Abstract
Purpose:
EGFR exon 19 deletion (Ex19Del) mutations account for ~60% of lung cancer-associated EGFR mutations and include a heterogeneous group of mutations. Although they are associated with benefit from tyrosine kinase inhibitors (TKIs), the relative inhibitor sensitivity of individual Ex19Del mutations is unknown.
Experimental Design:
We studied the TKI sensitivity and structural features of common EGFR Exon 19 deletion mutations and the consequences for patient outcomes on TKI treatment.
Results:
We found that the L747-A750>P mutation, which represents about 4% of all Ex19Del mutations, displays unique inhibitor selectivity. L747-A750>P differs from other Ex19Del mutations in not being suppressed completely by erlotinib or osimertinib, yet is completely inhibited by low doses of afatinib. The HCC4006 cell line (with the L747-A750>P mutation) exhibited increased sensitivity to afatinib over erlotinib and osimertinib, and computational modeling suggests explanations for this sensitivity pattern. Clinically, patients with EGFR L747-A750>P mutant tumors showed inferior outcomes when treated with erlotinib than patients with E746-A750 mutant tumors.
Conclusions:
These results highlight important differences between specific Ex19Del mutations that may be relevant for optimizing TKI choice for patients.
Keywords: EGFR mutations, Exon 19 deletions, Drug Resistance, Tyrosine Kinase Inhibitors, Afatinib
Introduction
EGFR mutations associated with clinical response to EGFR tyrosine kinase inhibitors (TKIs) were discovered over a decade ago in non-small cell lung cancer (NSCLC), and five TKIs (erlotinib, gefitinib, afatinib, dacomitinib and osimertinib) are currently approved by the FDA for the first-line treatment of EGFR-mutant NSCLC [1–4]. Erlotinib and gefitinib, both 1st generation TKIs, bind reversibly to the ATP binding pocket of the receptor, whereas the 2nd generation TKIs afatinib and dacomitinib additionally react covalently with the side-chain of cysteine 797 (C797) in EGFR [5]. The major mechanism of resistance to 1st and 2nd generation TKIs is a secondary acquired T790M mutation. Osimertinib is a 3rd generation irreversible TKI which also reacts with C797, and can inhibit EGFR harboring the secondary T790M mutation. Osimertinib has been shown to be effective in the second line for patients with NSCLC harboring EGFR T790M mutations [6], and more recently was shown to improve progression-free survival (PFS) compared to 1st generation TKIs in the first-line setting [7]. Based on these findings, osimertinib now has FDA approval for first-line treatment of patients with metastatic NSCLC harboring EGFR mutations.
The most common EGFR alterations associated with TKI sensitivity are in-frame deletions in exon 19 and a point mutation in exon 21 (L858R). Together, these account for approximately 90% of all EGFR alterations [8]. The most frequently observed EGFR exon 19 deletion leads to elimination of 5 amino acids (E746-A750) [9] between the third β-strand of the EGFR tyrosine kinase domain and its key regulatory αC helix. However, a number of other exon 19 deletion mutations have also been observed in NSCLC between amino acids 745 and 753 [10]. Many of these deletions start at leucine 747, and are often complex insertion-deletions (indels) leading to replacement of the deleted amino acids with a non-native residue – such as the L747-A750>P and L747-P753>S variants, where proline and serine respectively are introduced [10]. Although it is well established that EGFR exon 19 deletion mutant tumors are sensitive to TKIs [11, 12], very little is known about potential differences in TKI sensitivity between individual EGFR exon 19 deletions. One recent study in Ba/F3 cells confirmed sensitivity of several EGFR exon 19 deletions and indels to 1st, 2nd and 3rd generation TKIs [13], but suggested subtle differences in TKI sensitivity of individual mutants that have not been explored in detail. The impact of these differences for patient responses has also not been examined, but recent data indicate that they may be clinically important [14]. Recent data also suggest that the type of EGFR exon 19 deletion mutation present at baseline is associated with the emergence of a specific osimertinib resistance mutation [15]. Here, we investigate differences in TKI sensitivity among the most common EGFR exon 19 deletion mutants. We identify one key variant (L747-A750>P) that is partly refractory to inhibition by erlotinib and osimertinib in engineered, patient-derived and established cell lines, but is strongly inhibited by afatinib. We also report analysis of a Yale patient cohort in which erlotinib-treated patients with tumors harboring the L747-A750>P mutation demonstrated significantly worse outcomes than those with tumors harboring other exon 19 deletions. Our cellular and clinical data thus underscore the importance of analyzing the specific exon 19 mutation present in lung cancers at the time of diagnosis.
MATERIALS AND METHODS
Cell culture
Human lung adenocarcinoma cell lines (PC9, H1975, HCC827 and HCC4006) and the PDX-derived cell line were cultured in RMPI + L-glutamine (Thermo Fisher Scientific), supplemented with 10% heat-inactivated bovine serum (Thermo Fisher Scientific) and 1% penicillin/streptomycin (Thermo Fisher Scientific). Human embryonic kidney cells 293T cells were cultured in DMEM + L-glutamine (Thermo Fisher Scientific), supplemented with 10% heat-inactivated bovine serum (Thermo Fisher Scientific) and 1% penicillin/streptomycin (Thermo Fisher Scientific). CHO cells were cultured in DMEM/F12 medium (Gibco), supplemented with 10% FBS and 1% penicillin/streptomycin. PC9, H1975, HCC827, HCC4006, CHO, and 293T cell lines were purchased from ATCC. Cell lines were routinely tested for mycoplasma contamination. The lung adenocarcinoma cell lines were authenticated by testing for the presence of the expected EGFR mutation. The presence of the EGFR L747-A750>P alteration was confirmed in the PDX-derived cell line using a mutation specific TaqMan assay. 20 ng of DNA were used in the reaction using assay 12382 (EGFR c.2239_2248>C).
Functional analyses of EGFR mutations
Plasmids carrying cDNAs encoding EGFR mutations (EGFR E746-A750, EGFR L747-S752, EGFR L858R+T790M) and wild-type EGFR were as described previously [3, 16]. pcDNA3.1 (−) containing wild-type EGFR was used as a template to generate additional L747-A750>P and L747-P753>S EGFR mutated plasmids using primers designed through the QuickChange Primer design tool by Agilent Technologies (http://www.genomics.agilent.com/primerDesignProgram.jsp). The following primers were used to introduce the L747-A750>P mutation: forward 5’TAAAATTCCCGTCGCTATCAAGGAGCCAACATCTCCGAAAGCCAACAAGG-3’ and reverse 5’CCTTGTTGGCTTTCGGAGATGTTGGCTCCTTGATAGCGACGGGAATTTTA-3’. The following primers were used to introduce the L747-P753>S mutation: forward 5’ATTCCCGTCGCTATCAAGGAATCGAAAGCCAACAAG-3’ and reverse 5’CTTGTTGGCTTTCGATTCCTTGATAGCGACGGGAAT-3’. Mutations were introduced in full-length EGFR using QuickChange II Site-Directed Mutagenesis (Agilent Technologies). All mutated clones were confirmed using direct Sanger sequencing. The constructs were transiently expressed in 293T human embryonic kidney cells. Briefly, 293T cells were transfected with the plasmids for 36 hours, cells were then serum-starved for 24 hours. After starvation, cells were treated with varying concentrations of erlotinib, afatinib, osimertinib and dacomitinib for 1 hour and then harvested for immunoblot analysis. Erlotinib and afatinib (obtained from the Organic Synthesis Core Facility at MSKCC), osimertinib (AstraZeneca) and dacomitinib (Selleck) were dissolved in DMSO.
