Abstract
Purpose:
In glioma surgery, it is critical to maximize tumor resection without compromising adjacent non-cancerous brain tissue. Optical Coherence Tomography (OCT) is a non-invasive, label-free, real-time, high-resolution imaging modality that has been explored for glioma infiltration detection. Here we report a novel artificial intelligence (AI) assisted method for automated, real-time, in situ detection of glioma infiltration at high spatial resolution.
Experimental Design:
Volumetric OCT datasets were intraoperatively obtained from resected brain tissue specimens of 21 patients with glioma tumors of different stages and labeled as either non-cancerous or glioma-infiltrated based on histopathology evaluation of the tissue specimens (gold standard). Labeled OCT images from 12 patients were used as the training dataset to develop the AI assisted OCT-based method for automated detection of glioma-infiltrated brain tissue. Unlabeled OCT images from the other 9 patients were used as the validation dataset to quantify the method detection performance.
Results:
Our method achieved excellent levels of sensitivity (~100%) and specificity (~85%) for detecting glioma-infiltrated tissue with high spatial resolution (16 μm laterally) and processing speed (~100,020 OCT A-lines/second).
Conclusions:
Previous methods for OCT-based detection of glioma-infiltrated brain tissue rely on estimating the tissue optical attenuation coefficient from the OCT signal, which requires sacrificing spatial resolution to increase signal quality, and performing systematic calibration procedures using tissue phantoms. By overcoming these major challenges, our AI-assisted method will enable implementing practical OCT-guided surgical tools for continuous, real-time and accurate intra-operative detection of glioma-infiltrated brain tissue, facilitating maximal glioma resection and superior surgical outcomes for glioma patients.
INTRODUCTION
Gliomas are the most common and aggressive primary brain cancers in adults [1, 2]. It is well established that maximal glioma surgical resection can lead to both prolonged survival and delayed cancer recurrence [1, 3–6]. The challenge, however, lies in the limited ability of neurosurgeons to differentiate cancerous versus non-cancerous brain tissue during resection surgery. The standard of care, which is interpreted as the surgeon’s perception of cancer based on gross appearance and all available intraoperative surgical navigational systems, has shown to have 100% sensitivity and 40 to 50% specificity [7]. Overcoming this surgical challenge will enable both maximizing cancer resection and minimizing damage of healthy brain tissue, thus significantly improving both the overall survival rate (OS) and progression-free survival (PFS). [8–10]
Several imaging techniques are currently being evaluated or already adopted as image-guided surgical tools to assist with brain cancer resection. Magnetic resonance imaging (MRI) provides excellent visualization of soft tissue, but it is not sensitive at detecting microscopic diseases at tumor margin, even when used intraoperatively [11]. Intraoperative computed tomography (iCT) allows assessing for residual cancer, but has low resolution at the tumor periphery [12]. In addition, these imaging modalities are time-consuming, costly (upwards of $1 million dollars to adopt), and do not provide continuous real-time intraoperative guidance. Intraoperative ultrasound imaging (iUS) enables real-time imaging, but it has limited contrast and spatial resolution for brain cancer detection [13]. Intraoperative fluorescence imaging of 5-aminolevulinic acid (5-ALA) induced protoporphyrin-IX (PpIX) has shown a good correlation between fluorescence distribution and the presence of high-grade glioma [14], but it has shown limited sensitivity and specificity for detecting cancer-infiltrated brain tissue and low-grade gliomas [15, 16]. Raman spectroscopy and imaging have been broadly applied for brain tissue biochemical differentiation [17] and glioma infiltration detection by providing subcellular resolution and label-free imaging capabilities [18–22]. Unfortunately, several limitations are associated to these techniques, including the intrinsic weakness of the Raman signal, limited imaging depth and field of view (FOV), and slow imaging speed [23–25]. In addition, the capability of Raman spectroscopy and/or imaging for detecting cancer-infiltrated brain tissue intraoperatively has not yet fully demonstrated [26, 27]. More recently, coherent Raman Scattering (CRS) and Stimulated Raman Scattering Microscopy (SRS) have been explored for brain tumor margin differentiation; however, a definite intraoperative Computer-Aided Diagnosis system for human brain tissue differentiation has not been reported [23, 26, 28, 29]. In summary, there is still an urgent need for image-guided tools capable of providing continuous, in situ and accurate assessment of brain cancer infiltration during brain tumor resection surgery.
Optical coherence tomography (OCT) is a non-invasive medical imaging technique capable of continuous, label-free, high-resolution, 2D and 3D imaging of biological tissues [30]. Since the imaging depth of OCT (1.5 – 3 mm) is similar to the resection depth of cancer infiltrated brain regions, OCT has been evaluated as an image-guided tool for brain tumor resection surgery [7, 31–35]. One common limitation of previous studies, however, is the lack of adequate computational methods for rapid, automated and accurate intraoperative detection of cancer-infiltrated brain tissue at high spatial resolution, particularly for glioma resection.
Previous computational methods for OCT-based detection of glioma-infiltrated brain tissue rely on estimating the tissue optical attenuation coefficient from the OCT signal, which require averaging multiple A-line signals to reduce the noise and thus sacrificing spatial resolution. In addition, previous methods also require performing calibration procedures using tissue phantoms [7, 31, 33, 36]. To overcome these major challenges, we have developed a novel artificial intelligence (AI) based computational method in which each depth-dependent OCT intensity measurement (or A-line) is modeled as a linear combination of underlying characteristic intensity-depth profiles. As a result, this method enables identifying and quantifying intensity-depth signatures specific to A-lines from glioma-infiltrated brain tissue, which can be utilized as discriminative features within machine learning algorithms to detect glioma-infiltrated brain tissue. The method was successfully developed using a database of OCT 3D images taken from freshly resected human non-cancerous and glioma-infiltrated brain tissue samples, and its performance was robustly quantified using an independent validation database. Owing to its demonstrated accuracy, low computational cost and high spatial resolution, this method has the potential to enable the development of OCT-guided surgical tools for continuous, real-time and accurate in situ intra-operative detection of glioma infiltration.
