Abstract
The kinase, LKB1, is a critical tumor suppressor in sporadic and familial human cancers, yet the mechanisms by which it suppresses tumor growth remain poorly understood. To investigate the tumor-suppressive capacity of four canonical families of Lkb1 substrates in vivo, we employed CRISPR/Cas9-mediated combinatorial genome editing in a mouse model of oncogenic Kras-driven lung adenocarcinoma. We demonstrate that members of the salt-inducible kinase (Sik) family are critical for constraining tumor development. Histological and gene expression similarities between Lkb1- and Sik-deficient tumors suggest that Siks and Lkb1 operate within the same axis. Furthermore, a gene expression signature reflecting Sik deficiency is enriched in LKB1 mutant human lung adenocarcinomas and is regulated by LKB1 in human cancer cell lines. Together, these findings reveal a key Lkb1-Sik tumor-suppressive axis and underscore the need to redirect efforts to elucidate the mechanisms through which LKB1 mediates tumor suppression.
Keywords: LKB1, salt-inducible kinases, tumor suppressor, lung adenocarcinoma, CRISPR/Cas9
INTRODUCTION
Recurrently altered tumor suppressors represent key nodes in the programs that constrain tumorigenesis (1). Uncovering the molecular context in which these genes function can aid in understanding how these alterations derail normal cellular physiology and identifying therapeutic opportunities (2). However, the upstream regulators and downstream effectors of a given tumor suppressor are not always identifiable from human cancer genomics data alone, and molecular profiling along with biochemical studies only reveal interactions that could play a role in tumor suppression. Thus, functional genomics studies are ultimately required to directly interrogate the importance of interactors within networks radiating from bona fide tumor suppressors (3).
LKB1 (also known as serine/threonine kinase 11; STK11) is frequently inactivated in multiple human cancer types, and germline LKB1 mutations play a causative role in the familial cancer susceptibility disorder Peutz-Jeghers syndrome (4,5). Notably, in lung adenocarcinoma, LKB1 is mutationally inactivated in 15–30% of cases (6). Lkb1 inactivation dramatically accelerates tumor growth in mouse models of lung cancer and broadens the spectrum of histological subtypes that emerge (7). In a complex with the pseudokinase STRAD and scaffolding protein MO25, LKB1 phosphorylates and activates members of several subfamilies of AMPK-related kinases (AMPK, BRSK, MARK, NUAK, and SIK) through which it governs multiple cellular processes that could, in principle, contribute to tumor suppression (8,9).
LKB1 and its substrates have been implicated in a broad range of biological processes, including aging, neuronal differentiation, glucose homeostasis, and cell polarity adhesion (10–15). However, despite multiple lines of evidence supporting functional parallels between LKB1 and several of its substrates, AMPK is conventionally viewed as the focal point of tumor suppressor activity downstream of LKB1 (9,16,17). If AMPK were indeed the principal tumor suppressor downstream of LKB1, its ablation in vivo would enhance tumor growth. However, Ampk activity has recently been shown to be a requirement for oncogenic Kras-driven tumorigenesis in the lung, suggesting that other substrates likely suppress tumor growth downstream of Lkb1 (18). Thus, the key tumor-suppressive effectors downstream of LKB1 and the contexts in which they operate remain poorly defined.
Here, through CRISPR/Cas9-mediated combinatorial genome editing within a genetically engineered mouse model of lung adenocarcinoma, we investigate the tumor-suppressive capacity of canonical Lkb1 substrate families in vivo. We identify Sik1 and Sik3 as potent tumor suppressors and uncover multiple parallels between Sik family inactivation and Lkb1 deficiency that underscore the importance of an Lkb1-Sik tumor-suppressive axis.
