Abstract
ARID1A, encoding a subunit of the SWI/SNF complex, is the most frequently mutated epigenetic regulator in human cancers and is mutated in over 50% of ovarian clear cell carcinoma (OCCC), a disease that currently has no effective therapy. Inhibition of histone deacetylase 6 (HDAC6) suppresses the growth of ARID1A-mutated tumors and modulates tumor immune microenvironment. Here we show that inhibition of HDAC6 synergizes with anti-PD-L1 immune-checkpoint blockade in ARID1A-inactivated ovarian cancer. ARID1A directly repressed transcription of CD274, the gene encoding PD-L1. Reduced tumor burden and improved survival was observed in ARID1Aflox/flox/PIK3CAH1047R OCCC mice treated with the HDAC6 inhibitor ACY1215 and anti-PD-L1 immune-checkpoint blockade as a result of activation and increased presence of interferon-gamma positive CD8 T cells. We confirmed that the combined treatment limited tumor progression in a cytotoxic T-cell-dependent manner as depletion of CD8+ T cells abrogated these antitumor effects. Together, these findings indicate that combined HDAC6 inhibition and immune-checkpoint blockade represents a potential treatment strategy for ARID1A-mutated cancers.
Introduction
ARID1A encodes a subunit of the SWI/SNF chromatin-remodeling complex and functions as a tumor suppressor (1). SWI/SNF complexes are multi-subunit complexes that remodel chromatin in an ATP-dependent manner (1). In addition to core subunits such as SNF5 that are present in all SWI/SNF complexes, other subunits are only present in certain complexes. For example, the mutually exclusive ARID1A and ARID1B subunits are only associated with BRG1-associated factor (BAF) complexes, while ARID2, PBRM1 and BRD7 subunits are specific for polybromo BAF (PBAF) complexes (1). The ARID1A containing SWI/SNF complex epigenetically activates or represses gene expression via controlling gene accessibility (1,2).
ARID1A is amongst the most frequently mutated genes in human cancer (3). In addition to inactivating mutations, ARID1A shows deletions in many tumor types in the cBioPortal datasets. Notably, inactivating mutations in ARID1A occur frequently in ovarian clear cell carcinomas (OCCC; >50%) (4). Over 90% of ARID1A mutation in OCCCs are either frameshift or nonsense that led to loss of ARID1A protein expression (4). There is an unmet need for effective treatment modalities for ARID1A-mutated OCCCs. OCCC is generally refractory to standard agents used to treat ovarian cancers, and when diagnosed in advanced stages, OCCC carries the worst prognosis of all ovarian cancer histosubtypes (5).
Emerging evidence supports the idea that the SWI/SNF complexes play a critical role in tumor immunity (2). For example, in the SWI/SNF catalytic subunit SMARCA4-mutated small cell carcinoma of the ovary, hypercalcemic type (SCCOHT), PD-L1 is expressed in both tumor and stromal cells, and strong T cell infiltration was observed in the majority of tumors (6). Emerging clinical evidence suggests that checkpoint blockades such as anti-PD1 are effective in SCCOHTs (6). In addition, in clear cell renal cell cancer (ccRCC), patients who responded positively to anti-PD1/anti-PD-L1 therapy often carry a loss of function mutation in the PBRM1 subunit of the PBAF complex (7). Likewise, inactivation of the PBAF subunits BRD7, ARID2, and PBRM1 confers susceptibility to T cell-mediated killing in melanoma (8). Finally, ARID1A mutation correlates with an increase of PD-L1 expression (9). However, the mechanism by which ARID1A regulates PD-L1 expression remains not fully understood. Notably, published literature show that anti-PD-L1 treatment only has a modest effect on improving the survival of mice bearing ARID1A-inactivated tumors (9). This suggests that checkpoint blockade-based combination therapeutic strategies are necessary for treating ARID1A-mutated cancers.
The tumor microenvironment contains a variety of immune modulating cells such as T lymphocytes (10,11). These cells play a critical role in shaping the immune response against tumors. Therefore, agents that promote functional changes in T cells may alter immune microenvironment to affect tumor progression (11). HDAC6 inhibition suppresses the growth of ARID1A-mutated cancer (12). Although most translational studies on HDAC6 inhibitors have focused on their effects on tumor cells, emerging evidence suggests that HDAC6 inhibitors have immunomodulatory effects on tumor immune microenvironment (13-15). Notably, the complete genetic knockout of HDAC6 does not impair normal cell function (16). Consistently, the clinically applicable HDAC6 inhibitor ACY1215 was proven safe (17).
Here we show that ARID1A represses CD274 (encoding PD-L1) gene and HDAC6 inhibition synergizes with anti-PD-L1 in ARID1A-inactivated ovarian cancer. The combination depends on cytotoxic T cell activity to limit tumor progression in vivo. Our findings establish that HDAC6 inhibition and immune-checkpoint blockade combination represents a treatment strategy for ARID1A-mutated cancers.