MATERIALS AND METHODS
Database of OCT scans from fresh brain tissue surgical samples
Intraoperative, fresh brain tissue samples were obtained from the edge of the surgical cavity based on neurosurgeon visual interpretation and image-guided navigation in 21 surgical glioma patients. The imaging protocol was approved by the Institutional Review Board at Johns Hopkins University, which follows the Belmont Report ethical guidelines. Informed written consent was obtained from each subject or each subject’s legal guardian. The tissue samples corresponded to either non-cancerous or glioma-infiltrated brain regions. A number of OCT volumes were acquired from different locations within each brain tissue sample. The measured OCT lateral (i.e. horizontal) and axial (i.e. vertical or depth) resolutions were approximately 16.0 μm and 6.4 μm (in tissue), respectively [7]. Each volume consists of a series of 10 OCT cross-sectional images or B-scans of 1024 pixels (2 mm) laterally by 2048 pixels (2.5 mm) in depth, where each B-scan was acquired at 0.5 mm intervals, resulting in a volume of 5×2×2.5 mm3 (W × L × D). In order to divide the database of OCT volumes into training and validation sets, the 21 patients were randomly divided into two groups, one with 12 and another with 9 patients. All the volumes from the group of 12 patients were assigned to the training set, while all the volumes from the group of 9 patients were assigned to the validation set. All the samples underwent histopathological processing and evaluation by a neuropathologist [7]. The histopathological distributions of the OCT volumes in the training and validation sets are summarized in Table 1.
Table 1 –
Training set (12 patients, 194 OCT volumes, 1940 B-scans) and validation set (9 patients, 295 OCT volumes, 2950 B-scans).
| Patient # | (Location) | # Non-cancerous OCT Volumes | # Glioma-infiltrated OCT Volumes | Grade of Cancer | |
|---|---|---|---|---|---|
| TRAINING SET | Patient-1 | 1A | 3 | - | - |
| Patient-2 | 2A | 4 | - | - | |
| Patient-3 | 3A | 3 | - | - | |
| Patient-4 | 4A | 13 | - | - | |
| 4B | 8 | - | - | ||
| Patient-5 | 5A | 10 | - | - | |
| Patient-6 | 6A | - | 8 | Grade II | |
| 6B | - | 9 | Grade II | ||
| 6C | 11 | Grade II | |||
| 6D | - | 13 | Grade II | ||
| 6E | - | 16 | Grade II | ||
| Patient-7 | 7A | - | 5 | Grade II | |
| 7B | - | 11 | Grade II | ||
| Patient-8 | 8A | - | 8 | Grade II | |
| 8B | - | 10 | Grade II | ||
| 8C | - | 11 | Grade II | ||
| Patient-9 | 9A | 18 | - | - | |
| Patient-10 | 10A | - | 5 | Grade IV | |
| 10B | - | 5 | Grade IV | ||
| 10C | - | 5 | Grade IV | ||
| Patient-11 | 11A | - | 8 | Grade IV | |
| Patient-12 | 12A | 10 | - | - | |
| Total in Training Set | 69 | 125 | |||
| VALIDATION SET | Patient-13 | 13A | - | 13 | Grade II |
| 13B | - | 17 | Grade II | ||
| 13C | - | 14 | Grade II | ||
| Patient-14 | 14A | 18 | - | - | |
| Patient-15 | 15A | 15 | - | - | |
| 15B | 9 | - | - | ||
| 15C | 9 | - | - | ||
| Patient-16 | 16A | 16 | - | - | |
| 16B | 8 | - | - | ||
| 16C | 16 | - | - | ||
| Patient-17 | 17A | 23 | - | - | |
| 17B | 16 | - | - | ||
| Patient-18 | 18A | 21 | - | - | |
| Patient-19 | 19A | - | 22 | Grade IV | |
| 19B | - | 14 | Grade IV | ||
| Patient-20 | 20A | 12 | - | - | |
| 20B | - | 17 | Grade II | ||
| Patient-21 | 21A | 5 | - | - | |
| 21B | - | 15 | Grade IV | ||
| 21C | - | 15 | Grade IV | ||
| Total in Validation Set | 168 | 127 |
OCT B-scans preprocessing
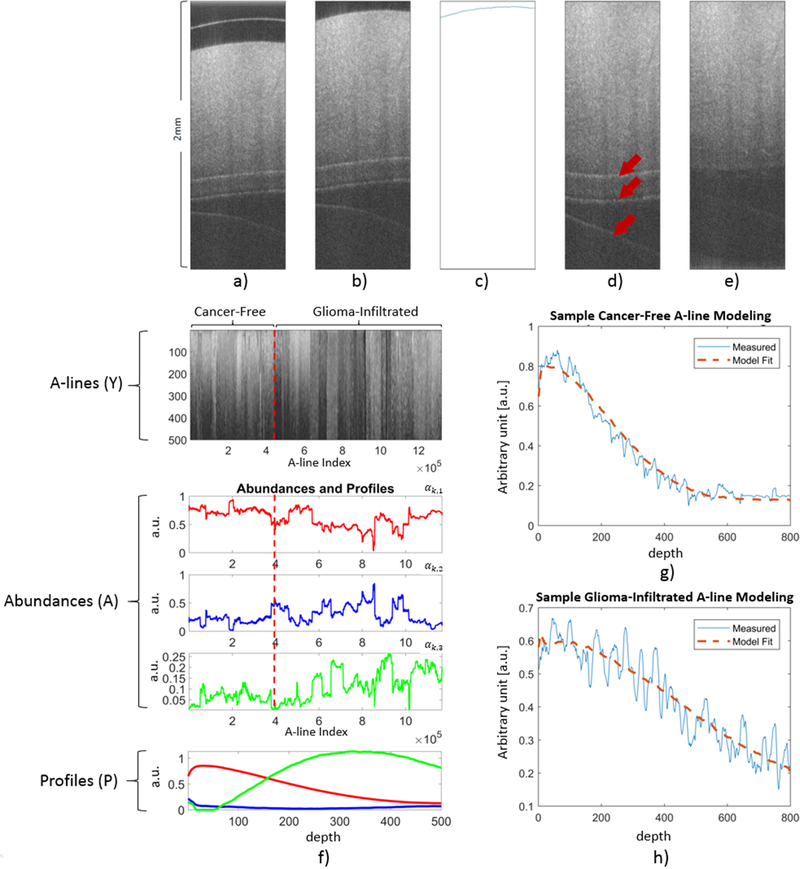
Each original OCT B-scan was preprocessed following the procedure described in (Figure 1). First, the original B-scan (Figure 1.a) was cropped to remove artifacts from above the tissue surface using a predefined fixed crop (Figure 1.b). Then, the tissue surface was detected from the cropped B-scan using the Canny Edge Detection algorithm (Figure 1.c) [37], and the B-scan was warped using a circle-shifting upward method in order to flatten the surface (Figure 1.d). Finally, in order to eliminate reflection artifacts within the tissue region caused by the cover glass or the saline surface (arrows in Figure 1.d), a peak detection algorithm was applied and the regions of the A-line around the detected peaks were smoothed using a 2D entropy filter of order 5×5. Although, these preprocessing steps do not guarantee the absolute elimination of all artifacts, the resulting preprocessed B-scans (Figure 1.e) were adequate for the application of the A-line modeling method described in the following section.