RESULTS
Prioritization of Lkb1 substrates for functional interrogation
To select candidate Lkb1 substrates for characterization in vivo, we examined their expression in normal murine lung epithelium and purified cancer cells from murine lung tumors at different stages of malignant progression (19). The brain-specific substrates, Brsk1 and Brsk2, as well as Mark1 were expressed at very low levels in normal lung as well as lung tumors and were thus excluded (TPM<2; Fig. 1A and Supplementary Fig. S1A). We also examined the expression of these substrates in human lung adenocarcinoma and normal human lung (20). The relative expression of Lkb1 substrates in human lung adenocarcinoma largely agreed with their expression in murine tumors, including the low expression of BRSK1/2, the lower expression of AMPKA2 relative to AMPKA1, the lower expression MARK1 relative to the other MARKs, and the higher expression of SIK1 relative to the other SIKs (Fig. 1B and Supplementary Fig. S1B). Beyond expression, analysis of the frequency and nature of mutations in LKB1 substrates in human lung adenocarcinoma did not highlight any substrates as candidate tumor suppressors (Supplementary Fig. S1C). From these analyses, we proceeded to interrogate the tumor-suppressive capacity of the expressed members of the Ampk, Mark, Nuak, and Sik families.
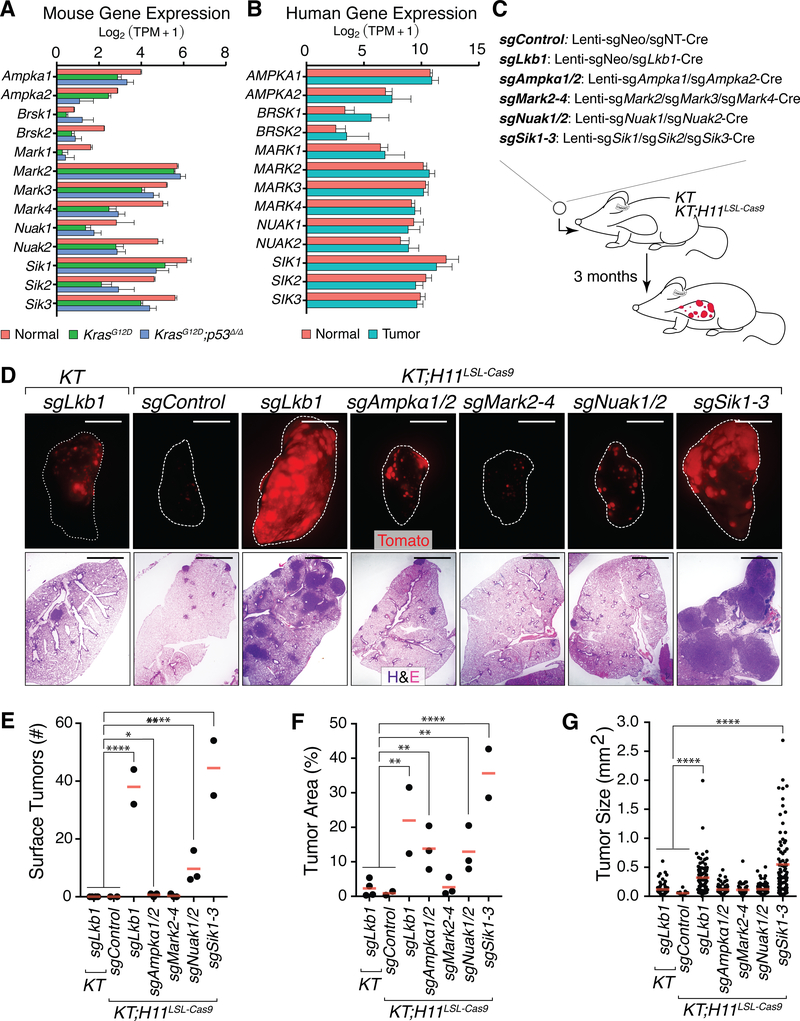
Figure 1. CRISPR/Cas9-mediated inactivation of Lkb1 substrate families in vivo identifies the Sik family as tumor suppressors in lung adenocarcinoma.
A. mRNA expression of Lkb1 substrates in normal mouse lung and in neoplastic cells isolated from lung tumors of KrasLSL-G12D/+;R26LSL-Tomato (KrasG12D, KT) and KrasLSL-G12D/+;p53flox/flox;R26LSL-Tomato (KrasG12D;p53Δ/Δ, KPT) mice. Mean +/− SD is shown.
B. Expression of LKB1 substrates in normal human lung and lung adenocarcinoma (20). Mean +/− SD is shown.
C. Outline of CRISPR/Cas9-mediated inactivation of Lkb1 substrate families to assess tumor-suppressive function. Tumors were initiated in KT and KT;H11LSL-Cas9 mice with the indicated Lenti-sgRNA/Cre vectors targeting the expressed members of each family.
