Abstract
Background: Crohn's disease is one of the most significant intestinal disorders and is known as inflammatory bowel disease; Campylobacter spp. are one of the leading causes of bacterial gastroenteritis in humans.
Methods: In this study, 60 tissue samples, including 30 cases with Crohn’s disease and 30 cases with no inflammatory bowel disease, were collected. Patients were referred to Taleghani hospital and Behboud clinic between March 2015 and May 2016. Biopsies were used for DNA extraction and assessment of Campylobacter jejuni in patients with Crohn's disease and controls using polymerase chain reaction and quantitative real-time polymerase chain reaction. All positive amplified fragments were sequenced. The gene encoding 16S rRNA, specific to Campylobacter genus, was amplified.
Results: The results were positive for Campylobacter genus in patients with Crohn's disease compared to healthy individuals. The quantitative real-time PCR showed a significantly higher prevalence of Campylobacter jejuni, particularly in hippurate hydrolase in tissue specimens of patients with Crohn's disease compared to control group. The correlation between Campylobacter jejuni and diarrhea symptoms in patients with Crohn's disease and controls was investigated. One positive case of Campylobacter jejuni found in patients without diarrhea was compared with 13 patients with diarrhea.
Conclusion: The present study demonstrated the alarmingly high rate of Campylobacter jejuni prevalence in Crohn's disease patients with diarrhea symptoms. However, further investigation is needed to determine the possible causing factors of this disease.
Keywords: Crohn disease, Campylobacter genus, Campylobacter jejuni, Real-time PCR, Inflammatory bowel disease
Introduction
Crohn's disease (CD) is a chronic inflammatory disorder of the gastrointestinal tract, which is considered as inflammatory bowel disease (IBD). The cause of IBD is still unknown. A variety of factors are implicated in this disorder, including intestinal microbiota, genetic, immunological and environmental factors (1). In addition, a bacterial component, such as Campylobacter spp., in humans and animals is responsible for the pathogenesis of IBD (2).
Moreover, the most common isolated Campylobacter species from colonic biopsies and human diarrheal feces samples are composed of C. jejuni and C. jejuni (3).
Chickens, farmed animals, such as cattle, pigs, and contaminated food or water are reservoirs of this organism (4). C. jejuni is the leading cause of severe diarrhea and gastrointestinal distress worldwide. The symptoms of infected humans are mild to acute diarrhea (5).
Cytotoxic effects and/or host-cell invasion of C. jejuni may be responsible for cellular damage (6). The cell cycle is usually arrested by producing cytolethal distending toxin (CDT) from C. jejuni. Consequently, this toxin can lead to apoptotic and nonapoptotic death of lymphocytes, monocytes, and endothelial cells, respectively (7). In terms of the ability of invasion of C. jejuni, it seems that these bacteria can cause intestinal damage through invasion in the mucosa.
The invasive action of C. jejuni in epithelial cells is correlated with increased permeability of human intestinal cells (8). In addition to C. jejuni, a leading cause of gastroenteritis, other members of Campylobacter genus consisting of C. concisus, C. coli, C. gracilis, C. upsaliensis, and C. lari, may also play a role in intestinal disease (9). The ability of hydrolase hippurate encoded by hipOgene in C. jejuni can distinguish this bacterium from other Campylobacter spp. (10). In addition to IBD, irritable bowel syndrome (IBS) is another type of chronic disease of the intestine that was recently shown to be associated with post infectious events.
Campylobacteriosis has been identified as one of the most usual risk factors for IBS. The induction of intestinal pathology occurs by pathological mechanism, such as damage to the epithelial cells, interruption of adherents junctions, and host cell death (11).
The aim of this study was to determine the prevalence of C. jejuni with hipOgene in intestinal biopsy samples, collected from patients with CD and control participants using molecular assay.