Sequencing
PCR products were sequenced by Sanger sequencing and sequences and chromatograms were manually reviewed in the forward and reverse directions. The presence of EGFR exon 19 deletions and the EGFR T790M mutation in exon 20 were evaluated using the following primers: EGFR-2074F: 5′-cttacacccagtggagaagc-3′, EGFR-2502R: 5′-caccaagcgacggtcctcca-3′. The presence of the EGFR L858R mutation in exon 21 was confirmed with the following primers: EGFR-2445F: 5′-caactggtgtgtgcagatcg-3′, EGFR-3616R: 5′-cactgcttggtggcgcgac-3′.
Immunoblotting
293T cells were serum-starved for 24 hours and treated with indicated concentration of TKIs. HCC4006, PC9, H1975 and the YLR086PDX-derived cell lines were treated with TKI for 6 hours before western blot analysis. Cells were lysed in ice-cold RIPA lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific #78440). Equal amounts of total protein were separated by SDS-PAGE and probed as indicated. Signals were detected using either SuperSignal West Pico or Femto chemiluminescent substrates (Thermo Scientific). Antibodies for immunoblotting against EGFR (#2232), phospho-EGFR-Y1068 (#3777), and the secondary anti-rabbit HRP antibody (#7074) were from Cell Signaling Technology (CST). The β-actin antibody (#47778) was from Santa Cruz. All antibodies were used at the dilutions suggested by manufacturer.
Cell viability assays
Cells were plated at 2,000 cells/well (PC9 and H1975), 8,000 cells/well (HCC4006 and HCC827) or 10,000 cells/well (YLR086) in a 96 well-plate. After 24h, cells were exposed to the relevant drugs in media containing serum for 72h. Cell viability was measured using 20μl/well of CellTiter Blue Reagent (Promega, G8081).
EGFR depletion assay in CHO cells following Hsp90 inhibition
Stable CHO cell lines expressing different EGFR exon 19 deletion mutations were obtained after ~2 months of G418 selection following electroporation with the relevant pcDNA 3.1(−) derivatives using Ingenio electroporation solution (Mirus Bio LLC), and maintained with 200 μg/ml G418. To analyze EGFR depletion following Hsp90 inhibition, cells were treated with 50–500 nM geldanamycin for 24 hours, adding the drug from a 1 mM stock in DMSO. After washing with ice-cold PBS buffer, cells were lysed by scraping in cell lysis buffer (Cell Signaling Technology) supplemented with Complete protease inhibitor (Roche) and PhosSTOP phosphatase inhibitor (Roche) on ice. Whole cell lysates were cleared by centrifugation at 14,000 rpm for 10 minutes at 4˚C, and analyzed by immunoblotting using antibodies against EGFR, Hsp90, and Grb2 as loading control. Anti-EGFR (D38B1:cat#4267), and Grb2 (cat#3972) antibodies were from Cell Signaling Technology, and Hsp90 monoclonal antibody (AC88:cat#ADI-SPA-830) was purchased from Enzo Lifescience. All primary antibodies were diluted 1:1000. Chemiluminescence detection using HRP-conjugated secondary antibodies was performed using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific) and a Kodak Image station 440CF (Kodak Scientific).
Molecular dynamics studies
A starting model was generated using the crystal structure of the EGFR kinase domain (aa 695–1022: (PDB ID: 1M17) in complex with erlotinib [17]. This structure was initially subjected to the ‘protein preparation wizard’ available in the Schrödinger Suite 2017 [18], to remove water molecules and fill in the missing amino acid residues as well as missing hydrogen atoms. Geometry minimization of the subsequent structure (verified by convergence), with and without erlotinib, yielded two corresponding structures that were then used as starting points for generating subsequent models for the wild-type kinase and exon 19 deletion mutants bound to the relevant inhibitors. Full geometry optimizations on the model systems were performed using the Optimized Potentials for Liquid Simulations (OPLS-2005) force field with simulated water (VSGB 2.0 solvation model) available under the Schrödinger Suite 2017 running on Macintosh platform [18, 19]. To verify convergence and consistency of the optimizations, a number of examples were re-optimized from multiple starting points; energetic variations of 0.1 kcal/mol or less were found among these calculated structures.
Geometry-optimized structures were used as starting points for Molecular Mechanics/ Generalized Born Surface Area (MM-GBSA) modeling studies in the Schrödinger Suite 2017 [18], considering non-covalent interactions only. No structural constrains were imposed on the binding pocket, (extending to residues 5.0 Å beyond the ligands) so that geometry re-optimization was possible. MM-GBSA calculations can be used to estimate relative binding affinities for different ligands – although absolute values do not typically agree with experimental binding affinities.
Clinical Data
Patients enrolled in Institutional Review Board (IRB) approved protocols at Yale Cancer Center with a diagnosis of NSCLC whose tumors harbored EGFR exon 19 mutations (E746-A750, L747-P753>S or L747-A750>P) were identified. Patients who received erlotinib as first-line therapy for advanced disease (as this was the TKI most commonly used during the time period observed), either alone or in combination with an agent not considered to have meaningful clinical activity were included in this analysis. Patients were excluded if they received systemic therapy for early-stage disease in the year prior to starting erlotinib, had previously received an EGFR TKI at any time, had discontinued TKI treatment due to toxicity, or had insufficient data.
Clinical, pathologic and molecular characteristics were retrospectively obtained from extraction of medical records. Duration of treatment (DOT) was defined as time from treatment initiation to discontinuation of erlotinib due to any reason other than toxicity. Progression-free survival (PFS) was defined as time from treatment initiation to clinical disease progression (as determined by the treating oncologist) or death, and overall survival (OS) was defined as time from treatment initiation to death. Primary resistance was defined as progression of disease on first imaging after initiation of erlotinib.
Quantification and Statistical Analysis
PFS, DOT and OS were compared using a log-rank test. Cox proportional hazard models were used to derive hazard ratios (HRs) and 95% confidence intervals (CI) comparing exon 19 mutation groups. Kaplan-Meier estimates were used to construct survival curves and calculate PFS, DOT and OS. Statistical analysis was performed with GraphPad Prism 7.0 software and R version 3.3.2.