Materials and Methods
Cell lines and culture conditions
The ovarian clear cell carcinoma cell line OVCA429 and mouse ovarian ID8-Defb29/Vegf cancer cells were cultured in RPMI 1640 (Corning Life Sciences) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) and 1% penicillin/streptomycin. The ovarian clear cell carcinoma cell line RMG1 was cultured in 1:1 Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Corning Life Sciences) supplemented with 10% FBS and 1% penicillin/streptomycin. RMG1 cells were obtained from the Japanese Collection of Research Bioresources in 2015. OVCA429 cells were obtained from I.M. Shih in 2015. ID8-Defb29/Vegf cells were obtained from J. R. Conejo-Garcia in 2015. All cells were used within 10 passages. The viral packaging Phoenix and 293FT cells were cultured in DMEM (Corning Life Sciences) supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were incubated at 37°C in humidified atmosphere containing 5% CO2. Cell lines were authenticated at the Wistar Institute Genomics Facility using short tandem repeat profiling using AmpFLSTR Identifiler PCR Amplification kit (Life Technologies) right before experiments. Mycoplasma infection was monthly tested with LookOut Mycoplasma PCR detection (Sigma-Aldrich) right before experiments.
Quantitative reverse-transcriptase PCR (qRT-PCR)
Total RNA was extracted using TRIzol (Invitrogen), and then purified with DNase treatment (Qiagen). RNA expression was determined using the QuantStudio 3 Real-Time PCR System (Thermo Fisher) and iTaq Universal SYBR Green One-step kit (Bio-Rad Laboratories). The primers sequences are as following: human CD274 (forward, 5’-ATGGTGGTGCCGACTACAA-3’; reverse, 5’-TCCAGATGACTTCGGCCTT-3’), mouse Cd274 (forward, 5’-GCCACTTCTGAGCATGAACTA-3’; reverse, 5’-GACACTTCTCTTCCCACTCAC-3’). The expression was normalized using human 18s (forward, 5’-AACTTTCGATGGTAGTCGCCG-3’; reverse, 5’-CCTTGGATGTGGTAGCCGTTT-3’) expression, or mouse Ppia (forward, 5’-GGGTTCCTCCTTTCACAGAA-3’; reverse, 5’-GATGCCAGGACCTGTATGCT-3’).
Reagents and antibodies
ACY1215 (Cat. No: S8001) was purchased from Selleckchem. Anti-PD-L1 (Cat. No: BE0101, clone: 10F.9G2) and anti-mouse CD8 (Cat. No: BE0117, clone: YTS 169.4) antibodies were purchased from Bio X Cell. Interferon-gamma was purchased from Thermo Fisher (Cat. No: PHC4031) or from ProSpec (Cat. No: CYT-358). The following antibodies were purchased from the indicated suppliers: rabbit anti-ARID1A (Cell Signaling, Cat. No: 12354, 1:1000), rabbit anti-ARID1B (Cell Signaling, Cat. No: 92964, 1:1000), mouse anti-ARID1B (Santa Cruz, Cat. No: sc-32762, 1:1000), mouse anti-β-actin (Sigma-Aldrich, Cat. No: A5441, 1:10,000), mouse anti-FLAG (Sigma-Aldrick, Cat. No: F1804), rabbit anti-PD-L1 (Cell Signaling, Cat. No: 13684S, 1:1000, Abcam, Cat. No: ab213480, 1:1000). For flow cytometric analysis, APC/CY7 anti-CD69 (Cat. No: 104525), BV711 anti-CD3 (Cat. No: 100349), APC anti-CD4 (Cat. No: 100516), PE anti-CD8 (Cat. No: 100708), FITC anti-Granzyme B (Cat. No: 372206), PE/Cy7 anti-interferon-gamma (Cat. No: 505825) antibodies were purchased from Biolegend and used at 1:150 dilutions. Anti-FOXP3 antibody (Cat. No: 563902, 1:150) was purchased from BD Biosciences. Zombie yellow dye (Biolegend, Cat. No: 423103, 1:200) was used as a viability staining.
Immunoblotting
Protein was isolated using RIPA buffer (50mM Tris pH 8.0, 150mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate and 1mM PMSF). The concentrations of protein samples were measured using Bradford assay. Protein samples were separated by SDS-PAGE gel and transferred to polyvinylidene fluoride membrane (Millipore). Membranes were blocked in TBS/0.1% Tween 20 with 5% non-fat milk (Bio-Rad), and then incubated sequentially with primary and secondary antibodies.
Generation of endogenously FLAG-tagged ARID1A and ARID1A knockout (ARID1A KO) cells by CRISPR
To generate endogenously FLAG-tagged ARID1A, PX458 (Addgene #48138) and pFETCh-donor (Addgene #63934) constructs were obtained from Addgene. Guide RNA sequence (5’-TGTCCCACGGCTGTCATGAC-3’) targeting terminal codon of ARID1A was inserted into PX458. About 500 bps homologous arms at both sides of guide RNA targeting site were cloned and inserted into pFETCh-donor. ARID1A endogenously tagged clones were isolated after 200 μg/ml G418 selection and validated by immunoblot.