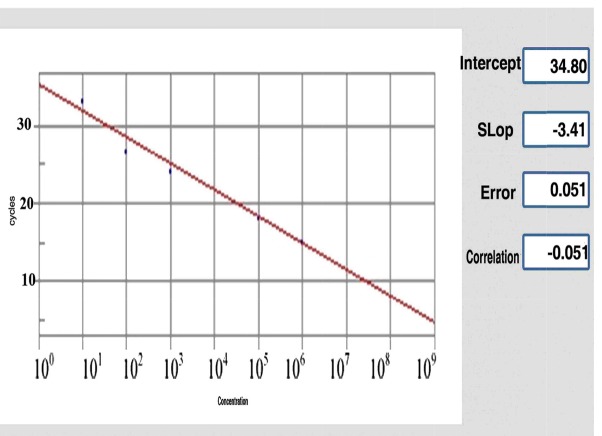
Fig. 2. Standard curve of DNA extracted from C. jejuni by quantitative real-time PCR assay. In standard curve, X and Y showed the concentration of C. jejuni and number of cycles respectively for control positive sample.
Fig. 2.
Standard curve of DNA extracted from C.jejuni by quantitative real-time PCR assay. In standard curve, X and Y showed the concentration of C.jejuni and number of cycles respectively for control positive sample.
Methods
Study participants and biopsy collection
In this study, 60 samples, including 30 samples of patients with CD and 30 without IBD, were collected from ileum, caecum, descending colon, ascending colon and rectum during colonoscopy. Patients were referred to Taleghani hospital and Behboud clinic between March 2015 and May 2016. The study was approved by the ethics committee of Tehran University of Medical Sciences. All the CD biopsy samples were taken from lesions of colon, terminal ileum. Moreover, the control biopsy samples were collected from healthy areas. CD diagnosis was based on clinical symptoms, laboratory evaluations, and colonoscopy finding which was confirmed by histological assessment. The control samples were taken from people with noninflammatory IBD (nIBD). None of the cohorts in this study used any antibiotics or probiotics for at last 3 months. All the samples were transferred in sterile plastic containing thioglycollate medium (Merck, Germany).
DNA extraction from biopsies
DNA was extracted from active ulcer and normal mucosa of individuals with CD and control participants. Biopsies were crushed and DNA was extracted by RTP® Mycobacteria kit (Berlin, Germany) and stored at −20 °C for PCR and real-time PCR assays.
Statistical analysis
Data were analyzed using chi-squared tests and Kruskal-Wallis test for the presence of Campylobacter jejuni. P value <0.05 was considered statistically significant. The positive samples from the experimental infection were evaluated in parallel with the real-time PCR assays. All data analyses were performed with Microsoft Excel.
Polymerase chain reaction (PCR)
All samples were tested for the presence of Campylobacter genus using PCR. PCR was performed in 12.5μL comprising of 5μL master mix (Amplicon, Pishgam), 0.5 μL of each primer, 2 μL of the DNA template (50 ng) and 4.5μL of ddH2O. Subsequently, the PCR was performed using the following thermal cycling conditions: 300 second at 94°C and 25 cycles of amplification consisting of 60 seconds at 95°C, 55 seconds at 58°C, followed by 60 seconds at 72°C, and 300 second at 72°C for the final extension. PCR products were examined by electrophoresis on a 1% agarose gel in 1X TBE buffer [10.8 g Tris and 5.5 g Boric acid, 0.5 M Na2EDTA (pH 8.0)] (12).
Real-time PCR assay
The presence of hipOgene was evaluated using real-time PCR assay. Therefore, quantitative real-time PCR was performed on a Line GeneK bioerr biosystem using Taq Man Probe. At the 5’end, the hipOgene probe was linked to the fluorophore FAM. For the real-time PCR, 12.5 µl Taq Man Master Mixes, 0.5 µL of each primer and probe, 10 µL ddH2O and 2 µL of the DNA template was added to a final volume of 25 µL.The thermocycler condition for hipOgene was 95°C for 10 minutes for the initial duration, followed by 40 cycles of a 2-stage temperature profile of 95°C for 10 seconds and 62°C for 1 minute. Cycle threshold (Ct) was assessed to provide standard curves for the quantification of hipOgene. PCR amplification efficiency (E) was estimated by the slope of the standard curve using the following formula E = 10(-1/ slope)-1. A reaction with 100% efficiency will generate a slope of -3.32.
Real-time PCR primers and probes
The nucleotide sequence of primers for Campylobacter genus-specific 16SrRNA was designed manually and checked with Primer 3 plus software (http:// www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi); also, the primer and probe for C. jejuni with hipOgene were used as described by Toplak et al (13). Oligoanalyzer (http://eu.idtdna.com/home/home.aspx) and VNTI database were used to check the specificity of primer target sites. Primers and probes are presented in Table 1.