Results
Uncommon mutations represent about 30% of EGFR exon 19 deletion mutations
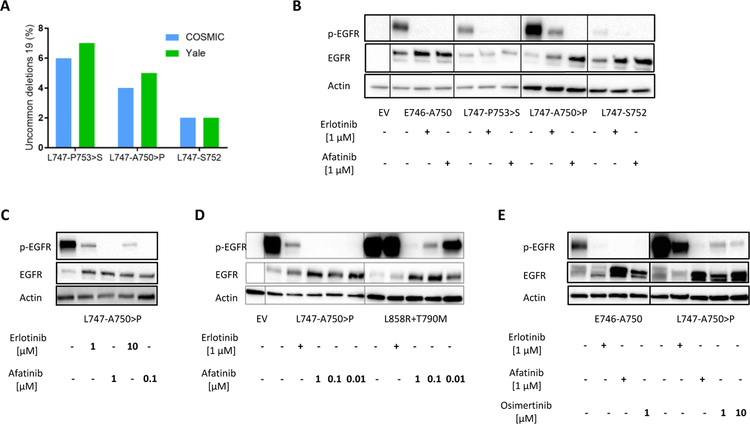
To study the frequency of uncommon EGFR exon 19 deletion mutations in lung cancer, we first analyzed data from the Catalog of Somatic Mutations in Cancer (COSMIC) that includes information on 4476 lung tumors with EGFR exon 19 deletions between residues K745 and I759 (http://cancer.sanger.ac.uk/cosmic/). As expected, in this cohort the majority of tumors had the classical EGFR exon 19 deletion mutation E746-A750 (69%). The remaining 31% represent a wide variety of EGFR exon 19 deletion mutations, including several complex indels. In decreasing order of frequency, uncommon deletions in exon 19 (reported in at least 40 samples) were: L747-P753>S (6%), L747-T751 (5%), L747-A750>P (4%), E746-S752>V (3%), L747-S752 (2%), E746-T751>A (1%) and L747-T751>P (1%) (Supplementary Table 1). We compared the mutational frequencies reported in COSMIC to those in a Yale cohort of 86 patients with EGFR mutant lung cancer enrolled on an IRB-approved repeat biopsy protocol at our institution. We found that the frequency of EGFR exon 19 deletions was consistent between the two datasets. Of 86 patients in the Yale cohort, 51 (59%) had tumors with EGFR exon 19 deletions, and information regarding the specific exon 19 deletion was available for 44 of these. The common exon 19 deletion (E746-A750) accounted for 34 (77%) of the tumors, whereas 10 tumors (23%) had one of the uncommon deletions in EGFR exon 19 (Supplementary Table 2). The most frequently observed indels were L747-P753>S and L747-A750>P, found in 3 and 2 patients, respectively. Collectively, these are the most commonly observed infrequent EGFR exon 19 indels across both COSMIC and Yale data (Figure 1A), so we proceeded to investigate their properties further. We also studied the L747-S752 infrequent deletion found in both cohorts.
Figure 1. The second-generation TKI afatinib effectively suppresses EGFR phosphorylation of the exon 19 L747-A750>P variant.
A. Bar graph illustrating the frequency of uncommon EGFR exon 19 deletions (L747-P753>S, L747-A750>P, L747-S752) reported in COSMIC and in a Yale patient cohort. B. Western blotting analysis of lysates from 293T cells transiently transfected with constructs encoding the E746-A750, L747-P753>S, L747-A750>P and L747-S752 deletion mutations and treated with 1 μM erlotinib or afatinib as indicated. Immunoblotting was performed with antibodies to detect phosphorylated EGFR (p-EGFR), EGFR, and actin. The empty vector (EV) was used a control; C. Western blotting analysis of lysates from 293T cells transfected with a plasmid encoding L747-A750>P EGFR, and treated with increasing concentrations of erlotinib (1 and 10 μM) or decreasing concentrations of afatinib (1 and 0.1 μM), and probed with antibodies to detect p-EGFR, EGFR, and actin; D. Western blotting analysis of lysates from 293T cells transfected with constructs encoding the L747-A750>P or L858R+T790M EGFR variants. Cells were treated with erlotinib (1 μM) or with decreasing concentrations of afatinib (1, 0.1 and 0.01 μM), revealing a dose-dependent inhibition by afatinib of L747-A750>P EGFR; E. Western blotting analysis of lysates from 293T cells transfected with constructs encoding the canonical exon 19 deletion variant E746-A750 or the uncommon L747-A750>P alteration, treated with erlotinib, afatinib or osimertinib at the noted concentrations, and immunoblotted to detect p-EGFR, EGFR, and actin. All Western blots are representative of at least 2 repeats.
The EGFR L747-A750>P variant exhibits limited sensitivity to 1st and 3rd generation TKIs
To study the sensitivity of different EGFR exon 19 deletion mutants to 1st generation TKIs, we generated expression constructs encoding E746-A750, L747-P753>S, L747-A750>P, L747-S752 and L858R+T790M [3, 16]. The mutated receptors were transiently expressed in 293T cells, which have low levels of endogenous EGFR. Western blotting of cell lysates (Figure 1B) showed strong constitutive autophosphorylation of the mutated (activated) EGFR variants – although this was reduced in the case of L747-S752, despite similar expression levels, as reported previously [3]. Adding erlotinib at 1 μM completely inhibited autophosphorylation of the canonical deletion mutant E746-A750 as well as the L747-P753>S and L747-S752 variants – reducing EGFR phosphorylation to undetectable levels in each case (Figure 1B). Unexpectedly, however, 1 μM erlotinib only partly reduced phosphorylation of the L747-A750>P variant (Figure 1B), arguing that the ability of erlotinib to suppress phosphorylation differs depending on the specific exon 19 deletion.
Based on these results we also compared the ability of the 2nd generation irreversible TKI afatinib to inhibit each of the deletion mutants. In contrast to erlotinib, 1 μM afatinib suppressed phosphorylation completely in all exon 19 deletion mutants (Figure 1B). Importantly, afatinib fully suppressed phosphorylation of EGFR in the L747-A750>P variant despite its reduced sensitivity to erlotinib (Figure 1B). In further studies at different drug doses, we found that erlotinib could not inhibit this variant completely even when added at 10 μM (Figure 1C), whereas afatinib could completely inhibit L747-A750>P EGFR at concentrations of 0.1 μM (Figure 1C), and even at the clinically achievable dose of 0.01 μM [20] (Figure 1D). By contrast, the L858R+T790M mutant, whose phosphorylation can be suppressed by afatinib in vitro, but not at clinically achievable doses in vivo, retained a high level of phosphorylation after treatment with 0.1 or 0.01 μM afatinib (Figure 1D). The other irreversible 2nd generation TKI, dacomitinib, was also able to suppress phosphorylation of L747-A750>P EGFR completely, similar to afatinib (Supplementary Figure 1). Finally, we tested the sensitivity of the L747-A750>P and E746-A750 variants to the 3rd generation TKI osimertinib. The L747-A750>P variant retained substantial residual phosphorylation even at 10 μM osimertinib (Figure 1E), whereas the E746-A750 variant was completely inhibited at 1 μM osimertinib.