ARID1A KO cells were generated and validated as previously reported (12). For all the controls, parental cells transduced with empty vector packaged virus were used. Briefly, OVCA429 cells were transfected with pSpCas9 (BB)-2A-Puro (PX459) (Addgene, Cat. No: 62988) inserting the ARID1A guide RNA (5’-CGGGTTGCCCAGGCTGCTGGCGG-3’). ID8-Defb29/Vegf cells were transfected with lentiCRISPR v2 (Addgene, Cat. No: 52961) inserting the Arid1a guide RNA (5’-CACCGTCTCCGCGGACGAGACAGCG-3’). Lipofectamine 2000 was used following the manufacturer’s specifications, then selection using 1μg/ml puromycin was performed. Fugene6 transfection reagent (Promega) was used following manufacturer’s specifications, then selection using 1μg/ml puromycin was performed. Clonal populations for ARID1A knockout were screened using immunoblotting.
Flow cytometry
PD-L1 expression on the cell surface was analyzed as we previously described (18). Briefly, cells were harvested and washed in PBS. Cells were then centrifuged and incubated in 100 μl FACS buffer (PBS with 3%FBS) with 1:100 diluted PE anti-human CD274 (BD Bioscience, Cat. No: 557924, clone: MIH1) or APC anti-mouse Cd274 (Biolegend, Cat. No: 124311, clone: 10F.9G2) for 40 min on ice. Cells were then stained with 100 μl FACS buffer with 1:5 diluted 7AAD (BD Pharmingen Cat. No: 51-68981E) for 10 min on ice. Cells were washed with PBS, suspended in 400μl PBS, and then submitted for analysis. At least 20,000 events were collected on flow cytometry, and then the data was analyzed with FlowJo version 7 software module. An isotype-matched immunoglobulin G (IgG) was used as a negative control.
Tumor cells were extracted using enzymatic cocktail from mouse tumor dissociation kit (Miltenyi Biotec, Cat. No: 130-096-730) according to manufacturer’s instructions. After dissociation, cells were mashed through a 70μM cell strainer and used for flow cytometric analysis. For peritoneal wash, peritoneal cavity of mice was washed three times with 5ml PBS and incubated in red blood cell lysis buffer before proceeding to staining.
Intracellular interferon-gamma (IFNγ) staining was performed by culturing tumor cell suspensions or peritoneal washes with protein transport inhibitor (BD Biosciences, Cat. No: 554724) and stimulated with phorbol 12-myristate 13-acetate (Sigma, Cat. No: P8139, 0,5 μg/ml) and ionomycin (Sigma, Cat. No: I0634, 1μg/ml) for 24 hours followed by surface and viability staining. Cells were fixed and permeabilized using Cytofix/Cytoperm kit (BD Biosciences, Cat. No: 554714) according to manufacturer’s instructions followed by intracellular staining.
Chromatin immunoprecipitation (ChIP) and CUT & RUN analysis
For ChIP analysis, cells were crosslinked with 1% formaldehyde, and the reaction was quenched by 125 mM glycine. Fixed cells were lysated using lysis buffer 1 (50 mM HEPES-KOH, pH7.5, 140 mM NaCl, 1 mM EDTA, pH8.0, 1% Triton X-100, 0.1% DOC) and lysis buffer 2 (10 mM Tris pH8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). Samples were digested with MNase in digestion buffer (10 mM Tris 8.0, 1 mM CaCl2, 0.2% Triton X-100) and the nucleus was broken down using one pulse of bioruptor with high output. Chromatin was incubated with antibodies overnight, and then protein A+G Dynabeads were added. After 1.5 hours incubation, chromatin was eluted, treated with proteinase K and then purified with a Gel extraction kit (Qiagen, Cat. No: 28706). The following antibodies were used to perform ChIP: rabbit anti-SNF5 (Bethyl, Cat. No: A301-087A), mouse anti-ARID1B (Abgent, Cat. No: AT1190a), rabbit anti-ARID1B (Cell Signaling, Cat. No: 92964), rabbit anti-H3K4me3 (Active Motif, Cat. No: 39159), mouse anti-RNA polymerase II (Santa Cruz, Cat. No: sc-47701) and mouse anti-FLAG (Sigma-Aldrick, Cat. No: F1804). An isotype-matched IgG was used as a negative control. ChIP DNA samples were analyzed by quantitative PCR against the promoter of the human CD274 gene (forward, 5’-GCCGATTTCACCGAAGGTC-3’; reverse, 5’-CAGCTGCTCAGCGTTGC-3’) or of the mouse Cd274 gene (forward, 5’-GCCACTTCTGAGCATGAACTA-3’; reverse, 5’-GACACTTCTCTTCCCACTCAC-3’).