Table 1. The primers and probe used for quantitative real-time PCR reaction .
| Gene name | Primer (5´-3´) | Probe | Product size |
| 16S rRNA | AATCTAATGGCTTAACCATTA GTAACTAGTTTAGTATTCCGG |
- | 850 |
| hipO | AATGCACAAATTTGCCTTATAAAAGC
TNCCATTAAAATTCTGACTTGCTAAATA |
FAM‐ACATACTACTTCTTTATTGCTTG-BHQ1 | 123 |
Results
Clinical information of patients and controls
Thirty samples were collected from CD patients (15 males and 15 females; mean age: 36.5 years, range: 18-64) and 30 samples were taken from nIBD (15 males and 15 females; mean age: 56.5 years, range: 25-76).
Detecting Campylobacter genus-specific 16S rRNA in tissue samples of cases with CD and controls using PCR assay
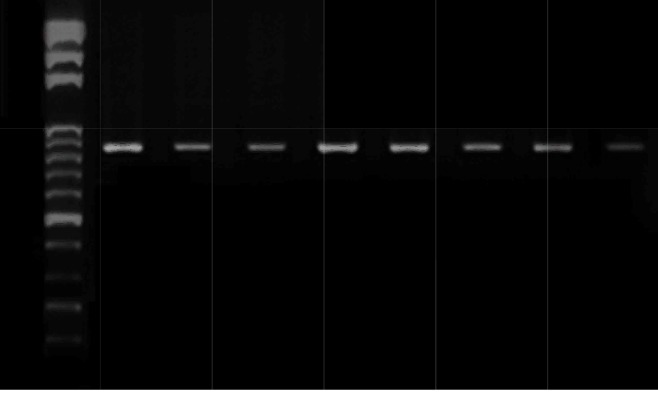
In this study, a Campylobacter genus-specific 16S rRNA PCR assay was used to examine the presence of Campylobacter genus in the current populations with CD and controls (Fig. 1). Of 30 control samples examined, 16.6% were positive and 83.4% were negative, and among 30 samples of patients with CD, 63.3% were positive and 36.7% were negative (Table 2).
Fig. 1.
Agarose gel electrophoresis of PCR amplification of Campylobacter genus-specific (16S rRNA). Lanes: 1, 100-bp DNA Ladder; 2-8, Crohn's disease samples; 9, Water (negative control).
Table 2. The prevalence of Campylobacter genus-specific 16S rRNA and Campylobacter jejuni hipOgene in CD and control subjects by PCR and real-time PCR .
| Group | PCR | Real time PCR | ||
| 16S rRNA | hipO | |||
| Positive | Negative | Positive | Negative | |
| Crohn’s disease | ||||
| Count | 19 | 11 | 21 | 9 |
| CD (%) | 63.3 | 36.7 | 70 | 30 |
| nIBD | ||||
| Count | 5 | 25 | 2 | 28 |
| nIBD (%) | 16.6 | 83.3 | 6.6 | 93.3 |
| p | 0.043* | 0.007* | ||
*Significantly higher as compared with the controls (p<0.05)
Molecular detection of C. jejuni in biopsies using real-time PCR
The specificity of each primer-probe for the identification of hipOgene in C. jejuni was tested. No signal was observed for any of the other Campylobacter species that could be present in biopsy samples. A serial dilution of bacterial DNA of C. jejuni and biopsy DNA samples was tested with the specific primers and FAM-labeled specific probes; and a strong linear correlation (R2 values were all equal to 0.98) was observed. The threshold cycle (Ct) values were defined in quantitative real-time PCR. Figure 2 displays the standard curves of the template DNA for C. jejuni with FAM-labeled specific probes and specific primers. Among 30 patients with CD and 30 individuals without IBD, 70% (n=21) and 6.6% (n=2) were positive for C. jejuni gene, respectively. Therefore, of the 60 tissues examined, 23 (38.33%) were positive and 37 (61.6%) were negative for hipO, and the difference between patients with CD and control participants were statistically significant (p<0.05) (Table 2).
In this study, there were 30 nIBD controls, of whom 26.6% (8/30) had diarrhea. One (1/8) was C. jejuni positive and 73.3% (22/30) did not have diarrhea, and none of them (0/22) was C. jejuni positive. Therefore, the differences between the percentages of C. jejuni positive in non-IBD controls with diarrhea, compared to those without diarrhea, was not statistically significant (p= 0.53).