The L747-A750>P alteration exhibits increased sensitivity to afatinib
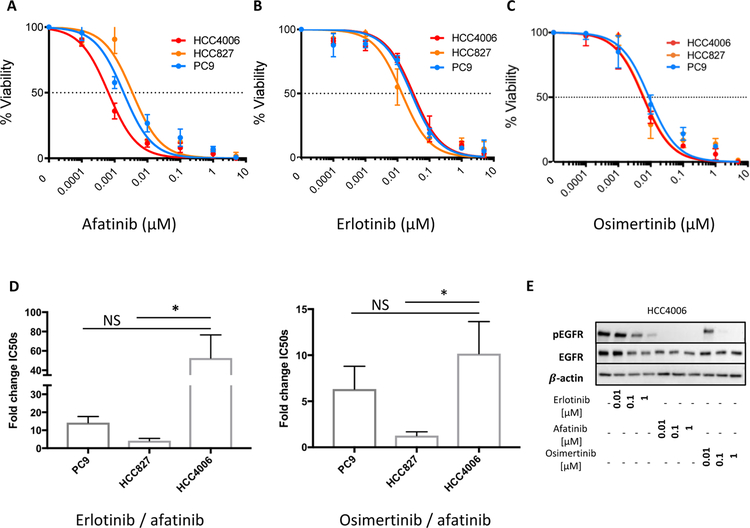
To explore functional consequences of the L747-A750>P mutation further, we investigated in vitro TKI sensitivity of an established human lung adenocarcinoma cell line HCC4006 that carries this specific exon 19 deletion, specifically comparing it with two lung cancer cell lines that carry the canonical E746-A750 exon 19 deletion (PC9 and HCC827). Consistent with the transient transfection studies, HCC4006 cells showed increased sensitivity to afatinib compared with PC9 (HCC4006 vs PC9 at 0.001μM, p=0.0455) and HCC827 cells (HCC4006 vs HCC827 at 0.001μM, p=0.0094) (Figure 2A), with an IC50 value of 0.0006 μM – which is 5-fold smaller than seen for HCC827 cells (IC50 = 0.004 µM) and 3.3-fold smaller than for PC9 cells (IC50 = 0.002 µM). All three cell lines showed similar sensitivities to the 1st generation TKI erlotinib (Figure 2B). In order of reduced sensitivity, IC50 values were 0.015 μM (HCC827 cells), 0.027 µM (PC9 cells), and 0.033 μM (HCC4006 cells). The sensitivity of HCC4006 cells to the 3rd generation inhibitor osimertinib (IC50 = 0.006 μM) was also within the same range seen for cells with an E746-A750 deletion (Figure 2C), which had IC50 values of 0.006 µM (HCC827 cells) and 0.009 µM (PC9 cells). Figure 2D reports the ratio of IC50 values for erlotinib/afatinib (left) and osimertinib/afatinib (right) as a measure of relative efficacy. Whereas afatinib is 14 times more effective at a given dose in PC9 cells and 4 times more effective in HCC827 cells, the ratio is increased to 53-fold in HCC4006 cells with the L747-A750>P mutation (Figure 2D, left). Afatinib is also 10 fold more effective than osimertinib in HCC4006 cells, while it is only 6 fold more effective in PC9 cells and 1.3 fold more effective in HCC827 cells (Figure 2D, right). Overall, as also shown by Western blotting analysis of phosphorylated EGFR levels in Figure 2E, HCC4006 cells are most sensitive to afatinib compared to the other TKIs.
Figure 2. Afatinib significantly inhibits cell viability in a cell line carrying the uncommon L747-A750>P exon 19 mutation.
A. Viability of EGFR mutant lung adenocarcinoma cell lines exposed for 72h to increasing concentrations of afatinib. Data for PC9 and HCC827 cells (which have the E746-A750 deletion) are shown in blue and orange, respectively, whereas data for the HCC4006 cell line (with a L747-A750>P mutation) are shown in red (HCC4006 vs PC9 at 0.001μM p=0.0455; HCC4006 vs HCC827 at 0.001μM, p=0.0094); B. Cell viability of PC9, HCC827 and HCC4006 cells treated with increasing concentrations of erlotinib for 72h (blue, orange and red, respectively); C. Cell viability of PC9, HCC827 and HCC4006 cells treated for 72h with increasing concentrations of osimertinib (blue, orange and red, respectively). All viability assays are representative of at least 3 repeats and p-values were calculated using t-tests at the indicated concentrations; D. Fold change IC50 bar graph (erlotinib/afatinib, left and osimertinib/afatinib, right) for PC9, HCC827 and HCC4006 cell lines (left: HCC4006 vs HCC287 p=0.0249; right: HCC4006 vs HCC827 p=0.0115). The p-values were calculated using t-tests not corrected for multiplicity (* p ≤ 0.05). NS: not significant. E. Western blotting analysis of lysates from the HCC4006 cell line treated with TKIs at indicated concentrations and immunoblotted for p-EGFR, EGFR and actin. The Western blot is representative of 3 repeats.
To further assess the increased afatinib sensitivity of EGFR harboring the L747-A750>P exon 19 alteration we studied the case of a patient enrolled on the Yale rebiopsy protocol. One patient, YLR086, whose tumor harbored a L747-A750>P exon 19 alteration, did not have a durable response to erlotinib. The patient, a 60-year old female never-smoker, was diagnosed in 2013 with stage IV adenocarcinoma of the lung. Molecular analysis of the diagnostic tumor biopsy specimen revealed the presence of the uncommon EGFR exon 19 deletion (L747-A750>P). The patient received first line systemic therapy with erlotinib, resulting in significant regression of disease in the lung, but progression in the brain. She then received palliative radiotherapy to the hip and brain. Further progression was observed within 2 months, so treatment with chemotherapy (carboplatin and pemetrexed) was initiated, alongside continuation of erlotinib. Imaging after 3 cycles of chemotherapy demonstrated further regression of disease in the lung, but multiple new osseous sclerotic lesions consistent with either treatment response or progressive disease. Imaging after two additional cycles of chemotherapy revealed new liver lesions consistent with metastatic disease, and progression of the osseous lesions. Biopsy of a liver mass confirmed the presence of the EGFR exon 19 L747-A750>P deletion mutation (Supplementary Figure 2A). She received no further TKI therapy.
Further analysis of the liver specimen from this patient revealed that it lacked known potential mechanisms of resistance to TKIs like the EGFR T790M mutation, or amplification of ERBB2 or MET. We were able to generate a cell line derived from a PDX of the liver biopsy sample and confirmed the presence of the EGFR L747-A750>P alteration (Supplementary Figure 2B). We then investigated erlotinib sensitivity of the YLR086 PDX-derived cell line. Erlotinib failed to completely suppress EGFR phosphorylation in YLR086 cells even at 5 μM (Supplementary Figure 2C), whereas 0.1 μM erlotinib was sufficient for complete suppression in PC9 (EGFR E746-A750) lung adenocarcinoma cells. H1975 lung adenocarcinoma cells (which harbor EGFR L858R and T790M mutations) are shown as a (resistant) negative control (Supplementary Figure 2C). The effect of erlotinib on cell viability was also substantially blunted in YLR086 PDX cells compared with PC9 cells, as shown in Supplementary Figure 2D, with IC50 increased by approximately 20-fold. Afatinib suppressed EGFR phosphorylation more effectively than erlotinib in YLR086 cells at concentrations down to 0.01 μM (Supplementary Figure 2E), and inhibited cell viability more effectively than erlotinib with an IC50 of ~0.06 μM (Supplementary Figure 2F, G). YLR086 cells also exhibited significant resistance to osimertinib (Supplementary Figure 2E, F, H), which suppressed EGFR phosphorylation only when added at 1 μM (Supplementary Figure 2E) and had an IC50 value in cell viability assays of ~0.2 μM (Supplementary Figure 2F, H). Therefore although the YLR086 PDX-derived cells were more resistant to TKIs overall compared to the other EGFR mutant cell lines studied, afatinib was the most effective TKI in this cell line. Altogether, our data indicate that the L747-A750>P mutation exhibits partial resistance to both erlotinib and osimertinib, but – importantly – not to afatinib.