Figure 1 –
Preprocessing steps applied to every B-scan in both the training and validation sets. a) Unprocessed B-scan. b) Cropped B-scan. c) Surface detection using the Canny Edge Detection algorithm. d) Warped B-scan to generate a flat surface. Arrows indicate reflection artifacts. e) Peak detection to identify locations of reflection artifacts, and entropy filtering around the detected peaks to obtain a completely preprocessed B-scan. f) Results of the BEAE analysis applied to the training set. Top panel: All the A-lines (Y) from the training set included in the BEAE analysis. Middle panels: Estimated abundances (A) for each A-line analyzed. Bottom panel: Estimated profiles (P) common to all the A-lines included in the BEAE analysis. g) Sample A-line from a non-cancerous volume and its fit modeled as the linear combination of the estimated common profiles (P). h) Sample A-line from a glioma-infiltrated volume and its fit modeled as the linear combination of the estimated common profiles (P).
Model-based OCT A-line feature extraction
The main idea behind our method for automated classification of non-cancerous vs. glioma-infiltrated brain regions is to model every A-line yk of any OCT B-scan as a linear combination of N profiles or end-members pn (n = 1,…,N):
| (1) |
The profiles pn are assumed to be the same for all the A-lines of the available data, while the linear coefficients or abundances αk,n, are assumed to be unique to each A-line yk. The profiles pn were first estimated from the training set, consisting of 1,940 B-scans (Table 1). To accelerate the estimation of the profiles, only every other A-line in each B-scan of the training set were used (512 out of 1,024 A-lines per B-scan). All the selected training A-lines (1,940 × 512 = 993,280) were arranged into a matrix Y = [y1 …yK] of size L × K, where L is the length of each A-line (1,024) and K is the total number of A-lines in the training set (993,280). The N unknown profiles pn were arranged into a matrix P = [p1 …pn] of size L × N, where L is the length of each profile (equal to the A-line length) and N is the number of profiles. The unknown abundances were arranged into a matrix A = [α1 …αK] of size N × K, respectively, where the abundance column vector at the kth A-line is denoted as αk = [αk,1 …αk,N]′. Using this matrix notation, the modeling of all the A-lines, based on Eq. (1), can be expressed as: Y = PA.
The simultaneous estimation of the unknown profile P and abundance A matrices from the training OCT A-line data Y can be formulated as a nonlinear quadratic optimization problem, with the following specific constrains: a) the profiles can have positive values only (P ≥ 0), b) the abundances can have positive values only (A ≥ 0), and c) the values of the abundances for a given A-line should add to one, since they represent the relative contribution of each profile to that A-line (AT1 = 1). The resolution of this constrained nonlinear quadratic optimization problem can be performed by applying our recently developed and validated blind end-member and abundance estimation (BEAE) method [38, 39], which minimizes the following cost function:
| (2) |
The first term is directly related to the quadratic optimization approach in Eq. (1). The second term is a regularization term that penalizes the distance between profiles by using a regularization parameter ρ > 0. The third term is a regularization parameter for the abundances which ensures low entropy conditions among A-lines by using μ > 0. Once the profiles P have been estimated from the training data, the abundances for any new set of A-lines can be directly estimated by solving Eq. (1) using a constrained linear least square approach. This estimation is computationally fast, as it only involves solving a system of linear equations with positivity constrain on the abundances.
Classifier training
The abundances αk,n estimated for each A-line can be used as discriminative features within a machine learning algorithm designed to classify each A-line as from either a non-cancerous or a glioma-infiltrated brain region. Due to the number of features (N − 1) vs. the number of training data (K), a simple logistic regression classifier was chosen over other more complex methods, such as support-vector machines and neural networks [40]. Since each whole OCT volume in the training set was annotated as either non-cancerous or glioma-infiltrated brain tissue (Table 1), all the A-lines in a given volume were labeled based on their volume annotation. The resulting abundances A from the 993,280 labeled A-lines in the training set (194 volumes × 10 B-Scans/volume × 512 A-lines/B-Scan, see Table 1) were then used to optimize the logistic regression classifier. Since the logistic regression classifier was trained to classify each A-line of a new OCT volume as either from a non-cancerous or a glioma-infiltrated brain region, not all the A-lines from the new OCT volume would necessarily be classified to the same class. Therefore, in order to classify a whole new OCT volume as either from a non-cancerous or a glioma-infiltrated brain region, a threshold on the percentage of A-lines classified as from a glioma-infiltrated region in that volume was used. This threshold was determined by performing a receiver-operating characteristic curve (ROC) analysis following a Leave-One-Patient-Out-Cross-Validation (LOPOCV) classification performance estimation strategy with the OCT volumes of the training set.
Classification performance estimation
The overall machine learning classification computational scheme optimized with the training set was applied to the validation set. First, the same profiles P estimated from the training set were used to directly estimate the abundances of each of the A-lines in the validation set. Then, the resulting abundances A from the 3,020,800 A-lines in the validation set (295 volumes × 10 B-Scans/volume × 1024 A-lines/B-Scan, see Table 1) were used to classify each A-line as either from a non-cancerous or a glioma-infiltrated brain region using the same logistic regression classifier optimized with the training set. Finally, each of the 295 OCT volumes in the validation set were classified as either from a non-cancerous or a glioma-infiltrated brain region using the same threshold on the volume percentage of A-lines classified as from a glioma-infiltrated region, previously optimized with the training set. The classification performance obtained from the validation set was quantified in terms of overall classification accuracy, sensitivity and specificity.
RESULTS
OCT A-line model performance
The BEAE method was applied using a model order of N=3 (number of profiles) to the A-lines of the OCT volumes in the training set, as illustrated in (Figure 1.f). A detailed description of the method used to determine the optimal BEAE order value of N=3 is provided as Supplemental Material. All the 1940 B-scans analyzed are shown stacked up next to each other horizontally in the top panel. No clear distinction between A-lines from non-cancerous and glioma-infiltrated brain regions (separated by the red dashed line) can be observed from the stacked B-scans. The estimated abundances A of the 993,280 A-lines from the training set included in the BEAE analysis are shown in the middle panel. The three profiles P estimated from the training set, shown in the bottom panel, have complementary shapes and positive amplitude values as expected due to the positivity optimization constrain. To illustrate the capability of the BEAE method to model A-lines as a linear combination of the three estimated common profiles (P), sample A-lines and their model fits are shown in (Figure 1.g) (non-cancerous volume) and (Figure 1.h) (glioma-infiltrated volume). It can be observed that the model fits capture the shape of the A-line without overfitting the noise in the OCT signal.