CUT & RUN for ARID1A’s association with the CD274 promoter analysis was performed as we previously described (19). Briefly, cells were washed with Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine and a Roche complete EDTA-free tablet (Sigma-Aldrich) per 50 ml) and centrifuged. Cell pellets were incubated in Antibody Buffer (antibody with 1:100 dilution in wash buffer supplemented with 0.05% digitonin and 2mM EDTA) for 2 hours. After centrifugation, cell pellets were washed with Dig-Wash Buffer (Wash Buffer with 0.05% digitonin), and incubated in Protein A-MNase at a final concentration of 700 ng/ml in Dig-Wash Buffer for 1 hour. Cells were suspended in 100 μl Dig-Wash Buffer, and then mixed with 2 μl 100 mM CaCl2. After 30 min incubation, reactions were stopped using STOP buffer (340 mM NaCl, 20 mM EDTA pH8, 4 mM EGTA, 0.05% digitonin, 50 μg/ml RNase A, 50 μg/ml glycogen), then centrifuged. The supernatant containing DNA was collected and purified using phenol–chloroform–isoamyl alcohol and chloroform extraction and ethanol precipitation. Rabbit anti-ARID1A (Abcam, Cat. No. ab182560) was used to perform CUT&RUN. An isotype-matched IgG was used as a negative control. CUT&RUN DNA samples were analyzed by quantitative PCR against the promoter of the human CD274 gene (forward, 5’-GCCGATTTCACCGAAGGTC-3’; reverse, 5’-CAGCTGCTCAGCGTTGC-3’) or of the mouse Cd274 gene (forward, 5’-GCCACTTCTGAGCATGAACTA-3’; reverse, 5’-GACACTTCTCTTCCCACTCAC-3’).
Arid1a−/−/Pik3caH1047R genetic ovarian clear cell ovarian carcinoma (OCCC) mouse model
All experimental protocols were approved by the Wistar Institutional Animal Care and Use Committee (IACUC). The transgenic mice were generated as we previously described (12). Briefly, Arid1aflox/flox mice (kindly provided by Dr. Wang, U. Michigan) were crossed with R26-PikcaH1047R mice (Jackson Laboratory, Jax No: 016977). Intrabursal adenovirus -Cre injection was used to induce ovarian clear cell carcinoma formation. Mice were randomized into four groups four weeks after adeno-Cre injection and treated with vehicle control (isotype control IgG and 2% DMSO/30% PEG 300/ddH2O), ACY1215 (50 mg/kg, daily), anti-PD-L1 antibody (10mg/kg, twice a week) or a combination for 21 days. ACY1215 was suspended in 2% DMSO/30% PEG 300/ddH2O, and anti-PD-L1 antibody was suspended in PBS. At the end of treatments, mice were euthanized and tumors were surgically dissected, or followed for survival analysis. Tumor burden was calculated based on tumor weight. The Wistar Institute IACUC guideline was followed in determining the time for ending the survival experiments (tumor burden exceeds 10% of body weight).
In Vivo CD8 T cell Depletion
An anti-CD8 antibody (BioXCell, Cat. No: BE0117, clone: YTS 169.4, 10 mg/kg) was used to deplete CD8+ T cells. An isotype matched IgG (Bio X Cell, Cat. No: BE0090, clone: LTF-2, 10 mg/kg) was used as a negative control. Antibodies were administered 3 days before starting the combination treatment and then twice a week until completion of the study. The depletion was confirmed by flow cytometry analysis of blood cells collected via the retro-orbital vein.
Statistical analysis and reproducibility
Statistical analysis was conducted using GraphPad Prism 6 software (GraphPad). Experiments were performed in three independent experiments unless otherwise stated, and representative results were shown. Quantitative data are expressed as mean ± S.E.M. unless otherwise stated. To improve data normality and homogeneity of variance, some data (e.g., tumor weight, ascites, and % of specific cell counts) were log-transformed before statistical test. A two-tailed t-test was conducted for two group comparison. ANOVA with post-hoc Tukey’s multiple comparisons test was used for experiment with 4 independent groups. For experiment with 8 independent groups, ANOVA with post hoc multiple t-tests and Benjamini and Hochberg adjusted P-values were reported. P < 0.05 was considered significant.
Data availability
Previously published ARID1A ChIP-seq in ARID1A wildtype RMG1 cells are available at the Gene Expression Ominibus (GEO) under access code GSE104545.
Results
CD274 is a direct ARID1A target gene.
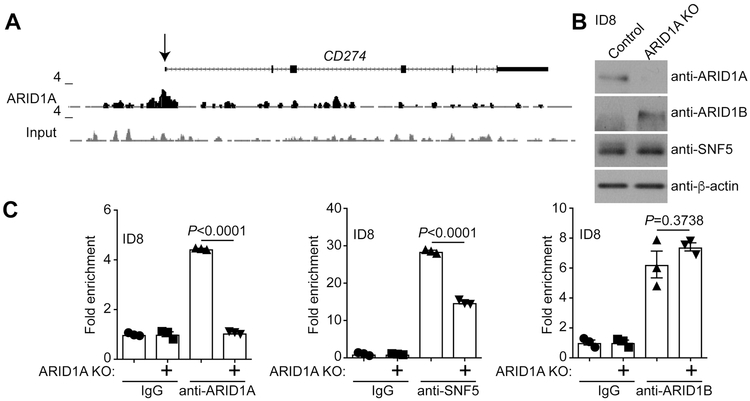
ARID1A chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) analysis revealed that ARID1A was associated with the PD-L1 encoding CD274 gene promoter in ARID1A wildtype OCCC cells (20) (Figure 1A). We validated the binding of ARID1A to the Cd274 gene promoter by ChIP in the mouse ovarian ID8-Defb29/Vegf cells (Figure 1B-C) in which PD-L1 is implicated (18). As a negative control, ARID1A binding to the Cd274 promoter was reduced to a level observed in IgG controls in ARID1A knockout ID8-Defb29/Vegf cells (Figure 1C). Notably, SNF5, a core subunit of the SWI/SNF complex, was also associated with the Cd274 promoter and its association was reduced by ARID1A knockout (Figure 1C). Expression of ARID1B, the mutually exclusive subunit of the SWI/SNF complex with ARID1A, was upregulated in ARID1A knockout ID8-Defb29/Vegf cells (Figure 1B) (21). Although ARID1B was also associated with the Cd274 promoter, ARID1A knockout did not affect the association of ARID1B with the Cd274 promoter (Figure 1C). This suggests that ARID1B is unable to compensate for ARID1A loss on the Cd274 promoter. Similar observations were also made in the ARID1A wildtype human OCCC cell lines OVCA429 and RMG1 cells (Figure S1), indicating that the association of ARID1A with the CD274 promoter is not a cell line-specific effect. Together, we conclude that CD274 is a direct ARID1A target gene.