D. Representative fluorescence (top) and H&E (bottom) images of lungs from KT and KT;H11LSL-Cas9 mice after tumor initiation with the indicated Lenti-sgRNA/Cre vectors. Lung lobes are outlined in white. Top scale bars = 5 mm. Bottom scale bars = 2 mm.
E-G. Number of Tomatopositive surface tumors (>1 mm in diameter; E) detected via fluorescence microscopy, tumor area (F) and size (G) from histology of KT and KT;H11LSL-Cas9 mice with tumors initiated with Lenti-sgRNA/Cre as indicated. In E and F, each dot represents a mouse. In G, each dot represents a tumor. The crossbars indicate the mean. *p-value < 0.05, ** p-value < 0.01, *** p-value <0.001 ****p-value <0.0001.
Generation of Lenti-sgRNA/Cre vectors targeting Lkb1 substrates
Genetically engineered mouse models enable the genetics of human cancers to be functionally interrogated within autochthonous tumors that recapitulate many histological and molecular features of human malignancies (21). The recent integration of CRISPR/Cas9-mediated somatic genome editing into these models has accelerated the rate at which genes can be functionally investigated in vivo (3). Additionally, the capacity of CRISPR/Cas9 to simultaneously target multiple loci should enable the study of gene families wherein there may exist functional redundancy. To inactivate Lkb1 substrates in vivo using CRISPR/Cas9, we identified sgRNAs that efficiently target each of the 10 expressed Lkb1 substrates (Supplementary Fig. S2A, Supplementary Table S1). To address potential functional redundancy between paralogs, we designed lentiviral vectors encoding up to three sgRNAs that simultaneously target the expressed members of the Ampk, Mark, Nuak, and Sik families. In cell culture, we confirmed that the multi-sgRNA design led to gene targeting efficiencies comparable to vectors encoding single sgRNAs (Supplementary Fig. S2B–D).
CRISPR/Cas9-mediated targeting of the Siks in vivo leads to rapid lung tumor growth
To assess the tumor-suppressive function of the Ampk, Nuak, Mark, and Sik families of kinases in KrasG12D-driven lung tumors in vivo, we initiated tumors in KrasLSL-G12D/+;R26LSL-Tomato;H11LSL-Cas9 (KT;H11LSL-Cas9) mice with each double or triple Lenti-sgRNA/Cre vector (Fig. 1C). Three months after tumor initiation, negative control KT mice (which lack the H11LSL-Cas9 allele) transduced with Lenti-sgLkb1/Cre and KT;H11LSL-Cas9 mice transduced with a dual-sgRNA control vector (Lenti-sgControl/Cre) had very few large surface tumors as detected by fluorescence imaging (Fig. 1D,E). In contrast, positive control KT;H11LSL-Cas9 mice transduced with Lenti-sgLkb1/Cre developed many large tumors (Fig. 1D,E). Strikingly, KT;H11LSL-Cas9 mice transduced with Lenti-sgSik1–3/Cre also developed many large tumors, while KT;H11LSL-Cas9 mice transduced with lentiviral vectors targeting the Mark, Nuak, and Ampk families developed fewer large tumors. Quantification by histology confirmed that KT;H11LSL-Cas9 mice transduced with Lenti-sgSik1–3/Cre had high overall tumor burden and many large tumors, indicating that Siks are tumor suppressors (Fig. 1F,G and Supplementary Fig. S3A).
To confirm that indels were generated at the targeted loci in vivo, we FACS-isolated Tomatopositive neoplastic cells from large individual tumors and bulk tumor-bearing lungs. PCR amplification and sequencing of the targeted regions confirmed that neoplastic cells from tumors initiated with Lenti-sgRNA/Cre vectors targeting the Ampk, Nuak, and Mark families harbored indels at each targeted locus (Supplementary Fig. S3B–D). Using this approach, we also observed high-frequency indels (>70%) in Sik1 and/or Sik3 in large Lenti-sgSik1–3/Cre-initiated tumors, indicating that they may be the predominant tumor suppressors of the Sik family (Fig. 2A). In contrast, indels in Sik2 were infrequent, which agrees with previous reports of pro-tumorigenic functions of SIK2 in other cancer types (22–24). Consistent with indels in Sik1 and Sik3, immunohistochemical staining for Sik1 and Sik3 revealed reduced protein expression in tumors initiated by Lenti-sgSik1–3/Cre in KT;H11LSL-Cas9 mice (Fig. 2B,C and Supplementary Fig. S4A).