Model-based OCT A-line feature extraction
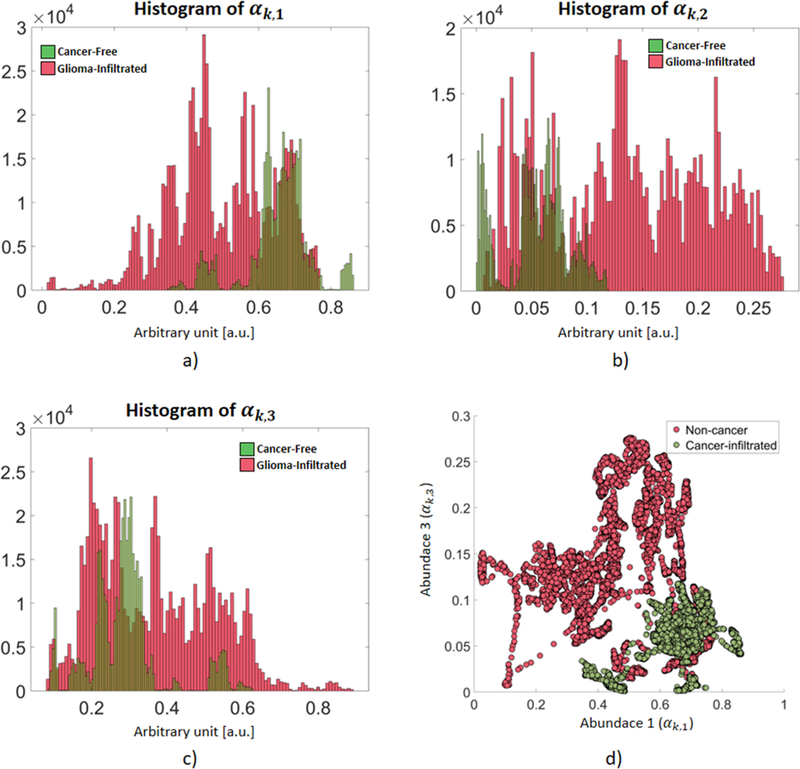
An important consequence of modeling any A-line yk as a linear combination of a set of commons profiles pn is the resulting unique representation of each A-line in terms of its abundances αk,n. Since these unique sets of abundances parameterize the unique shape of each A-line, they can be utilized as feature vectors within a machine learning classification algorithm. Since the abundances of each A-line add to 1, only (N-1) abundances are independent. Since the OCT training A-lines were modeled using three abundances (N=3), only two of them could be used as classification features. The distributions of each of the three abundances for the non-cancerous and glioma-infiltrated A-lines from the training set are shown in (Figure 2). It can be observed that the first abundance is distributed at lower values for the glioma-infiltrated A-lines (Figure 2.a), while the opposite can be observed for the third abundance (Figure 2.c). In addition, the distributions of the first and second abundances are roughly the reflection of each other, providing redundant information (Figure 2.a,b). Following an exhaustive feature selection approach, the first and third abundances (αk,1, αk,3) were chosen for the optimal feature vector xk = [αk,1, αk,3] used for training the logistic regression classifier aiming to identify an A-line as either from a non-cancerous or a glioma-infiltrated brain region. In Figure 2.d, the distributions of these feature vectors for the non-cancerous and glioma-infiltrated A-lines of the training set are shown in the two-dimensional space (αk,1, αk,3). A detailed description of the method used for feature selection is provided as Supplementary Material.
Figure 2 –
Distributions of the three abundances of the non-cancerous and glioma-infiltrated A-lines in the training set. a) The first abundance (αk,1) is distributed at lower values for the glioma-infiltrated A-lines. b) The distribution of the second abundance (αk,2) roughly mirrors that of the first abundance. c) The third abundance (αk,3) is distributed at higher values for the glioma-infiltrated A-lines. d) Distributions of the feature vectors xk = [αk,1, αk,3] for the non-cancerous and glioma-infiltrated A-lines (only 0.02 % of the total training set is shown for clarity).
Classifier training
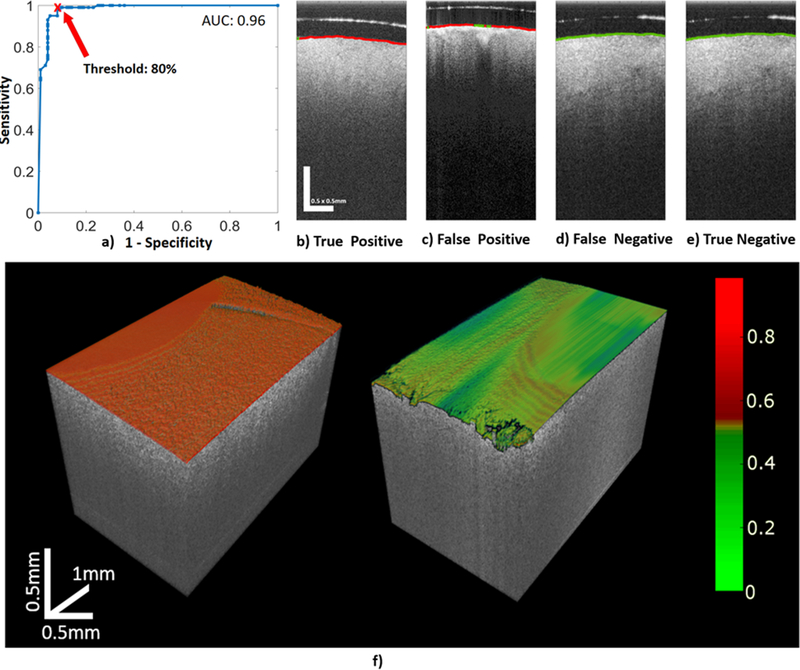
The logistic regression classifier was trained to classify any A-line as either from a non-cancerous or a glioma-infiltrated brain region using the 194 OCT volumes in the training set. To classify a whole new OCT volume as either non-cancerous or glioma-infiltrated, the threshold on the percentage of A-lines classified as from a glioma-infiltrated region in that volume was determined by applying ROC analysis following a LOPOCV performance estimation strategy in all the 194 OCT volumes of the training set. The corresponding area under the ROC curve (AUC) was AUC=0.96, which indicates a very promising classification performance. Since the clinical emphasis is to obtain maximal glioma-infiltrated tissue resection while preserving as much healthy tissue as possible, sensitivity for detecting glioma-infiltrated tissue was prioritized over specificity. A threshold of 80% was selected in order to maximize the sensitivity for detecting glioma-infiltrated region (99.15%) while maintaining as much non-cancerous tissue as possible (86.21%). The results of the ROC analysis are shown in (Figure 3.a).