Exon 19 deletions do not significantly affect EGFR stability
We hypothesized that the reduced apparent sensitivity of L747-A750>P EGFR to erlotinib might reflect its destabilization by introduction of a rigid proline between the β3 strand and αC helix – contrasting with the serine introduced instead in the L747-P753>S variant. Erlotinib cycles on and off the EGFR kinase domain very rapidly, limiting its occupancy time in the active site [21] – which in turn defines efficacy. Destabilizing the kinase would reduce this occupancy time further by accelerating conformational fluctuations in the kinase. It was reported that several lung cancer mutations destabilize EGFR, as evidenced by increased dependence on Hsp90 for their stability [22–24]. Such destabilization could explain why L747-A750>P EGFR shows reduced sensitivity to erlotinib (which rapidly cycles on/off) but not to afatinib (which binds covalently to EGFR). To test this possibility, we compared levels of different exon 19 deletion variants over a range of concentrations of the Hsp90 inhibitor geldanamycin as described [22]. As shown in Supplementary Figure 3, although several exon 19 deletion variants did increase sensitivity of EGFR levels to geldanamycin treatment compared with wild-type EGFR, the effect was not significantly greater for the L747-A750>P mutant. The effects were small, and the most destabilized variant appeared to be E746-A750 EGFR, which retains normal erlotinib sensitivity (Figure 1B). IC50 values for geldanamycin were 106 ± 20 nM for wild-type EGFR degradation, 44 ± 6 nM for E746-A750 EGFR, and 98 ± 38 nM for L747-A750>P. These results argue that stability per se cannot explain the altered inhibition profile of this variant.
Structural basis for reduced erlotinib sensitivity and preserved afatinib sensitivity in L747-A750>P EGFR
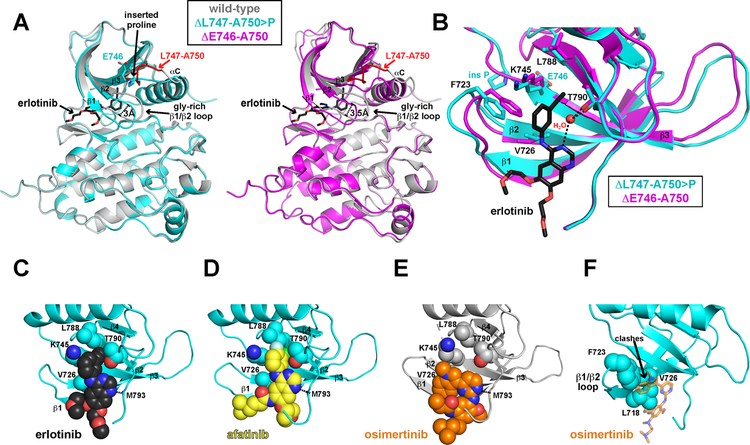
Since reduced EGFR stability does not appear to explain the reduced erlotinib sensitivity of L747-A750>P EGFR, we explored structural consequences of this deletion mutant using molecular dynamics (MD) simulations (see Methods). We performed simulations of the wild-type EGFR kinase domain plus the E746-A750 and L747-A750>P exon 19 deletion variants – both with and without bound TKIs (erlotinib, afatinib, and osimertinib). As shown in Figure 3A, the most notable structural change in the deletion mutants is a downward displacement of the glycine-rich (β1/β2) loop by about 3 Å in the L747-A750>P mutant (cyan) and 3.5 Å in the E746-A750 (magenta) mutant compared with wild-type (grey). This change arises from steric clashes of the β1/β2 loop with the β3/αC loop that do not occur in wild-type EGFR. The different displacement in the two deletion mutants arises because of different β3/αC loop conformations, reflecting the effective insertion of ‘Glu-Pro’ into this loop of L747-A750>P compared with E746-A750 (Figure 3A). Associated with this change is a small displacement of several residues that interact with the bound erlotinib molecule as described by Stamos et al. [17] (Figure 3B). The L788 side-chain is moved away from erlotinib slightly (by ~1 Å) in L747-A750>P compared with E746-A750 – taking it out of van der Waal’s contact with the drug. The F723 side-chain makes van der Waal’s contact with erlotinib in the E746-A750/erlotinib complex, but is displaced by about 2 Å in the L747-A750>P/erlotinib complex so that this contact is lost. The K745 side-chain is also slightly shifted, but other interactions observed by Stamos et al. remain. We infer that this slight overall structural modification at the drug/ATP-binding site of the EGFR kinase domain in the L747-A750>P deletion mutant impairs erlotinib binding. The diminution of erlotinib binding strength is relatively small, however, as evidenced by the fact that significant inhibition of L747-A750>P EGFR is still clearly seen in Figure 1B (it is just not complete at 1 μM).
Figure 3. Structural consequences of L747-A750>P mutation for inhibitor binding.
A. Overlay of geometry-minimized wild type EGFR kinase domain (residues 696–988) bound to erlotinib (grey) with the geometry minimized L747-A750>P kinase domain model shown in cyan (residues 696–988) at left, and the geometry minimized E746-A750 kinase domain model shown in magenta at right. The proline inserted in the β3/αC loop of L747-A750>P EGFR is marked, as is the retained E746 residue. The altered β3/αC loop clashes with the glycine-rich β1/β2 loop to displace it in both deletion mutants, downwards in the figure, by ~3 Å for L747-A750>P and ~3.5 Å for E746-A750 as marked, to influence the ATP-binding site (as well as β1 and β2 strand positions). B. Close-up overlay of erlotinib bound geometry-minimized EGFR L747-A750>P (cyan) and E746-A750 (magenta) kinase domains, with bound erlotinib shown in stick representation (black). Residues involved in erlotinib binding are marked and labeled. There is no significant change in the binding mode of erlotinib, and only relatively minor changes in the positions of key interacting residues (see also Supplementary Figure 4A). Notably, the L788 side-chain is slightly displaced (and taken out of van der Waal’s contact with drug), and the F723 side-chain is shifted by ~2 Å so that it makes van der Waal’s contact with erlotinib in the E746-A750 complex but – importantly – not in the L747-A750>P complex. The K745 side-chain is also slightly shifted. C, D. Erlotinib (black) and afatinib (yellow) are shown (in sphere representation) bound to geometry-minimized L747-A750>P EGFR kinase domain, shown in cyan. The aniline ring of each inhibitor can be seen to nestle into a cavity formed by the side-chains of K745, L788, T790, with more intimate packing of the afatinib 3-chloro-4-fluoroanilene ring than the erlotinib 3-ethynylaniline ring – plus halogen bonding (-OH···Cl) of the chlorine (green) with the T790 side-chain. The side-chain of K728 (not shown – see Supplementary Figure 4B) also interacts uniquely with the tetrahydropyranyl ring of afatinib only in the L747-A750>P variant (bottom right of D). E. Osimertinib (orange) shown (in sphere representation) bound to wild-type EGFR kinase domain, from PDB entry 4ZAU (Yosaatmadja et al., 2015) shown in grey. The cavity formed by the side-chains of K745, L788, T790 is empty in this complex. Instead, the phenyl ring of osimertinib packs against the side-chain of L718, and its indole ring packs against the F723 and V726 side-chains in β1, β2, and the β1/β2 loop. F. Depiction of how the displaced β1/β2 loop (and β1 and β2 strands) causes clashes between the L718, F723 and V726 side-chains (shown as spheres) and the bound osimertinib (orange sticks). The cyan ribbon is the geometry minimized L747-A750>P EGFR kinase domain, and bound osimertinib is overlaid from the coordinates of PDB entry 4ZAU, in which the drug is bound to the wild-type kinase (Yosaatmadja et al., 2015). The location of key clashes is marked, which presumably reduce osimertinib efficacy with L747-A750>P EGFR.