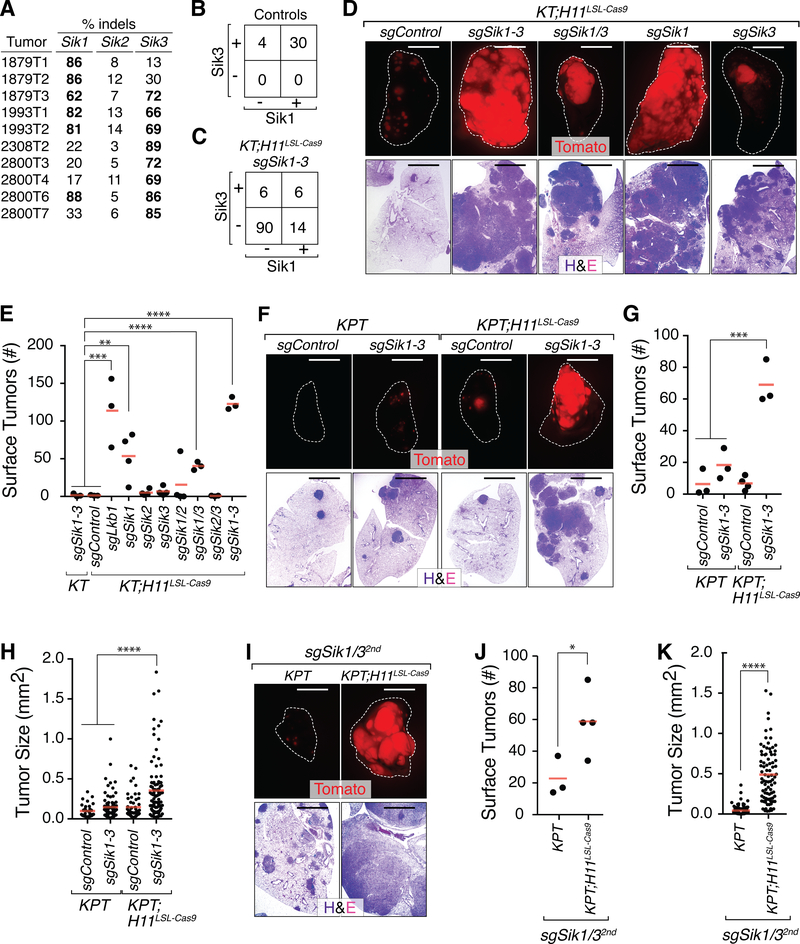
Figure 2. Sik1 and Sik3 constrain lung tumor growth in vivo.
A. Percent indels at the target sites in Sik1, Sik2, and Sik3 in cancer cells sorted from individual Lenti-sgSik1–3/Cre-initiated tumors from KT;H11LSL-Cas9 mice.
B,C. Immunohistochemistry for Sik1 and Sik3 on tumors from Control mice (KT mice with Lenti-sgSik1–3/Cre-initiated tumors and KT;H11LSL-Cas9 mice with Lenti-sgControl/Cre-initiated tumors; B) and KT;H11LSL-Cas9 mice with Lenti-sgSik1–3/Cre-initiated tumors (C).
D. Representative fluorescence (top) and H&E (bottom) images of lungs from KT;H11LSL-Cas9 mice after tumor initiation with Lenti-sgRNA/Cre vectors as indicated. Lung lobes are outlined with a white dashed line. Top scale bars = 5 mm. Bottom scale bars = 2 mm.
E. Number of Tomatopositive surface tumors (>1 mm in diameter) in KT and KT;H11LSL-Cas9 mice with tumors initiated with Lenti-sgRNA/Cre vectors as indicated. Each dot represents a mouse. The crossbars indicate the mean.