Figure 3 –
a) Results of the ROC analysis performed on the training set to select the threshold in the volume percentage of A-lines classified as from a glioma-infiltrated region used to classify the whole volume as either from a non-cancerous or a glioma-infiltrated region. The AUC=0.96 indicates a very promising classification performance. A threshold of 80% was selected in order to maximize the sensitivity (99.15%) at the best specificity level possible (86.21%) for classifying volumes from a glioma-infiltrated region. Sample classified B-scans from the validation set, in which the surface was color-coded based on each A-line classification (red: glioma-infiltrated; green: non-cancerous): b) True Positive, c) False Positive, d) False Negative, and e) True Negative. f) Sample OCT 3D rendered images of brain regions in which the surface of the imaged brain tissue is color-coded using a colormap proportional to the estimated post-probability of each A-line being from a glioma-infiltrated brain region. For the sample OCT volume of a glioma-infiltrated brain region (left panel), 100 % of the A-lines were correctly classified as from glioma-infiltrated brain region. For the sample OCT volume of a non-cancerous brain region (right panel), 92 % of the A-lines were correctly classified as from non-cancerous brain region.
Classification performance estimation
The performance of the trained logistic regression classifier was estimated blindly on the totally independent validation set as follows. First, the abundances of each A-line in the validation set were estimated using the same profiles already estimated from the training set (Figure 1.f, bottom panel). The abundance estimation is computationally fast, due to the matrix operation approach, allowing computing a B-Scan (1024 A-lines) in 30 ms using MATLAB in a Core i7 4790K 4GHz processor. Once the abundances have been estimated, each A-line in the validation set was classified using the independently trained logistic regression classifier. After the A-line level classification, each OCT volume was finally classified as either from a non-cancerous or a glioma-infiltrated region using the previously selected threshold on the percentage of A-lines in that volume classified as from a glioma-infiltrated region. The results on the classification of the OCT volumes in the validation set are summarized in Table 2. The applied double-blinded validation indicated promising levels of sensitivity (>90%) and specificity (>80%) for discriminating low-grade and/or high-grade glioma-infiltrated tissue from non-cancerous tissue. Nevertheless, ~15% of all volumes were misclassified, probably due to significant intra-class variability and extra-class similarity observed among the OCT images, as illustrated in (Figure 3.b–e). It is worth noting that this validation set was completely independent (collected from 9 different patients) and the validation was blindly performed, meaning that the validation set was provided unlabeled for the described performance estimation procedure.
Table 2 –
Confusion matrix of the blind validation classification results. Left column: results for the classification of low-grade glioma-infiltrated vs. non-cancerous brain tissue. Middle column: results for the classification of high-grade glioma-infiltrated vs. non-cancerous brain tissue. Right column: results for the classification of low/high-grade glioma-infiltrated vs. non-cancerous brain tissue.
| Low-Grade vs. Non-cancerous | High-Grade vs. Non-cancerous | Low/High-Grade vs. Non-cancerous | ||||
|---|---|---|---|---|---|---|
| Predicted | Sensitivity 90.16% | Specificity 80.95% | Sensitivity 95.45% | Specificity 82.14% | Sensitivity 90.55% | Specificity 82.73% |
| 55 | 32 | 63 | 30 | 115 | 29 | |
| 6 | 136 | 3 | 138 | 12 | 139 | |
In order to demonstrate the potential of our computational framework for real-time accurate detection and volumetric visualization of cancerous and non-cancerous brain tissue, two unlabeled high-resolution OCT volumes (5×2×2.5 mm3; 256×2048×2048 pixels) from a non-cancerous and a glioma-infiltrated brain region were blindly processed with our trained computational framework. The computational speed for processing and classifying each A-line was >100,000 A-lines per second. In order to visualize the classification results, the OCT volumetric data was 3D rendered, and the surface of the imaged brain region was color-coded using a colormap proportional to the estimated post-probability of each A-line being from a glioma-infiltrated brain region (Figure 3.f). For the OCT volume of a glioma-infiltrated brain region (Figure 3.f, left panel), 100 % of the A-lines were correctly classified as from glioma-infiltrated brain region. For the OCT volume of a non-cancerous brain region (Figure 3.f, right panel), 92 % of the A-lines were correctly classified as from non-cancerous brain region.
DISCUSSION
Maximal tumor resection both improves the overall survival and delays cancer recurrence in low-grade and high-grade glioma patients [6, 41–43]. The limited ability of neurosurgeons to differentiate non-cancerous versus cancer-infiltrated brain tissues during resection surgery is the main challenge preventing higher rates of maximal tumor resection. Although several imaging techniques have been utilized routinely to assist brain cancer surgeries [7, 13, 29, 44, 45], there are significant limitations to these modalities. An effective image-guided tool for brain cancer resection surgery should be capable of providing high-resolution, accurate, continuous, and real-time in situ discrimination between non-cancerous and cancer-infiltrated brain tissue from intra-operative volumetric brain images.
Optical imaging modalities are well suited to enable such capabilities. Among them, intraoperative imaging of 5-ALA induced PpIX brain tissue fluorescence is perhaps the most extensively evaluated approach. Unfortunately, its performance for identifying glioma-infiltrated brain regions has not been fully demonstrated, with different studies reporting a wide range of sensitivity (50–100%) and specificity (70–100%) [46]. One major limitation of this approach is its dependency on the sufficient and specific 5-ALA uptake by the glioma tissue, which can be affected by many factors, including blood-brain barrier permeability, cellular/vascular proliferation and glioma grade [16, 47]. Another limitation is the lack of quantitative methods to image the 5-ALA induced PpIX fluorescence, which has prevented successfully moving from a subjective to a more objective and accurate interpretation of 5-ALA induced PpIX brain tissue fluorescence images [48].