Comparing Figures 3C and 3D (and Supplementary Figure 4) provides one suggestion as to why afatinib binding might be retained more effectively than erlotinib binding in L747-A750>P EGFR. The most notable difference is that the 3-ethynylaniline ring of erlotinib appears to be less intimately ‘packed’ between the side-chains of K745, L788 and T790 in Figure 3C than does the equivalently located 3-chloro-4-fluoroanilene ring of afatinib in Figure 3D. Moreover, the position of afatinib’s 3-chloro-4-fluoroanilene moiety is stabilized by halogen bonding with the T790 side-chain in both deletion variants. The tetrahydropyranyl ring unique to afatinib (bottom right in Figure 3D, far left in Supplementary Figure 4B) is also predicted to form a hydrogen bond with the K728 side-chain in EGFR, but only in models of the L747-A750>P variant (Supplementary Figure 4B). This interaction is also seen in the crystal structure of the afatinib-bound EGFR kinase [25], but cannot form with erlotinib [17] or osimertinib [26]. The resulting increased set of interactions made between the L747-A750>P variant and afatinib, alongside the more intimate aniline ring packing outlined above, argues that slight rearrangement of the drug binding site in this variant has less negative impact on afatinib binding than on erlotinib binding – as we see experimentally. As a result, the non-covalent afatinib/EGFR complex presumably can last long enough with the L747-A750>P variant for covalent bonding to C797 to proceed efficiently, allowing effective inhibition to be retained. MM-GBSA calculations (see Methods) further suggested that L747-A750>P EGFR makes a more extensive set of non-covalent interactions with afatinib than with erlotinib (by ~3 kcal/mol), whereas results for afatinib and erlotinib binding were very similar (within 1 kcal/mol) for wild-type EGFR and other exon 19 deletions.
Structural basis for reduced osimertinib sensitivity in L747-A750>P EGFR
In parallel, we considered osimertinib binding to the L747-A750>P EGFR variant. As shown in Figure 3E, osimertinib does not utilize the cavity in the EGFR kinase domain into which the aniline rings of erlotinib and afatinib project, so alterations in this cavity cannot explain the reduced inhibition seen in Figure 1E. However, movement in the L747-A750>P variant of the β1/β2 loop (Figure 3A), will reposition residues L718, F723, and V726, which all make van der Waal’s contacts with osimertinib to promote binding. Following the 3 Å displacement of the β1/β2 loop marked in Figure 3A, these three side-chains will clash sterically with bound osimertinib (Figure 3F) unless it also moves by an equivalent amount – which in turn would break other interactions and reduce binding and covalent interaction with EGFR. Just as moving the β1/β2 loop (including L718) toward the bound osimertinib (in L747-A750>P EGFR) reduces the effectiveness of this inhibitor (data in Figure 1E) through steric clashes, so does a recent report suggest that ‘retracting’ the L718 side-chain by mutating it to valine impairs osimertinib binding and function [27].
The EGFR L747-A750>P mutation is associated with inferior outcomes in patients treated with erlotinib
To correlate these in vitro and in silico findings with clinical observations, we investigated responses to erlotinib in patients with tumors carrying EGFR exon 19 deletions. We included patients with tumors harboring an EGFR exon 19 mutation (E746-A750, L747-P753>S or L747-A750>P) who had received erlotinib as first-line therapy for advanced disease. Between June 2008 and May 2016, 32 patients met criteria for analysis: twenty-four patients harbored the common E746-A750 mutation, 4 had the L747-P753>S mutation, and 4 had the L747-A750>P mutation. Thirty of the 32 patients received erlotinib alone, and two received erlotinib in combination with hydroxychloroquine. At the time of analysis, all 32 patients had experienced progression of disease, 30 of 32 (94%) had discontinued erlotinib and 22 (69%) had died.
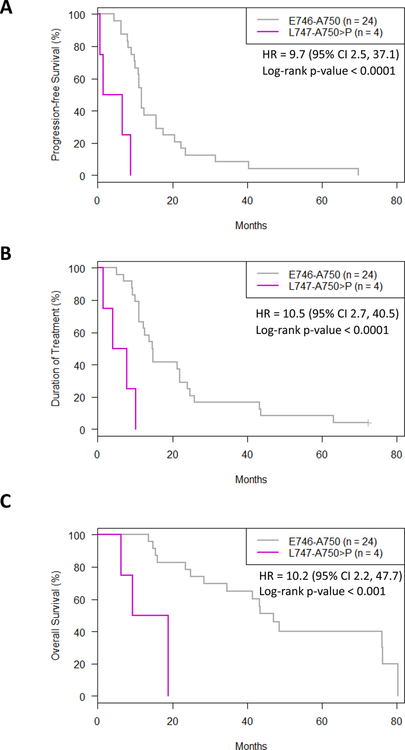
Erlotinib-treated patients with tumors harboring the L747-A750>P mutation demonstrated significantly worse outcomes than those with tumors harboring E746-A750 or L747-P753>S mutations (Figure 4, Supplementary Figure 5). PFS was significantly shorter for erlotinib-treated patients with tumors with the L747-A750>P mutation than those with tumors with the E746-A750 mutation (median 4.1 months and 11.7 months, respectively, HR = 9.7 (95% CI 2.5 – 37.1), p < 0.0001) (Figure 4A). Similarly, the duration of treatment (DOT) was shorter for patients with tumors harboring the L747-A750>P mutation than for those with tumors with the E746-A750 mutation (median 5.9 months and 14.8 months, HR = 10.5 (95% CI 2.7 – 40.5), p < 0.0001) (Figure 4B). Overall survival (OS) was also significantly shorter for erlotinib-treated patients with L747-A750>P mutant tumors than those with tumors with the E746-A750 mutation (median 14.1 months and 47.0 months, HR = 10.2 (95% CI 2.2 – 47.7), p < 0.001) (Figure 4C). There was no significant difference in outcomes between the E746-A750 and L747-P753>S groups (Supplementary Figure 5). Importantly, two of four patients with tumors harboring the L747-A750>P mutation demonstrated primary resistance to erlotinib (i.e. progressive disease as the best response), whereas no patients harboring the E746-A750 or L747-P753>S mutation exhibited primary resistance.