F, I. Representative fluorescence (top) and H&E (bottom) images of lungs from KPT and KPT;H11LSL-Cas9 mice with tumors initiated with Lenti-sgRNA/Cre vectors as indicated. Lung lobes are outlined in white. Top scale bars = 5 mm. Bottom scale bars = 2 mm.
G, J. Number of Tomatopositive surface tumors (>1 mm in diameter) in KPT and KPT;H11LSL-Cas9 mice with tumors initiated with Lenti-sgRNA/Cre vectors with the indicated sgRNAs. Each dot represents a mouse. The crossbars indicate the mean.
H, K. Tumor size as measured by histology in KPT and KPT;H11LSL-Cas9 mice with tumors initiated with Lenti-sgRNA/Cre vectors as indicated. Each dot represents an individual tumor. The crossbars indicate the mean. An independent set of sgRNAs targeting Sik1 and Sik3 was used in I, J, and K. * p-value < 0.05, ** p-value < 0.01, ***p-value < 0.001, ****p-value <0.0001.
Sik1 and Sik3 function as tumor suppressors in lung cancer
To assess the tumor-suppressive activity of each Sik paralog, we initiated lung tumors in KT;H11LSL-Cas9 mice with lentiviral vectors targeting each Sik family member individually, each pairwise combination of Siks, as well as all three Siks (Supplementary Fig. S4B). In agreement with our initial results, Lenti-sgControl/Cre-transduced KT;H11LSL-Cas9 mice and Lenti-sgSik1–3/Cre-transduced KT mice developed very few large tumors (Fig. 2D,E). KT;H11LSL-Cas9 mice transduced with vectors encoding sgSik1 or sgSik1 and sgSik3 had an increased number of large surface tumors, greater overall tumor burden, and larger individual tumors (Fig. 2D,E and Supplementary Fig. S4C–E). In line with our prior indel analyses, these findings indicated that Sik1 and Sik3 are the predominant tumor suppressors of the Sik family.
Given the fundamental importance of the p53 tumor suppressor in lung cancer and previous data suggesting that Sik1 promotes p53-dependent anoikis in transformed human mammary epithelial cells, we determined whether the tumor suppressor function of Siks in lung adenocarcinoma was mediated through p53 (20,25). To test this, we initiated tumors in KT;p53flox/flox (KPT) and KPT;H11LSL-Cas9 mice with Lenti-sgControl/Cre and Lenti-sgSik1–3/Cre vectors. Lenti-sgSik1–3/Cre-initiated tumors were significantly larger than tumors in any of the control cohorts, suggesting that Siks mediate tumor suppression even in the context of p53 deficiency (Fig. 2F–H and Supplementary Fig. S5A). Consistent with the KT;H11LSL-Cas9 context, Sik1 and Sik3 were frequently inactivated as determined by indel analysis and immunohistochemistry (Supplementary Fig. S5B,C). To confirm that enhanced tumor growth was driven by on-target effects, we used a dual Lenti-sgRNA/Cre vector encoding a second set of sgRNAs targeting Sik1 and Sik3 (Lenti-sgSik1/32nd/Cre) to initiate tumors in KPT;H11LSL-Cas9 mice. KPT;H11LSL-Cas9 mice transduced with Lenti-sgSik1/32nd/Cre had more large tumors and greater overall tumor burden than KPT mice transduced with the same viral vector (Fig. 2I–K and Supplementary Fig. S5D). Collectively, these findings demonstrate that Siks, particularly Sik1 and Sik3, have tumor-suppressive function that is at least partially independent of p53 activity in our model.
To enable quantitative and multiplexed analysis of tumor suppressors in vivo, we recently integrated CRISPR/Cas9-based genome editing with tumor barcoding and high-throughput barcode sequencing (Tuba-seq) (26,27). To measure tumor size, Lenti-sgRNA/Cre vectors are diversified with a vector-specific identifier (sgID) and a random barcode (BC) which stably label each transduced cell and their progeny within the resulting clonal tumors (Fig. 3A and Supplementary Fig. S6A). High-throughput sequencing of the two-component barcodes amplified from bulk genomic DNA of tumor-bearing lungs enables pet-tumor measurements of cancer cell number (Fig. 3B)(26). Analysis of the number of cancer cells in tumors of different genotypes relative to tumors initiated with vectors encoding inert sgRNAs can uncover the magnitude of tumor suppression.