OCT can be seemingly implemented as a hand-held surgical tool and/or integrated into standard surgical microscopes to provide label-free, high-resolution and fast volumetric tissue imaging. These capabilities and its relatively inexpensive implementation cost make OCT an ideal imaging modality to enable continuous real-time guidance during brain cancer resection surgery. However, for OCT to become an impactful image-guided tool for brain cancer resection surgery, computer-aided diagnostic (CAD) systems are needed to enable in situ intra-operative automated, objective and accurate detection of cancer-infiltrated brain tissue, as well as real-time volumetric visualization of cancerous and non-cancerous brain tissue during tumor resection surgery.
Previously reported approaches for OCT-based detection of cancer-infiltrated brain tissue rely on estimating the tissue optical attenuation coefficient from the OCT signal, which is used as a discriminative feature for identifying cancerous or cancer-infiltrated brain tissue [7, 34, 49]. For instance, Kut et al. recently introduced a computationally efficient method to estimate the optical attenuation coefficient of brain tissue from OCT scans, and demonstrated the potential of this estimated optical parameter to detect glioma-infiltrated brain tissue (sensitivity ~90%, specificity ~80%) using an independent validation set of 59 brain tissue samples [7].
One major limitation of all these OCT quantitative methods, however, is the need to spatially average neighboring OCT A-lines in order to attain signal-to-noise ratio levels that are adequate for estimating the tissue optical attenuation coefficient. For example, in the study by Kut et al., a B-scan of 1024 A-lines was divided in three adjacent regions, and all the ~341 A-lines in each region were averaged to estimate three optical attenuation coefficient values per B-scan; thus, only three regions per B-scan could be classified using their approach [7]. In contrast, A-line averaging is not needed in our method; thus, each A-line can be classified, resulting in an improvement in spatial resolution of ~341:1 compared to the method by Kut et al. [7]. Moreover, since the spatial resolution of the classification map provided by our method is equal to the lateral optical resolution of the OCT instrument used (16.0 μm for this study), the resolution of the classification map can be as good as the best optical resolution possible with the available state-of-the-art OCT instrumentation technology. The demonstrated superior spatial resolution enabled by our method is particularly relevant for the accurate detection of glioma-infiltrated brain tissue, which is characterized by showing a wide range in the degree of cancer infiltration at the tumor margins.
Furthermore, the previously reported approaches for OCT-based detection of cancer-infiltrated brain tissue, including the method by Kut et al. [7], is the need to perform calibration procedures which could be cumbersome. In comparison, our method reported here only requires a training set of labeled OCT brain tissue scans obtained with the same or a similar OCT instrument to estimate the depth profiles pn used to model each A-line and train the logistic regression classifier.
The classification performance of our computational framework for detecting glioma-infiltrated brain tissue (sensitivity: >90%; specificity: >82%) was quantified following a robust and unbiased double-blinded validation strategy using an independent validation set of 295 brain tissue samples. Moreover, the methods adopted at each stage of our computational framework (preprocessing, feature extraction, classification) were also strategically chosen and designed to enable real-time processing of OCT volumetric images. Thus, another relevant feature of our computational framework is its high processing speed (100,020 A-lines per second, using MATLAB in a Core i7 4790K 4GHz processor), which would enable processing an arbitrary tissue volume of 5×5×2.5 mm3 (256×256×2048 pixels) in ~0.7 seconds. It should be noted that the processing speed of this novel computational framework can be significantly increased by implementing it using object-oriented programming languages and parallel programing and computing. Altogether, the demonstrated classification accuracy and processing speed of our computational framework, once embedded within intra-operative OCT imaging instruments, would enable developing clinically relevant CAD systems for automated, accurate, real-time in situ detection of glioma infiltration during tumor resection surgery.
Study limitations
Although the results of this preliminary study are quite encouraging, it still has some limitations. First, our computational framework showed a misclassification rate of ~15%, in part due to the noticed intra-class variability and extra-class similarity observed among the OCT images (Figure 3.b–e), which might indicate the need for using additional OCT features and/or more sophisticated classification methods. Furthermore, from the computational point of view, noisy labeling is expected in medical imaging data, and in our OCT brain tissue datasets in particular due to the heterogeneity of the glioma tissue samples. To overcome this problem, the use of classification methods with higher tolerance to noisy labels (e.g. weakly supervised learning models) might improve the performance of the model. Moreover, the current OCT databases, when analyzed at the A-line level, could be sufficiently large to allow exploring state-of-the-art classifiers such as those based on Convolutional Neural Networks (CNN), which might also improve the detection of glioma-infiltrated brain tissue.
In addition, it can be quite possible that a classification model designed and trained to detect infiltration of a specific tumor type and grade could outperform a more general model designed and trained to detect infiltration of a plurality of brain tumor types and/or grades. To investigate this alternative approach, two additional classification models were trained using either the available Low-Grade or High-Grade glioma-infiltrated samples, and their results are reported as Supplementary Material. Compared to the original more general classification model, these more targeted classifiers performed with lower sensitivity but better specificity. These observations suggest that once more comprehensive databases become available, more specific classification models can be developed and compared against more general ones to determine an optimal approach.
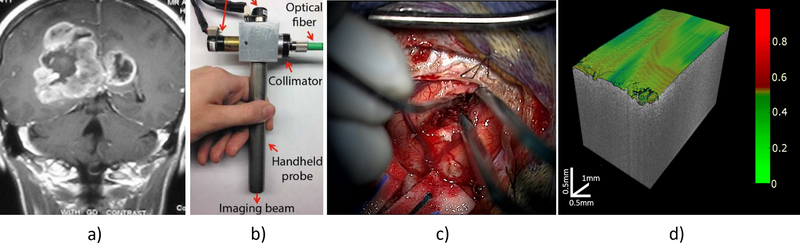
Another limitation is that the OCT datasets used for training and validating our computational framework were acquired from freshly resected non-cancerous, Low-Grade glioma-infiltrated and High-Grade glioma-infiltrated brain tissue samples from glioma patients undergoing tumor resection surgery. Since the training and validation datasets used in this study cannot be considered representative of the “universe” of tumor, the training of our computational framework needs to be repeated using more comprehensive databases of in vivo OCT volumes that include other tumor types. Once optimized, the computational framework would be embedded into a intraoperative OCT imaging instrument, and the performance of the resulting OCT-guided surgical tool for automated real-time in situ intraoperative detection of brain tumor infiltration will have to be quantified in a prospective clinical study, as depicted in (Figure 4).
Figure 4 –
Methodology for an OCT-based CAD system. a) After a preoperative MRI-imaging for evaluation, we will use a b) OCT probe for c) intraoperatively image guidance during surgery. Finally, after preprocessing, feature extraction and classification, e) the automated real-time classification using a volumetric color-coded map of the tissue will be displayed for real-time guidance.