Figure 1. CD274 is a direct ARID1A target gene.
(A) ARID1A ChIP-seq track on the CD274 gene locus in ARID1A wildtype RMG1 cells. (B) Expression of ARID1A, ARID1B, SNF5 and β-actin in the ARID1A wildtype control and ARID1A knockout mouse ovarian ID8-Defb29/Vegf cells. (C) The indicated ID8-Defb29/Vegf cells were subjected to ChIP analysis for the association of the indicated proteins with the Cd274 gene promoter using the indicated antibodies against ARID1A, SNF5, ARID1B or an isotype-matched IgG control. Error bars represent ± S.E.M. n= 3 independent experiments.
ARID1A represses CD274 gene transcription.
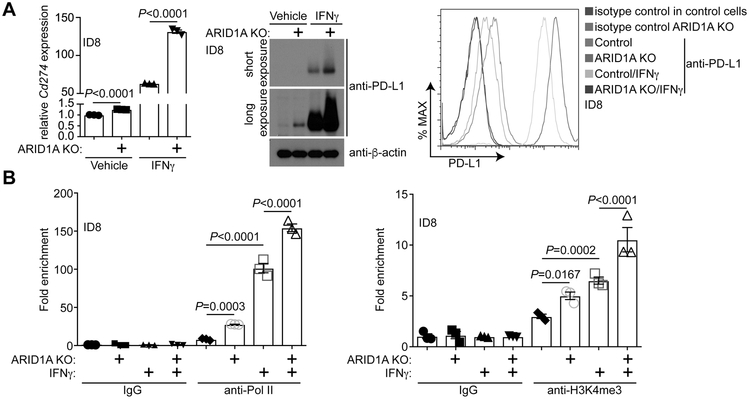
We next determined the effect of ARID1A status on changes in Cd274 mRNA and PD-L1 expression. Compared with ARID1A wildtype control ID8-Defb29/Vegf cells, Cd274 mRNA was increased by ARID1A knockout (Figure 2A). Consistently, PD-L1 expression measured by both immunoblot and fluorescence-activated cell sorting (FACS) analysis was upregulated upon ARID1A knockout (Figure 2A). Interferon-gamma (IFNγ) plays a major role in inducing PD-L1 expression (22). Thus, we examined the effects of ARID1A knockout on IFNγ-induced PD-L1 expression. ARID1A knockout significantly enhanced the upregulation of Cd274 mRNA and PD-L1 expression induced by IFNγ treatment (Figure 2A). Similar findings were made in both ARID1A wildtype mouse ID8-Defb29/Vegf cells and human OVCA429 and RMG1 cells with or without ARID1A knockout (Figure S2). We next examined the association of RNA polymerase II (Pol II) and lysine 4 trimethylated histone H3 (H3K4me3), a transcription active promoter epigenetic mark, with the Cd274 promoter. Consistent with changes observed in Cd274 mRNA and PD-L1 expression, ARID1A knockout enhanced the association of Pol II and H3K4me3 with the Cd274 promoter with or without IFNγ stimulation (Figure 2B). Together, we conclude that ARID1A represses CD274 gene transcription at both the basal levels and in response to IFNγ stimulation.
Figure 2. ARID1A transcriptionally represses Cd274.
(A) Expression of Cd274 mRNA and PD-L1 protein in ARID1A wildtype control and ARID1A knockout mouse ovarian ID8-Defb29/Vegf cells treated with or without 20 ng/ml IFNγ determined by qRT-PCR (left) or immunoblot (middle). The cell surface expression of PD-L1 in these cells was determined by flow cytometry analysis (right). (B) The indicated ID8-Defb29/Vegf cells treated with or without 20 ng/ml IFNγ cells were subjected to ChIP analysis for the Cd274 gene promoter using the indicated antibodies or an isotype-matched IgG control. Error bars represent ± S.E.M. n= 3 independent experiments.