Figure 4. Patients treated with erlotinib harboring tumors with the L747-A750>P alteration have worse outcomes compared to those with tumors harboring the more common E746-A750 deletion.
A. Progression-free survival (PFS) (Log-rank p-value<0.0001), B. Duration of treatment (DOT) (Log-rank p-value<0.0001) and, C. Overall survival (OS) (Log-rank p-value<0.001) graphs of patients with tumors harboring the different EGFR exon 19 deletion mutations treated with erlotinib. E746-A750 (grey); L747-A750>P (purple).
Discussion
The key finding described here, revealed by biochemical, signaling and cell viability studies, is that individual EGFR exon 19 deletion mutations can differ in their sensitivity for individual EGFR-targeted TKIs in a clinically significant way. Specifically, we find that the L747-A750>P variant exhibits limited sensitivity to both the 1st generation TKI erlotinib and the 3rd generation TKI osimertinib – yet retains high sensitivity to the 2nd generation TKIs afatinib and dacomitinib. Importantly, these observations are likely to underlie our finding that patients with EGFR L747-A750>P mutant tumors who were treated with erlotinib have inferior clinical outcomes compared with patients whose tumors harbor the common E746-A750 mutation.
Indications that there are differences in TKI sensitivity amongst EGFR exon 19 deletion mutations have begun to emerge from several experimental and clinical studies. In vitro cellular studies of a range of patient-derived EGFR variants have shown different transforming potential and TKI sensitivities [13, 28]. Moreover, patients with tumors harboring exon 19 deletions in which the LRE motif (between L747 and E749) was included in the deleted region were reported to have better clinical outcomes than those with mutations that do not eliminate this motif [29]. Further, when EGFR exon 19 deletion mutant tumors were subdivided into those with the deletion beginning at E746 versus those beginning at L747, the former were found to have better PFS than the latter in one study [30] - although a different study did not reach this conclusion [31]. A recent report also showed that the G724S osimertinib-resistance EGFR mutation emerges in the context of specific EGFR exon 19 deletion mutations [15]. Together with our results, these data argue that differences between individual exon 19 deletions in EGFR should be examined in detail. The findings also underscore the fact that not all EGFR mutations are the same – highlighting the importance of mutation-specific EGFR-TKI selection. Indeed, it has been shown that the precise nature of deletions in the structural equivalent (β3/αC loop) region of BRAF and ERBB2 affects drug selectivity [32], arguing that this may be a general property of therapeutically targetable oncogenic kinases.
In addition to our observations with the L747-A750>P variant, analyses of several previous clinical case studies suggest that variants in which L747 in the β3/αC loop of EGFR is replaced with a proline are associated with primary resistance to 1st generation TKIs [33–35]. Based on these clinical findings plus our experimental and modeling studies, it seems likely that (as in L747-A750>P) the presence of a proline at this position displaces the adjacent β1/β2 glycine-rich loop (Figure 3A, B) and thus impairs drug binding. Interestingly, EGFR insertions in exon 19 have been described that also include a proline mutation at position 747 [36]. Similar to our findings with the L747-A750>P mutation, exon 19 insertion mutations exhibit sensitivity to afatinib and partial sensitivity to erlotinib. In light of our studies, more in depth analyses of these different mutations and their TKI sensitivity patterns is warranted. Further, it is interesting that the L747-A750>P variant is most sensitive to a 2nd generation irreversible inhibitor (afatinib) and less sensitive to both a reversible inhibitor (erlotinib) and an irreversible inhibitor (osimertinib). Understanding the reduced sensitivity to erlotinib seems relatively straightforward – the altered binding site impairs kinase occupancy and thus inhibition. For the irreversible/covalent inhibitors, the difference between osimertinib and afatinib underscores the need for non-covalent interactions to drive kinase occupancy for a time sufficient to allow reaction of the inhibitor with the C797 side-chain. These findings have potential implications for drug design since they indicate that the types of covalent and non-covalent interactions formed by a drug with different mutant EGFR molecules need to be considered.
We demonstrated that patients with tumors with the L747–750>P mutation have inferior outcomes compared to those harboring the canonical E746-A750 mutation when treated with a 1st generation EGFR TKI. Limitations of our study include the small sample size and the lack of clinical outcome data for 2nd and 3rd generation EGFR TKIs in patients harboring the L747-A750>P mutation. Ongoing and future investigations attempting to validate these findings in larger cohorts including patients with tumors with uncommon exon 19 mutations treated with 2nd and 3rd generation EGFR TKIs will be important. An interesting question that remains is whether osimertinib is optimally effective in the context of the EGFR T790M mutation (which is the most common resistance mechanism for both 1st and 2nd generation EGFR TKIs) in tumors with uncommon exon 19 mutations.
In conclusion, our study demonstrates that the presence of the L747-A750>P deletion is associated with exquisite sensitivity to afatinib and reduced sensitivity to the 1st generation TKI erlotinib and 3rd generation TKI osimertinib. Our study provides evidence that the precise type of EGFR exon 19 deletion mutation influences sensitivity to different EGFR-targeted TKIs. Our findings argue in favor of analyzing the specific exon 19 mutation at the time of diagnosis. Moreover, particularly in light of recent studies demonstrating increased PFS with osimertinib treatment over 1st generation EGFR TKIs, our findings argue that there may be an important role for 2nd generation EGFR-targeted TKIs in patients with certain uncommon exon 19 deletions. Indeed, while all of these TKIs retain some activity against the L747-A750>P mutant (especially osimertinib), incomplete suppression of EGFR activity by 1st and 3rd generation TKIs could affect the long-term clinical benefit of these therapies. Further investigation of these issues will help define optimal treatment strategies for patients with tumors harboring various EGFR exon 19 deletion mutations.
Supplementary Material
Statement of Significance.
Tyrosine kinase inhibitors (TKIs) have transformed the paradigm for the treatment of EGFR mutant lung cancer, however differences in TKI sensitivity between specific EGFR mutations are increasingly being recognized. Here, we show that the specific type of EGFR exon 19 deletion affects the pattern of sensitivity to different TKIs. This has important clinical implications given that patients with tumors harboring specific exon 19 deletions may benefit from treatment with specific TKIs.
Acknowledgements
This work was supported by NIH/NCI grants P50 CA196530 (K. Politi, S. B. Goldberg, M. A. Lemmon, S. Gettinger and D. Zelterman) and R01 CA198164 (M. A. Lemmon), T32 CA193200 (J. Starrett), an Italian Association for Cancer Research (AIRC) fellowship (A. Truini), and an Arnold and Mabel Beckman Foundation Postdoctoral fellowship (K. Ashtekar).