Conclusion
In conclusion, we have introduced a novel computational method for OCT-based automated detection of glioma-infiltrated from non-cancerous brain tissue. Our method applies a modeling approach to parametrize the information encoded in the shape of each depth-dependent OCT intensity signal (A-line) and uses the A-line model parameters as features within a machine learning classification scheme. Since our method can process OCT images at their original high spatial resolution and does not require performing calibration procedures using tissue phantoms, it overcomes major challenges of previously reported methods. Due to its demonstrated detection accuracy, robustness and low computational cost, this method could enable developing faster, and more accurate OCT-guided surgical tools for continuous, real-time and accurate in situ intra-operative detection of any stage glioma infiltration, facilitating extensive glioma resection and improved surgical outcomes for glioma patients.
Supplementary Material
Translational Relevance:
Maximal tumor resection improves overall survival and delays cancer recurrence in glioma patients; however, the margins of highly infiltrating gliomas are often very difficult to delineate during glioma resection surgery. Various medical imaging modalities are used pre- and/or intra-operatively to assist in the delineation of glioma margins. Unfortunately, none of these technologies can provide quantitative, real-time, accurate and continuous guidance during glioma resection surgery. Optical Coherence Tomography (OCT) is a non-invasive, label-free, real-time, high-resolution volumetric imaging modality. Previous computational methods for OCT-based detection of glioma infiltration require sacrificing significantly spatial resolution and performing cumbersome calibration procedures. We have developed and validated an alternative accurate and fast artificial intelligence (AI) assisted computational method that overcomes these major limitations. Our method can be implemented within generic OCT instruments to enable real-time, high-resolution, automated, accurate, in situ, intraoperative detection of glioma infiltration, facilitating maximal tumor resection and improved surgical outcomes for glioma patients.
ACKNOWLEDGEMENTS
This research was partially supported by grants from the National Institutes of Health (grants R01CA218739, R01CA200399), the Cancer Prevention and Research Institute of Texas (grant RP180588), the Coulter H. Wallace Foundation, NSF-NCSA XSEDE ASC170017, FONDECYT-CONCYTEC Fellowship. AQH acknowledged the support by the William J. and Charles H. Mayo Professorship and a Mayo Clinician Investigator award. KLC acknowledged the support by the Mayo RACER award.
REFERENCES
- 1.Almeida JP, et al. , The value of extent of resection of glioblastomas: clinical evidence and current approach. Curr Neurol Neurosci Rep, 2015. 15(2): p. 517. [DOI] [PubMed] [Google Scholar]
- 2.Marko NF, et al. , Extent of resection of glioblastoma revisited: personalized survival modeling facilitates more accurate survival prediction and supports a maximum-safe-resection approach to surgery. J Clin Oncol, 2014. 32(8): p. 774–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaichana KL, et al. , Multiple resections for patients with glioblastoma: prolonging survival. J Neurosurg, 2013. 118(4): p. 812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lara-Velazquez M, et al. , Advances in Brain Tumor Surgery for Glioblastoma in Adults. Brain Sci, 2017. 7(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eseonu CI, et al. , Comparative volumetric analysis of the extent of resection of molecularly and histologically distinct low grade gliomas and its role on survival. J Neurooncol, 2017. 134(1): p. 65–74. [DOI] [PubMed] [Google Scholar]
- 6.Chaichana KL, et al. , When Gross Total Resection of a Glioblastoma Is Possible, How Much Resection Should Be Achieved? World Neurosurgery, 2014. 82(1–2): p. E257–E265. [DOI] [PubMed] [Google Scholar]
- 7.Kut C, et al. , Detection of human brain cancer infiltration ex vivo and in vivo using quantitative optical coherence tomography. Sci Transl Med, 2015. 7(292): p. 292ra100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGirt MJ, et al. , Association of surgically acquired motor and language deficits on overall survival after resection of glioblastoma multiforme. Neurosurgery, 2009. 65(3): p. 463–9; discussion 469–70. [DOI] [PubMed] [Google Scholar]
- 9.Chaichana KL, et al. , The butterfly effect on glioblastoma: is volumetric extent of resection more effective than biopsy for these tumors? J Neurooncol, 2014. 120(3): p. 625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman M, et al. , The effects of new or worsened postoperative neurological deficits on survival of patients with glioblastoma. J Neurosurg, 2017. 127(1): p. 123–131. [DOI] [PubMed] [Google Scholar]
- 11.Mehranian A, Arabi H, and Zaidi H, Quantitative analysis of MRI-guided attenuation correction techniques in time-of-flight brain PET/MRI. Neuroimage, 2016. 130: p. 123–33. [DOI] [PubMed] [Google Scholar]
- 12.Spivak CJ and Pirouzmand F, Comparison of the reliability of brain lesion localization when using traditional and stereotactic image-guided techniques: a prospective study. J Neurosurg, 2005. 103(3): p. 424–7. [DOI] [PubMed] [Google Scholar]
- 13.Rygh OM, et al. , Comparison of navigated 3D ultrasound findings with histopathology in subsequent phases of glioblastoma resection. Acta Neurochir (Wien), 2008. 150(10): p. 1033–41; discussion 1042. [DOI] [PubMed] [Google Scholar]
- 14.Valdes PA, et al. , delta-aminolevulinic acid-induced protoporphyrin IX concentration correlates with histopathologic markers of malignancy in human gliomas: the need for quantitative fluorescence-guided resection to identify regions of increasing malignancy. Neuro Oncol, 2011. 13(8): p. 846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ando T, et al. , Precise comparison of protoporphyrin IX fluorescence spectra with pathological results for brain tumor tissue identification. Brain Tumor Pathol, 2011. 28(1): p. 43–51. [DOI] [PubMed] [Google Scholar]
- 16.Montcel B, et al. , Two-peaked 5-ALA-induced PpIX fluorescence emission spectrum distinguishes glioblastomas from low grade gliomas and infiltrative component of glioblastomas. Biomed Opt Express, 2013. 4(4): p. 548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tashibu K, Analysis of water content in rat brain using Raman spectroscopy. No To Shinkei, 1990. 42(10): p. 999–1004. [PubMed] [Google Scholar]