Combination of HDAC6 inhibitor and anti-PD-L1 in the ARID1Aflox/flox/PIK3CAH1047R OCCC mouse model.
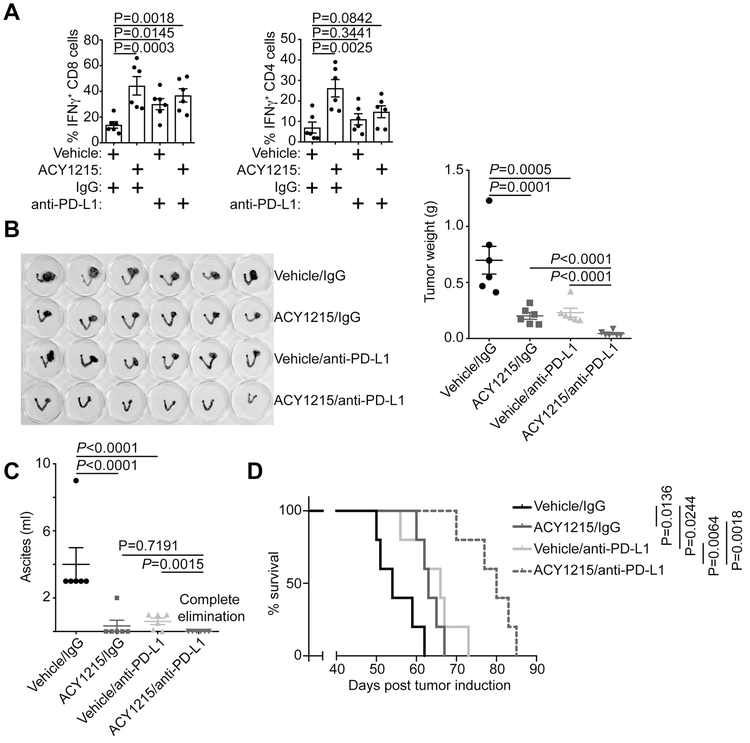
Given HDAC6 inhibitors’ role in immune modulation (13-15), we examined the effects of HDAC6 inhibitor ACY1215 in a conditional genetic ARID1Aflox/flox/PIK3CAH1047R OCCC mouse model (12,23) (Figure S3A). Notably, HDAC6 inhibitor ACY1215 significantly increased the CD69+ activated CD4 and CD8 T cells in the peritoneal wash (Figure S3B). Consistently, IFNγ+ CD4 and CD8 T cells were also significantly increased by ACY1215 treatment (Figure 3A). In contrast, ACY1215 did not significantly affect Granzyme B+ CD8 T cells or Foxp3+ regulatory T cells (Figure S3C). These findings suggest that HDAC6 inhibition may boost antitumor immunity. However, a combination of ACY1215 and anti-PD-L1 treatment only increased IFNγ+ CD8, but not CD4 T cells (Figure 3A). This suggests the implication of CD8 T cells in the combination treatment.
Figure 3. ACY1215 and anti-PD-L1 are synergistic in limiting tumor progression in vivo.
(A) ARID1Aflox/flox/PIK3CAH1047R OCCCs were induced by intrabursal adenovirus-encoded Cre injection and allowed to establish for four weeks. The mice were randomized into 4 indicated treatment groups and treated for an additional three weeks. At the end of treatment, percentage of IFNγ positive CD8 and CD4 T cells was assessed by flow cytometry in the peritoneal wash. (B) 6 mice from each of the indicated groups were euthanized after treatment. Images of dissected reproductive tracks with tumors were shown in the left panel. The weights of the dissected tumors were quantified as a surrogate for tumor burden on the right. (C) Same as (B), but quantified for the ascites produced. (D) After completing treatment, the mice were followed for survival and shown are the Kaplan–Meier survival curves for each of the indicated groups. Error bars represent ± S.E.M.
Since ARID1A directly represses PD-L1 and HDAC6 inhibition increases T cell activation and activity, we sought to determine the effects of HDAC6 inhibitor ACY1215 and anti-PD-L1 combination in ARID1A-inactivated OCCCs. Toward this goal, we first establish OCCCs in 6-8 weeks old ARID1Aflox/flox/PIK3CAH1047R female mice by intrabursally injecting adenovirus-Cre (12). Four weeks after the adenovirus-Cre injection, the mice were randomized into four treatment groups: 1) vehicle and IgG control; 2) ACY1215 (50 mg/kg daily by i.p.) and IgG control; 3) vehicle control and anti-PD-L1 antibody (10 mg/kg twice weekly by i.p.); and 4) ACY1215 and anti-PD-L1 antibody combination for an additional three weeks. At the end of treatment, orthotopic tumors were surgically removed (Figure 3B). The tumor weight was measured as a surrogate for tumor burden. As previously reported (9,12), both anti-PD-L1 antibody and ACY1215 significantly reduced the tumor weight in the OCCC model (Figure 3B). We also examined effects of the ACY1215 and anti-PD-L1 combination in reducing ascites produced in the Arid1aflox/flox/Pik3caH1047R OCCC model. Both ACY1215 and anti-PD-L1 single treatment significantly reduced the amount of ascites produced in this model (Figure 3C). The reduction in tumor weight and ascites production by ACY1215 or anti-PD-L1 single treatment correlated with an improvement of survival (Figure 3D). The HDAC6 inhibitor ACY1215 and anti-PD-L1 combination was synergistic in reducing the tumor burden and improving the survival of tumor-bearing mice (Figure 3B and D). Notably, the combination completely eliminated the ascites production (Figure 3C). The doses of ACY1215 and anti-PD-L1 used in this study did not significantly affect the body weight of treated mice (Figure S3D), suggesting that effective combination doses can be achieved without gross toxicity. Together, we conclude that HDAC6 inhibitor ACY1215 and anti-PD-L1 are synergistic in reducing tumor burden, which correlated with an improvement of survival of mice bearing ARID1A-inactivated OCCCs.