In the CD samples, 60% (18/30) had diarrhea, of which 72/2% (13/18) was C. jejuni positive. In the CD group, 40% (12/30) did not have diarrhea, of whom 25% were C. jejuni positive. Differences between the percentages of C. jejuni positive in the CD group with diarrhea, compared to those without diarrhea, was statistically significant (p=0.011) (Table 3). All positive amplified fragments were sequenced and confirmed for C. jejuni.
Table 3. C.jejuni in patients with and without diarrhea .
| Group | C.jejuni | |
| Positive | Negative | |
| CD | ||
|
Diarrhea No diarrhea Total p |
13 3 30 0.011 |
5 9 |
| nIBD | ||
|
Diarrhea No diarrhea Total |
1 0 30 |
8 22 |
| p | 0.53* | |
*Significantly higher as compared with the controls (p<0.05).
Quantification ofC. jejuni
The results of PCR confirmed the higher rate of C. jejuni with hipOgene in CD patients compared to the controls, which clearly indicated that the average number and std. The error of the mean (SEM) of C. jejuni was significantly higher in patients with CD (6.80E01± 2.90E01 bacteria per gram of tissue samples, p=0.004) compared to control participants (median 3.00E-02) (Table 4).
Table 4. Quantitative analysis of C.jejuni from CD and nIBD biopsy samples .
| Group | C.jejuni |
| Crohn’s disease | |
| Mean | 68 |
| N | 30 |
| SEM | 29 |
| nIBD | |
| Mean | 0.03 |
| N | 30 |
| SEM | 0.03 |
| p | 0.01* |
*The mean difference is significant at the 0.05 level.
SEM: Standard error of mean
Discussion
CD is the most common form of IBD. There is limited information about the etiology of this disease in Iran (14). However, it was reported that the prevalence of IBD is increasing in Asia and developing countries (15). In the past decades, IBD was diagnosed more than ever in various parts of Iran (16).
In Europe and North America, more information has been reported about the epidemiology, clinical symptoms and etiology of IBD, as a consequence of the high prevalence of the disease in these areas (17). However, it seems that combinations of genetic and environmental factors that can affect the immune responses are implicated in causing the disease. There is numerous information on the transfer of Campylobacter spp., particularly C. jejuni. Previous studies demonstrated that C. jejuni is one of the causing factors of human infections (18) and could be transmitted through poultry products, contaminated water (19), livestock, such as cattle (20), and unpasteurized dairy products (21).
A study investigated whether chicken (38% to 77%) and cattle (16% and 54%) are the first and second most common reservoir of Campylobacter infection, respectively (22).
Rahimi et al reported that C. jejuni (88.5%) was the most prevalent of Campylobacter species isolated from row duck and goose meat in Iran (23). In another study, Rahimi et al demonstrated that the high percentage of Campylobacter spp. (41.7%) was found in poultry meat in Esfahan, Iran (24).
Nevertheless, a higher prevalence of C. concisus was reported both in colonic biopsy samples from patients with ulcerative colitis (31%; 4/13) in the samples from CD patients (53%; 8/15) than in nIBD (18%; 6/33 individuals) (P<0.05) (25).
A recent study by our group addressed this issue by investigating the prevalence of C. jejuni with hipOgene in patients with CD in Tehran, Iran. The high positive rate of Campylobacter genus-PCR and intestinal prevalence of C. jejuni in patients with CD observed in this study are consistent with previous findings in CD patients (26). In the present study, C. jejuni was detected in 62.5% of CD patients with diarrhea and in 28.5% without diarrhea symptoms.
In a later study, Man SM et al detected a significantly higher prevalence of C. concisus in stool samples of children with CD (27).
Another finding of this study was the increased prevalence of C. jejuni in the patients with CD compared to the control participants. Some previous studies detected a high prevalence of C. jejuni in patients with gastroenteritis and C. concisus (28).
In a follow-up study, Zhang et al reported a high presence of C. concisus (75%) in oral samples with healthy individuals and 95% in intestinal specimens through PCR (29). In early 2015, it was reported that several factors and different gut microbiota may be associated with IBD (30). Moreover, our evidence revealed that C. jejuni may be an important predisposing factor for the initiation and development of CD.
In this study, C. jejuni was detected in a significantly larger proportion of CD patients compared to control populations.
