1318 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Listgarten M. A., The structure of dental plaque. Periodontol. 2000 5, 52–65 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Kolenbrander P. E., et al. , Bacterial interactions and successions during plaque development. Periodontol. 2000 42, 47–79 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Fine D. H., et al. , A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J. Clin. Microbiol. 51, 2850–2861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fine D. H., et al. , Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: Longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 45, 3859–3869 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karched M., Furgang D., Sawalha N., Fine D. H., Rapid identification of oral isolates of Aggregatibacter actinomycetemcomitans obtained from humans and primates by an ultrafast super convection based polymerase chain reaction. J. Microbiol. Methods 89, 71–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilian M., Rölla G., Initial colonization of teeth in monkeys as related to diet. Infect. Immun. 14, 1022–1027 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venketaraman V., et al. , Both leukotoxin and poly-N-acetylglucosamine surface polysaccharide protect Aggregatibacter actinomycetemcomitans cells from macrophage killing. Microb. Pathog. 45, 173–180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgess D., Huang H., Harrison P., Aukhil I., Shaddox L., Aggregatibacter actinomycetemcomitans in African Americans with localized aggressive periodontitis. JDR Clin. Trans. Res. 2, 249–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kram K. E., Hovel-Miner G. A., Tomich M., Figurski D. H., Transcriptional regulation of the tad locus in Aggregatibacter actinomycetemcomitans: A termination cascade. J. Bacteriol. 190, 3859–3868 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomich M., Planet P. J., Figurski D. H., The tad locus: Postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Brown S. A., Whiteley M., A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol. 189, 6407–6414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stacy A., et al. , Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 111, 7819–7824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faust K., Raes J., CoNet app: Inference of biological association networks using Cytoscape. F1000 Res. 5, 1519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T., Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 27, 431–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiFranco K. M., et al. , Leukotoxin (Leukothera®) targets active leukocyte function antigen-1 (LFA-1) protein and triggers a lysosomal mediated cell death pathway. J. Biol. Chem. 287, 17618–17627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ocon S., et al. , Transcription profiling reveals potential mechanisms of dysbiosis in the oral microbiome of rhesus macaques with chronic untreated SIV infection. PLoS One 8, e80863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colombo A. P., et al. , Clinical and microbiological parameters of naturally occurring periodontitis in the non-human primate Macaca mulatta. J. Oral Microbiol. 9, 1403843 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.