Footnotes
Declaration of Interests
The authors have the following conflicts to disclose: Research funding to the Institution from AstraZeneca, Bristol-Myers Squibb, Ariad/Takeda, Iovance, Roche/Genentech, Kolltan and Symphogen. Consulting/Advisory Role honoraria from AstraZeneca (KP, SBG), Merck (KP), Novartis (KP), Boehringer Ingelheim (SBG), Bristol-Myers Squibb (SBG, SG), Eli Lilly (SBG), Roche/Genentech (SBG), Amgen (SBG), Nektar (SG), Spectrum (SBG), Tocagen (KP), Maverick Therapeutics (KP) and Dynamo Therapeutics (KP). Royalties from IP licensed to Molecular MD from MSKCC (KP). AT is currently an employee of MSD, however, these studies were conducted when she was at Yale. MD is currently an employee of Regeneron Pharmaceuticals, however, these studies were conducted when she was at Yale.
References
- 1.Lynch TJ, et al. , Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med, 2004. 350(21): p. 2129–39. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, et al. , EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science, 2004. 304(5676): p. 1497–500. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, et al. , EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A, 2004. 101(36): p. 13306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan CS, Gilligan D, and Pacey S, Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol, 2015. 16(9): p. e447–e459. [DOI] [PubMed] [Google Scholar]
- 5.Peters S, Zimmermann S, and Adjei AA, Oral epidermal growth factor receptor tyrosine kinase inhibitors for the treatment of non-small cell lung cancer: comparative pharmacokinetics and drug-drug interactions. Cancer Treat Rev, 2014. 40(8): p. 917–26. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, et al. , Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med, 2017. 376(7): p. 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soria JC, et al. , Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med, 2017. [DOI] [PubMed]
- 8.Ladanyi M and Pao W, Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol, 2008. 21 Suppl 2: p. S16–22. [DOI] [PubMed] [Google Scholar]
- 9.Murray S, et al. , Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non-small cell lung cancer: an analytical database. J Thorac Oncol, 2008. 3(8): p. 832–9. [DOI] [PubMed] [Google Scholar]
- 10.Sharma SV, et al. , Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer, 2007. 7(3): p. 169–81. [DOI] [PubMed] [Google Scholar]
- 11.Maemondo M, et al. , Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med, 2010. 362(25): p. 2380–8. [DOI] [PubMed] [Google Scholar]
- 12.Sequist LV, et al. , Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol, 2013. 31(27): p. 3327–34. [DOI] [PubMed] [Google Scholar]
- 13.Kohsaka S, et al. , A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med, 2017. 9(416). [DOI] [PubMed] [Google Scholar]
- 14.Improta G, et al. , Uncommon frame-shift exon 19 EGFR mutations are sensitive to EGFR tyrosine kinase inhibitors in non-small cell lung carcinoma. Med Oncol, 2018. 35(3): p. 28. [DOI] [PubMed] [Google Scholar]
- 15.Brown BP, et al. , On-target resistance to the mutant-selective EGFR inhibitor osimertinib can develop in an allele specific manner dependent on the original EGFR activating mutation. Clin Cancer Res, 2019. [DOI] [PMC free article] [PubMed]
- 16.Pao W, et al. , Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med, 2005. 2(3): p. e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stamos J, Sliwkowski MX, and Eigenbrot C, Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem, 2002. 277(48): p. 46265–72. [DOI] [PubMed] [Google Scholar]
- 18.Banks JL, et al. , Integrated Modeling Program, Applied Chemical Theory (IMPACT). J Comput Chem, 2005. 26(16): p. 1752–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, et al. , The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins, 2011. 79(10): p. 2794–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wind S, et al. , Clinical Pharmacokinetics and Pharmacodynamics of Afatinib. Clin Pharmacokinet, 2017. 56(3): p. 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barkovich KJ, et al. , Kinetics of inhibitor cycling underlie therapeutic disparities between EGFR-driven lung and brain cancers. Cancer Discov, 2012. 2(5): p. 450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimamura T, et al. , Epidermal growth factor receptors harboring kinase domain mutations associate with the heat shock protein 90 chaperone and are destabilized following exposure to geldanamycins. Cancer Res, 2005. 65(14): p. 6401–8. [DOI] [PubMed] [Google Scholar]
- 23.Jorge SE, et al. , EGFR Exon 20 Insertion Mutations Display Sensitivity to Hsp90 Inhibition in Preclinical Models and Lung Adenocarcinomas. Clin Cancer Res, 2018. 24(24): p. 6548–6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pines G, Kostler WJ, and Yarden Y, Oncogenic mutant forms of EGFR: lessons in signal transduction and targets for cancer therapy. FEBS Lett, 2010. 584(12): p. 2699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solca F, et al. , Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J Pharmacol Exp Ther, 2012. 343(2): p. 342–50. [DOI] [PubMed] [Google Scholar]
- 26.Yosaatmadja Y, et al. , Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed. J Struct Biol, 2015. 192(3): p. 539–44. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, et al. , Acquired EGFR L718V mutation mediates resistance to osimertinib in non-small cell lung cancer but retains sensitivity to afatinib. Lung Cancer, 2018. 118: p. 1–5. [DOI] [PubMed] [Google Scholar]
- 28.Furuyama K, et al. , Sensitivity and kinase activity of epidermal growth factor receptor (EGFR) exon 19 and others to EGFR-tyrosine kinase inhibitors. Cancer Sci, 2013. 104(5): p. 584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung KP, et al. , Clinical outcomes in non-small cell lung cancers harboring different exon 19 deletions in EGFR. Clin Cancer Res, 2012. 18(12): p. 3470–7. [DOI] [PubMed] [Google Scholar]
- 30.Lee VH, et al. , Association of exon 19 and 21 EGFR mutation patterns with treatment outcome after first-line tyrosine kinase inhibitor in metastatic non-small-cell lung cancer. J Thorac Oncol, 2013. 8(9): p. 1148–55. [DOI] [PubMed] [Google Scholar]
- 31.Su J, et al. , Molecular characteristics and clinical outcomes of EGFR exon 19 indel subtypes to EGFR TKIs in NSCLC patients. Oncotarget, 2017. 8(67): p. 111246–111257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foster SA, et al. , Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2. Cancer Cell, 2016. 29(4): p. 477–493. [DOI] [PubMed] [Google Scholar]
- 33.Wang YT, et al. , Exon 19 L747P mutation presented as a primary resistance to EGFR-TKI: a case report. J Thorac Dis, 2016. 8(7): p. E542–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu G, et al. , EGFR mutation L747P led to gefitinib resistance and accelerated liver metastases in a Chinese patient with lung adenocarcinoma. Int J Clin Exp Pathol, 2015. 8(7): p. 8603–6. [PMC free article] [PubMed] [Google Scholar]
- 35.Coco S, et al. , Uncommon EGFR Exon 19 Mutations Confer Gefitinib Resistance in Advanced Lung Adenocarcinoma. J Thorac Oncol, 2015. 10(6): p. e50–2. [DOI] [PubMed] [Google Scholar]
- 36.He M, et al. , EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res, 2012. 18(6): p. 1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.