Conclusion
In summary, in this study, a significantly higher correlation of C. jejuni was detected in colonic biopsies from patients with CD as compared to control participants. However, the absence of C. jejuni in CD patients suggests that different bacterial genera may be associated with IBD. The present study proposed that infection with C. jejuni can be one of the significant reasons in the development of IBD.
Acknowledgments
We would like to thank the staff of Behbood Clinic for their kind cooperation in this study. This work was supported by Tehran University of Medical Sciences.
This study was supported in part by a grant (No: 32467) provided by Tehran University of Medical Sciences.
Ethical approval
In the present study, samples were collected following the approval by the ethics committee of Tehran University of Medical Sciences. (Ethics approval code:
IR.TUMS.MEDICINE.REC.1395.1178).
Conflict of Interests
The authors declare that they have no competing interests.
Cite this article as: Sarabi Asiabar A, Asadzadeh Aghdaei H, Zamani S, Bokaie S, Zali MR, Feizabadi MM. Molecular detection of Campylobacter jejuni in patients with Crohn’s disease in Iran. Med J Islam Repub Iran. 2019 (29 Jul);33:76. https://doi.org/10.34171/mjiri.33.76
References
- 1.Carter M, McGee R, Taylor B, Williams S. Health outcomes in Tontini GE, Vecchi M, Pastorelli L, Neurath MF, Neumann H Differential diagnosis in inflammatory bowel disease colitis: state of the art and future perspectives. World J Gastroenterol. 2015 Jan 7;21(1):21. doi: 10.3748/wjg.v21.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaban B, Ngeleka M, Hill JE. Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiol. 2010 Dec;10(1):73. doi: 10.1186/1471-2180-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. Global epidemiology of Campylobacter infection. Clin Microbiol Rev. 2015 Jul 1;28(3):687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epps SV, Harvey RB, Hume ME, Phillips TD, Anderson RC, Nisbet DJ. Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int J Environ Res Public Health. 2013 Nov 26;10(12):6292–304. doi: 10.3390/ijerph10126292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullman S, O'leary J, Corcoran D, Sleator RD, Lucey B. Molecular-based detection of non-culturable and emerging campylobacteria in patients presenting with gastroenteritis. Epidemiol Infect. 2012 Apr;140(4):684–8. doi: 10.1017/S0950268811000859. [DOI] [PubMed] [Google Scholar]
- 6.Humphrey S, Chaloner G, Kemmett K, Davidson N, Williams N, Kipar A. et al. Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. MBio. 2014 Aug 29;5(4):e01364–14. doi: 10.1128/mBio.01364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shenker BJ, Walker LP, Zekavat A, Dlakić M, Boesze‐Battaglia K. Blockade of the PI‐3K signalling pathway by the Aggregatibacter actinomycetemcomitans cytolethal distending toxin induces macrophages to synthesize and secrete pro‐inflammatory cytokines. Cell Microbiol. 2014 Sep 1;16(9):1391–404. doi: 10.1111/cmi.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backert S, Boehm M, Wessler S, Tegtmeyer N. Transmigration route of Campylobacter jejuni across polarized intestinal epithelial cells: paracellular, transcellular or both? Cell Commun Signal. 2013 Dec;11(1):72. doi: 10.1186/1478-811X-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Man SM. The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol. 2011 Dec;8(12):669. doi: 10.1038/nrgastro.2011.191. [DOI] [PubMed] [Google Scholar]
- 10.Holmberg M, Rosendal T, Engvall EO, Ohlson A, Lindberg A. Prevalence of thermophilic Campylobacter species in Swedish dogs and characterization of C jejuni isolates. Acta Vet Scand. 2015 Dec;57(1):19. doi: 10.1186/s13028-015-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Rowehl L, Krumsiek JM, Orner EP, Shaikh N, Tarr PI. et al. Identification of candidate adherent-invasive E coli signature transcripts by genomic/transcriptomic analysis. PloS One. 2015 Jun 30;10(6):e0130902. doi: 10.1371/journal.pone.0130902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Boer P, Rahaoui H, Leer RJ, Montijn RC, Van der Vossen JM. Real-time PCR detection of Campylobacter spp: a comparison to classic culturing and enrichment. Food Microbiol. 2015 Oct 1;51:96–100. doi: 10.1016/j.fm.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Toplak N, Kovač M, Piskernik S, Možina SS, Jeršek B. Detection and quantification of Campylobacter jejuni and Campylobacter coli using real‐time multiplex PCR. J Appl Microbiol. 2012 Apr 1;112(4):752–64. doi: 10.1111/j.1365-2672.2012.05235.x. [DOI] [PubMed] [Google Scholar]