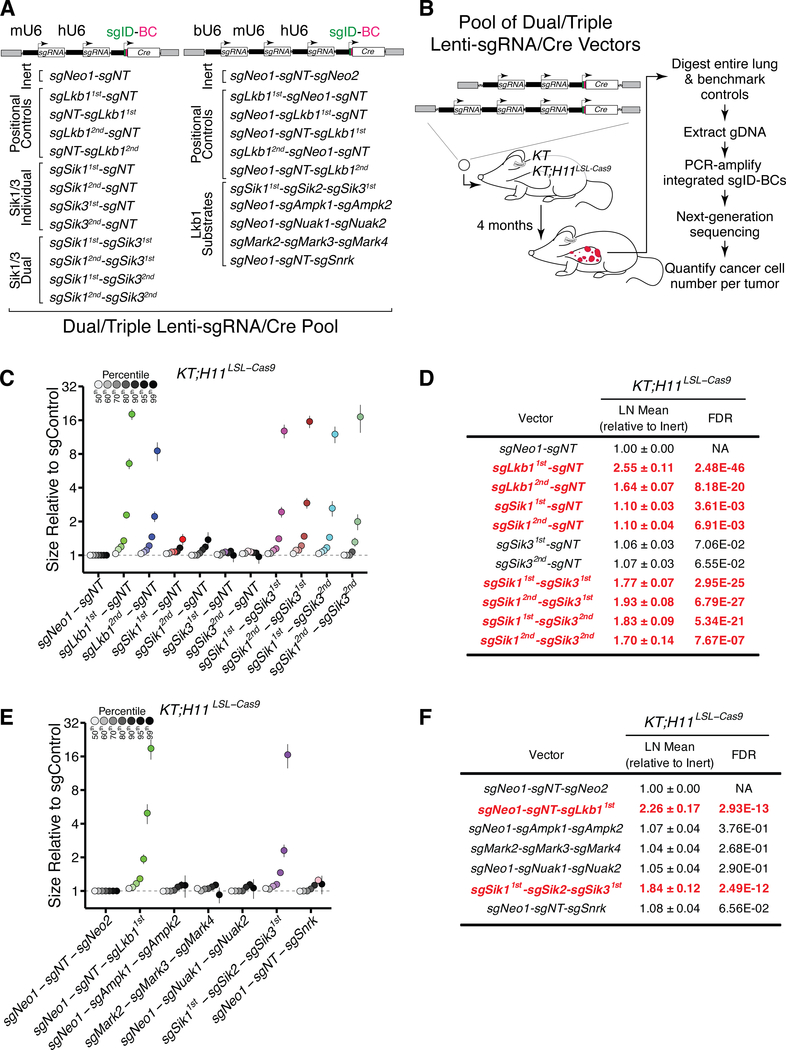
Figure 3. Quantitative analysis of tumor suppression uncovers functional redundancy between Sik1 and Sik3.
A. Summary of the Lenti-sgRNA/Cre vectors that were introduced into KT and KT;H11LSL-Cas9 mice in a pooled format. The sgRNA identifier (sgID; green) and random barcode (BC; pink) cassettes encoded within lentiviral vectors are denoted.
B. Outline of pipeline to quantify tumor suppression at high resolution in a multiplexed format. Five KT and 10 KT;H11LSL-Cas9 mice were transduced with the Lenti-sgRNA/Cre pool outlined in (A).
C,E. Analysis of Sik1/3-mediated tumor suppression (C) and tumor suppression across Lkb1 substrate families (E) in KT;H11LSL-Cas9 mice at 16 weeks post-initiation. Relative size of tumors at the indicated percentiles is merged data from 10 mice, normalized to the size of sgNeo1-sgNT (C) or sgNeo1-sgNT-sgNeo2 (E) tumors. Error bars denote 95% confidence intervals determined by bootstrap sampling. Percentiles that are significantly different from control vectors are colored.
D,F. Tables summarizing maximum likelihood estimates and corresponding 95% confidence intervals for mean tumor size, assuming a log-normal tumor size distribution (26). Emboldened, red-colored vectors significantly increased the LN mean. Benjamini-Hochberg-corrected, bootstrapped p-values are shown.