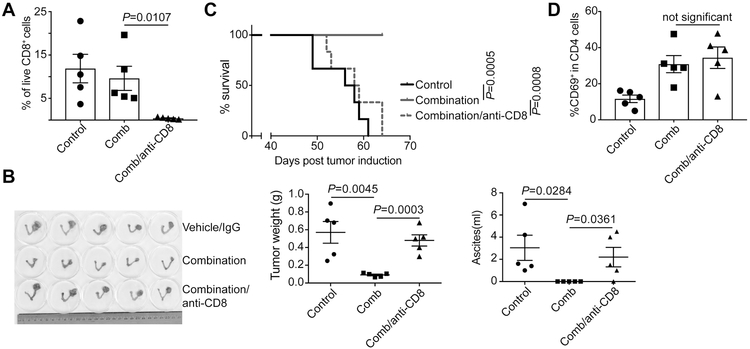
CD8+ T cell depletion abrogates the antitumor effects of ACY1215 and anti-PD-L1 combination.
Since ACY1215 and anti-PD-L1 combination increases IFNγ+ CD8, but not CD4, T cells (Figure 3A) and cytotoxic CD8 T cells play a critical role in mediating the antitumor effects of anti-PD-L1 treatment (10), we next sought to determine whether the combination limits the progression of ARID1A-mutated OCCCs through CD8 T cells. Towards this goal, we depleted CD8 T cells by treating the combination-treated mice with an anti-CD8 antibody (Figure 4A). Compared with IgG control-treated mice, anti-CD8 antibody significantly abrogated the observed reduction in tumor weight and ascites production induced by the combination (Figure 4B). Consistently, the improvement of survival observed in the combination treatment group was also abrogated by the anti-CD8 antibody (Figure 4C). However, anti-CD8 antibody did not significantly reduce the CD4 T cell activation (Figure 4D). This result indicates that T cell activation induced by ACY1215 is not merely a reflection of reduction in tumor burden in the treated mice. Together, these results support that the observed antitumor effects in ARID1A-inactivated OCCCs by ACY1215 and anti-PD-L1 combination is CD8 cytotoxic T cell dependent.
Figure 4. Depletion of CD8+ T cells abrogates the antitumor effects of ACY1215 and anti-PD-L1 combination.
(A) ARID1Aflox/flox/PIK3CAH1047R OCCCs were induced by intrabursal adenovirus-encoded Cre injection and allowed to establish for four weeks. The mice were randomized into three indicated experimental groups (n=5 mice/group). The depletion of CD8+ T cells was confirmed by flow cytometry analysis of blood cells collected via the retro-orbital vein. (B) Images of dissected reproductive tracks with tumors were shown in the right panel. The weights of the dissected tumors were quantified as a surrogate for tumor burden in the middle panel, and ascites produced were quantified in the right panel. (C) After three weeks treatment, the mice were followed for survival and shown are the Kaplan–Meier survival curves. (D) At the end of treatment, percentage of CD69 positive CD4 T cells was assessed by flow cytometry in the dissected tumors. Error bars represent ± S.E.M.
Discussion
Here we show that HDAC6 inhibitor ACY1215 activates both CD4 and CD8 T cells and increases IFNγ+ CD4 and CD8 T cells. In addition, HDAC6 inhibition suppresses the growth of ARID1A-mutated tumor in immunocompromised xenograft models (12). Thus, HDAC6 inhibitor may suppress ARID1A-mutated tumors by both targeting cancer cells and restoring antitumor immunity. The fact that depletion of CD8 T cells significantly abrogated the antitumor effects of HDAC6 inhibitor and anti-PD-L1 combination suggests that in this context the effects of the combination on tumor immune microenvironment played a major role in the observed antitumor effects.
Here we report that ARID1A directly represses CD274 gene transcription. In addition, it has been reported that ARID1A inactivation created a mutator phenotype. Indeed, ARID1A mutation correlated with an increase in PD-L1 expression (9) and there was a trend toward improved response rate in clear cell ovarian cancer in which ARID1A is mutated in >50% of cases (24). Further, there is evidence to suggest that inactivation of PBAF complex increased tumor cells sensitivity to IFNγ, resulted in enhanced secretion of chemokines that recruit effector T cells (7,8). Thus, inactivation of ARID1A containing BAF complex may increases PD-L1 expression directly at the CD274 gene promoter or indirectly through increasing mutation loads. In addition, IFNγ appears to play a central role in regulating immune checkpoint and effector T cell function by boosting PD-L1 expression when ARID1A-containing BAF complex is inactivated or through boosting IFNγ responsive genes when PBAF complex is inactivated (7,8).