- 14.Ng SC. Ng SCEpidemiology of inflammatory bowel disease: focus on Asia . Best Pract Res Clin Gastroenterol. 2014 Jun 1;28(3):363–72. doi: 10.1016/j.bpg.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra R, Turner K, Sonnenberg A, Genta RM. High prevalence of inflammatory bowel disease in United States residents of Indian ancestry. Clin Gastroenterol Hepatol. 2015 Apr 1;13(4):683–9. doi: 10.1016/j.cgh.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 16.Sadkowska-Todys M, Kucharczyk B. Campylobacteriosis in Poland in 2010. Przegl Epidemiol. 2012;66(2):255–8. [PubMed] [Google Scholar]
- 17.Nichols GL, Richardson JF, Sheppard SK, Lane C, Sarran C. Campylobacter epidemiology: a descriptive study reviewing 1 million cases in England and Wales between 1989 and 2011. BMJ Open. 2012 Jan 1;2(4):e001179. doi: 10.1136/bmjopen-2012-001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Little CL, Gormley FJ, Rawal N, Richardson JF. A recipe for disaster: outbreaks of campylobacteriosis associated with poultry liver pâté in England and Wales. Epidemiol Infect. 2010 Dec;138(12):1691–4. doi: 10.1017/S0950268810001974. [DOI] [PubMed] [Google Scholar]
- 19.Pires SM, Vigre H, Makela P, Hald T. Using outbreak data for source attribution of human salmonellosis and campylobacteriosis in Europe. Foodborne Pathog Dis. 2010 Nov 1;7(11):1351–61. doi: 10.1089/fpd.2010.0564. [DOI] [PubMed] [Google Scholar]
- 20.Boysen L, Rosenquist H, Larsson JT, Nielsen EM, Sørensen G, Nordentoft S. et al. Source attribution of human campylobacteriosis in Denmark. Infect Control Hosp Epidemiol. 2014 Aug;142(8):1599–608. doi: 10.1017/S0950268813002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zamani S, Zali MR, Aghdaei HA, Sechi LA, Niegowska M, Caggiu E. et al. Mycobacterium avium subsp paratuberculosis and associated risk factors for inflammatory bowel disease in Iranian patients. Gut Pathog. 2017 Dec;9(1):1. doi: 10.1186/s13099-016-0151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skarp CP, Hänninen ML, Rautelin HI. Campylobacteriosis: the role of poultry meat. Clin Microbiol Infect. 2016 Feb 1;22(2):103–9. doi: 10.1016/j.cmi.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Rahimi E, Alian F, Alian F. Prevalence and characteristic of Campylobacter species isolated from raw duck and goose meat in Iran. IPCBEE. 2011;9:171–5. [Google Scholar]
- 24.Rahimi E, Tajbakhsh E. Prevalence of Campylobacter species in poultry meat in the Esfahan city, Iran. J Vet Med B Infect Dis Vet Public Health. 2008 Dec 1;11(4):257–62. [Google Scholar]
- 25.Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis 2013 Feb. 4;19(3):481–8. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, Hold GL. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS One. 2011 Jun 27;6(6):e21490. doi: 10.1371/journal.pone.0021490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Man SM, Zhang L, Day AS, Leach ST, Lemberg DA, Mitchell H. Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn's disease. Inflamm Bowel Dis. 2010 Jun 1;16(6):1008–16. doi: 10.1002/ibd.21157. [DOI] [PubMed] [Google Scholar]
- 28.Mahendran V, Riordan SM, Grimm MC, Tran TA, Major J, Kaakoush NO. et al. Prevalence of Campylobacter species in adult Crohn's disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS One. 2011 Sep 23;6(9):e25417. doi: 10.1371/journal.pone.0025417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Budiman V, Day AS, Mitchell H, Lemberg DA, Riordan SM, Grimm M, Leach ST, Ismail Y. Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J Clin Microbiol. 2010 Aug 1;48(8):2965–7. doi: 10.1128/JCM.02391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015 Jan 1;1(37):47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]