Using Tuba-seq, we directly quantified the extent to which the targeting of Sik1 and/or Sik3 phenocopies Lkb1 loss in promoting tumor growth. To uncover potential additive or synergistic interactions, we initiated tumors in KT and KT;H11LSL-Cas9 mice with a pool of barcoded vectors encoding combinations of sgRNAs (Fig. 3A,B). Consistent with previous work, Lkb1 targeting dramatically increased tumor size (Fig. 3C,D and Supplementary Fig. S6B)(26,27). Permutation of the position of Lkb1-targeting sgRNAs within the dual- or triple-sgRNA configurations resulted in tumor size distributions that were highly correlated, suggesting that sgRNA activity is not strongly influenced by its position in these Lenti-sgRNA/Cre vectors (Supplementary Fig. S6C,D).
Targeting of Sik1 led to a modest increase in tumor size, while Sik3 targeting had little to no impact on tumor size (Fig. 3C,D and Supplementary Fig. S6B). Strikingly, concomitant targeting of Sik1 and Sik3 was synergistic, increasing tumor size to a magnitude comparable to Lkb1 targeting (Fig. 3C,D). This result was consistent across a series of combinations of two sets of sgRNAs targeting Sik1 and Sik3, suggesting that Sik1 and Sik3 operate as functionally redundant tumor suppressors (Fig. 3C,D). In contrast, targeting of Sik2 in addition to Sik1 and Sik3, did not significantly increase tumor size, thus reinforcing Sik1 and Sik3 as the predominant tumor-suppressive paralogs (Supplementary Fig. S6E,F). Finally, in line with our initial screen, targeting of the other families of Lkb1 substrates, including a more divergent member of the Ampk family, Snrk, did not dramatically increase tumor growth. (Fig. 3E,F and Supplementary Fig. S6G). Together, our findings establish the tumor-suppressive activity of the Sik family, with Sik1 and Sik3 serving as the primary contributors to tumor suppression.
Sik targeting recapitulates histological features of Lkb1-deficient tumors
To understand at the cellular level the driving force underlying the increased tumor burden resulting from Sik inactivation, we quantified proliferation and cell death within Sik-targeted and control tumors. Immunohistochemical staining for phosphorylated histone H3 (Ser10) and Ki-67 revealed a significant increase in the fraction of proliferating cells within Sik-targeted tumors (Fig. 4A–C). Cleaved caspase 3 and TUNEL staining revealed no dramatic change in cell death (Fig. 4A,D,E). Thus, increased proliferation in Sik-targeted tumors likely underlies their increase in size relative to control tumors.
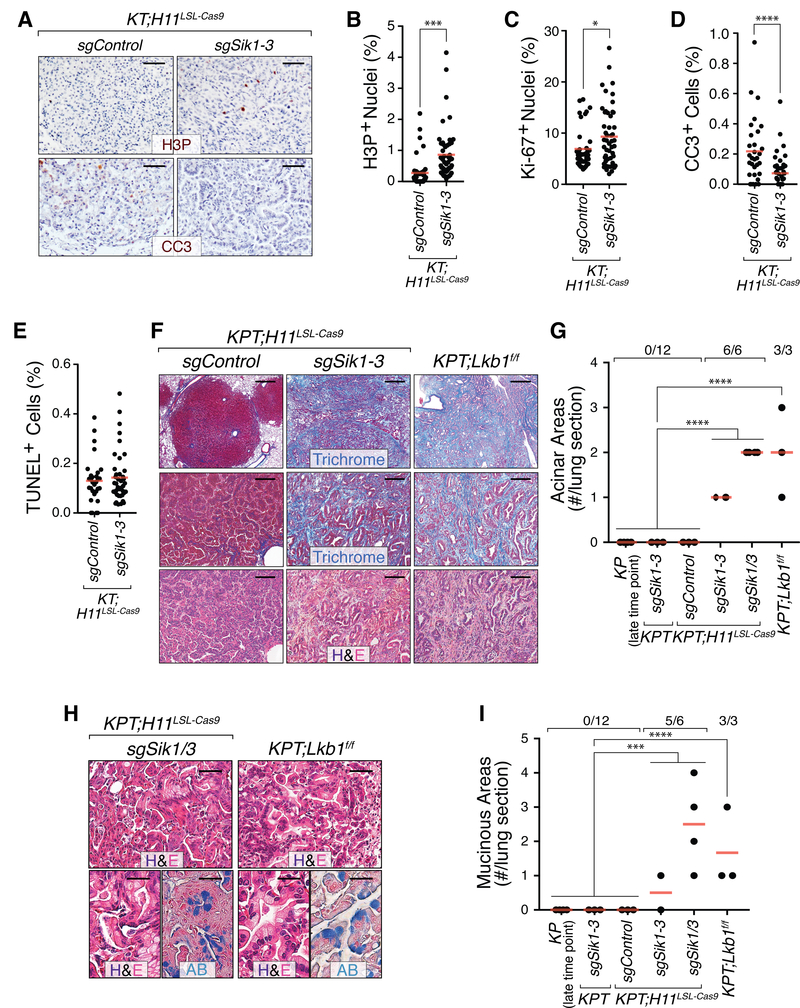
Figure 4. Sik targeting generates tumors with phenotypes characteristic of Lkb1 deficiency.