In summary, our findings identify a combination of HDAC6 inhibition and immune checkpoint blockade as an effective treatment strategy for ARID1A-inactivated tumors. Interestingly, ARID1A mutation predicts clinical response to pan-HDAC inhibition in urothelial carcinoma and specific HDAC6 inhibition was most potent in suppressing the growth of ARID1A-mutated urothelial cells (25). The HDAC6 inhibitor ACY1215 is now in clinical development for other cancer types, and anti-PD-L1 is FDA-approved. Thus, they are readily available for a combinational clinical application in ARID1A-mutated cancers.
Supplementary Material
Acknowledgements
This work was supported by US National Institutes of Health grants (R01CA160331, R01CA163377, R01CA202919, R01CA239128 and P50CA228991 to R. Z.) and US Department of Defense (OC150446 and OC180109 to R. Z.). The Honorable Tina Brozman Foundation for Ovarian Cancer Research (to R.Z.) and Ovarian Cancer Research Alliance (Collaborative Research Development Grant to R.Z. and D.G., and Ann and Sol Schreiber Mentored Investigator Award to S.W.). Support of Core Facilities was provided by Cancer Centre Support Grant (CCSG) CA010815 to The Wistar Institute.
Footnotes
Disclosure of Potential Conflicts of Interests: The authors declare no competing financial interests.
References
- 1.Helming KC, Wang X, Roberts CWM. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 2014;26:309–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukumoto T, Magno E, Zhang R. SWI/SNF Complexes in Ovarian Cancer: Mechanistic Insights and Therapeutic Implications. Mol Cancer Res 2018;16:1819–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;173:371–85 e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 2010;363:1532–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackay HJ, Brady MF, Oza AM, Reuss A, Pujade-Lauraine E, Swart AM, et al. Prognostic relevance of uncommon ovarian histology in women with stage III/IV epithelial ovarian cancer. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society 2010;20:945–52 [DOI] [PubMed] [Google Scholar]
- 6.Jelinic P, Ricca J, Van Oudenhove E, Olvera N, Merghoub T, Levine DA, et al. Immune-Active Microenvironment in Small Cell Carcinoma of the Ovary, Hypercalcemic Type: Rationale for Immune Checkpoint Blockade. J Natl Cancer Inst 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018;359:770–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol 2018;19:108–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bitler BG, Wu S, Park PH, Hai Y, Aird KM, Wang Y, et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell Biol 2017;19:962–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adeegbe DO, Liu Y, Lizotte PH, Kamihara Y, Aref AR, Almonte C, et al. Synergistic Immunostimulatory Effects and Therapeutic Benefit of Combined Histone Deacetylase and Bromodomain Inhibition in Non-Small Cell Lung Cancer. Cancer Discov 2017;7:852–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woan KV, Lienlaf M, Perez-Villaroel P, Lee C, Cheng F, Knox T, et al. Targeting histone deacetylase 6 mediates a dual anti-melanoma effect: Enhanced antitumor immunity and impaired cell proliferation. Mol Oncol 2015;9:1447–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Li Y, Liu S, Adeegbe DO, Christensen CL, Quinn MM, et al. NK Cells Mediate Synergistic Antitumor Effects of Combined Inhibition of HDAC6 and BET in a SCLC Preclinical Model. Cancer Res 2018;78:3709–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol 2008;28:1688–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santo L, Hideshima T, Kung AL, Tseng JC, Tamang D, Yang M, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 2012;119:2579–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, et al. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep 2016;16:2829–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 2018;13:1006–19 [DOI] [PubMed] [Google Scholar]
- 20.Trizzino M, Barbieri E, Petracovici A, Wu S, Welsh SA, Owens TA, et al. The Tumor Suppressor ARID1A Controls Global Transcription via Pausing of RNA Polymerase II. Cell Rep 2018;23:3933–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med 2014;20:251–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandler RL, Damrauer JS, Raab JR, Schisler JC, Wilkerson MD, Didion JP, et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat Commun 2015;6:6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamanishi J, Mandai M, Ikeda T, Minami M, Kawaguchi A, Murayama T, et al. Safety and Antitumor Activity of Anti-PD-1 Antibody, Nivolumab, in Patients With Platinum-Resistant Ovarian Cancer. J Clin Oncol 2015;33:4015–22 [DOI] [PubMed] [Google Scholar]
- 25.Gupta S, Albertson DJ, Parnell TJ, Butterfield A, Weston A, Pappas LM, et al. Histone Deacetylase Inhibition Has Targeted Clinical Benefit in ARID1A-Mutated Advanced Urothelial Carcinoma. Mol Cancer Ther 2019;18:185–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published ARID1A ChIP-seq in ARID1A wildtype RMG1 cells are available at the Gene Expression Ominibus (GEO) under access code GSE104545.