- 18.Koljenovic S, et al. , Discriminating vital tumor from necrotic tissue in human glioblastoma tissue samples by Raman spectroscopy. Lab Invest, 2002. 82(10): p. 1265–77. [DOI] [PubMed] [Google Scholar]
- 19.Kalkanis SN, et al. , Raman spectroscopy to distinguish grey matter, necrosis, and glioblastoma multiforme in frozen tissue sections. J Neurooncol, 2014. 116(3): p. 477–85. [DOI] [PubMed] [Google Scholar]
- 20.Kast R, et al. , Identification of regions of normal grey matter and white matter from pathologic glioblastoma and necrosis in frozen sections using Raman imaging. J Neurooncol, 2015. 125(2): p. 287–95. [DOI] [PubMed] [Google Scholar]
- 21.Kast RE, et al. , Raman molecular imaging of brain frozen tissue sections. J Neurooncol, 2014. 120(1): p. 55–62. [DOI] [PubMed] [Google Scholar]
- 22.Ji M, et al. , Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med, 2013. 5(201): p. 201ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desroches J, et al. , A new method using Raman spectroscopy for in vivo targeted brain cancer tissue biopsy. Scientific Reports, 2018. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, et al. , Accuracy of Raman spectroscopy in differentiating brain tumor from normal brain tissue. Oncotarget, 2017. 8(22): p. 36824–36831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans CL, et al. , Chemically-selective imaging of brain structures with CARS microscopy. Opt Express, 2007. 15(19): p. 12076–87. [DOI] [PubMed] [Google Scholar]
- 26.Jermyn M, et al. , Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci Transl Med, 2015. 7(274): p. 274ra19. [DOI] [PubMed] [Google Scholar]
- 27.Hollon TC, et al. , Rapid Intraoperative Diagnosis of Pediatric Brain Tumors Using Stimulated Raman Histology. Cancer Res, 2018. 78(1): p. 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, et al. , Ex vivo and in vivo imaging of myelin fibers in mouse brain by coherent anti-Stokes Raman scattering microscopy. Opt Express, 2008. 16(24): p. 19396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollon T, et al. , Improving the accuracy of brain tumor surgery via Raman-based technology. Neurosurg Focus, 2016. 40(3): p. E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujimoto JG, et al. , Optical coherence tomography: an emerging technology for biomedical imaging and optical biopsy. Neoplasia, 2000. 2(1–2): p. 9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boppart SA, et al. , Optical coherence tomography for neurosurgical imaging of human intracortical melanoma. Neurosurgery, 1998. 43(4): p. 834–41. [DOI] [PubMed] [Google Scholar]
- 32.Chong SP, et al. , Noninvasive, in vivo imaging of subcortical mouse brain regions with 1.7 mum optical coherence tomography. Opt Lett, 2015. 40(21): p. 4911–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohringer HJ, et al. , Imaging of human brain tumor tissue by near-infrared laser coherence tomography. Acta Neurochir (Wien), 2009. 151(5): p. 507–17; discussion 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kut C, et al. Real-Time, Label-Free Optical Property Mapping for Detecting Glioma Invasion with SSOCT for Potential Guidance of Surgical Intervention in Accepted for Oral Presentation at: 2015 SPIE Annual Meeting. 2015. San Diego, CA. [Google Scholar]
- 35.Bohringer HJ, et al. , Time-domain and spectral-domain optical coherence tomography in the analysis of brain tumor tissue. Lasers Surg Med, 2006. 38(6): p. 588–97. [DOI] [PubMed] [Google Scholar]
- 36.Bizheva K, et al. , Imaging ex vivo healthy and pathological human brain tissue with ultra-high-resolution optical coherence tomography. J Biomed Opt, 2005. 10(1): p. 11006. [DOI] [PubMed] [Google Scholar]
- 37.Canny J, A computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell, 1986. 8(6): p. 679–98. [PubMed] [Google Scholar]
- 38.Gutierrez-Navarro O, et al. , Blind end-member and abundance extraction for multispectral fluorescence lifetime imaging microscopy data. IEEE J Biomed Health Inform, 2014. 18(2): p. 606–17. [DOI] [PubMed] [Google Scholar]
- 39.Rico-Jimenez JJ, et al. , Automatic classification of atherosclerotic plaques imaged with intravascular OCT. Biomed Opt Express, 2016. 7(10): p. 4069–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang HH, Xu T, and Yang J, Comparing logistic regression, support vector machines, and permanental classification methods in predicting hypertension. BMC Proc, 2014. 8(Suppl 1): p. S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickel K, et al. , The patients’ view: impact of the extent of resection, intraoperative imaging, and awake surgery on health-related quality of life in high-grade glioma patients-results of a multicenter cross-sectional study. Neurosurg Rev, 2017. [DOI] [PubMed] [Google Scholar]
- 42.Chaichana KL, et al. , Establishing percent resection and residual volume thresholds affecting survival and recurrence for patients with newly diagnosed intracranial glioblastoma. Neuro Oncol, 2014. 16(1): p. 113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaichana KL, et al. , Supratentorial glioblastoma multiforme: the role of surgical resection versus biopsy among older patients. Ann Surg Oncol, 2011. 18(1): p. 239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrecca K, et al. , Failure pattern following complete resection plus radiotherapy and temozolomide is at the resection margin in patients with glioblastoma. J Neurooncol, 2013. 111(1): p. 19–23. [DOI] [PubMed] [Google Scholar]
- 45.Coburger J, et al. , Histopathological Insights on Imaging Results of Intraoperative Magnetic Resonance Imaging, 5-Aminolevulinic Acid, and Intraoperative Ultrasound in Glioblastoma Surgery. Neurosurgery, 2017. [DOI] [PubMed] [Google Scholar]
- 46.Mansouri A, et al. , The role of 5-aminolevulinic acid in enhancing surgery for high-grade glioma, its current boundaries, and future perspectives: A systematic review. Cancer, 2016. 122(16): p. 2469–78. [DOI] [PubMed] [Google Scholar]
- 47.Utsuki S, et al. , Possibility of using laser spectroscopy for the intraoperative detection of nonfluorescing brain tumors and the boundaries of brain tumor infiltrates - Technical note. Journal of Neurosurgery, 2006. 104(4): p. 618–620. [DOI] [PubMed] [Google Scholar]
- 48.Cordova JS, et al. , Semi-Automated Volumetric and Morphological Assessment of Glioblastoma Resection with Fluorescence-Guided Surgery. Mol Imaging Biol, 2016. 18(3): p. 454–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan W, et al. , Robust and fast characterization of OCT-based optical attenuation using a novel frequency-domain algorithm for brain cancer detection. Sci Rep, 2017. 7: p. 44909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.