A. Representative images of immunohistochemistry for phosphorylated histone H3 (Ser10; H3P) and cleaved caspase 3 (CC3) in Lenti-sgControl/Cre(sgControl)- and Lenti-sgSik1–3/Cre(sgSik1–3)-initiated tumors from KT;H11LSL-Cas9 mice. Scale bar = 50 μm.
B-E. Quantification of proliferation (B,C) and cell death (D,E) in Lenti-sgControl/Cre(sgControl)- and Lenti-sgSik1–3/Cre(sgSik1–3)-initiated tumors in KT;H11LSL-Cas9 mice. The dots represent individual tumors. The crossbar is the mean.
F. Sik targeting and Lkb1 ablation in a p53-deficient context results in the development of areas of acinar adenocarcinoma accompanied by abundant desmoplastic, collagen-rich stroma (highlighted by blue Masson’s Trichrome staining), and admixed poorly formed nests and single cancer cells. H&E (top) and Trichrome (bottom) staining highlight the stroma-rich nature and histology of these areas. Of note, while mild stromal reactions develop in oncogenic Kras-driven, p53-deficient tumors (Grade 4 tumors), these areas are never as large as those in Sik-targeted and Lkb1-deficient tumors. Top scale bars = 0.5 mm. All other scale bars = 100 μm.
G. Number of large, highly stromalized, acinar areas per lung section from the indicated genotypes of mice. Each dot represents a mouse and the crossbar is the mean. The fraction of mice in each group with at least one acinar area is indicated above the plot.
H. Representative histology of lung tumor areas with mucinous differentiation. Genotypes of mice are indicated. Alcian blue (AB) stains intracellular mucin. Top scale bars = 50 μm. Bottom scale bars = 25 μm.
I. Number of alcian blue-positive tumor areas with mucinous features in lung sections from the indicated genotypes of mice. Each dot represents a mouse and the crossbar is the mean. The fraction of mice in each group with at least one mucinous area is indicated above the plot. * p-value < 0.05, ***p-value < 0.001, ****p-value <0.0001.
Similar to their Sik-proficient counterparts, the majority of Lenti-sgSik1–3/Cre- and Lenti-sgSik1/3/Cre-initiated tumors in KT;H11LSL-Cas9 and KPT;H11LSL-Cas9 mice were adenomas and early adenocarcinomas. However, most KPT;H11LSL-Cas9 mice with tumors in which all Sik paralogs were targeted also harbored one or more large regions of invasive acinar adenocarcinoma (Fig. 4F,G). These areas were characterized by the presence of cancer cell clusters and glandular structures embedded within extensive desmoplastic stroma, resembling tumors in KPT;Lkb1flox/flox mice and previously reported acinar tumors in mouse models of Lkb1-deficient lung cancer (Fig. 4F)(28,29). Importantly, this histological pattern was not present in KP or KPT mice, even at very late time points, nor was it present in p53-proficient tumors in KT and KT;Lkb1flox/flox mice or Sik-targeted tumors in KT;H11LSL-Cas9 mice (Fig. 4G and Supplementary Fig. S7A). In KP tumors, Nkx2–1 and Hmga2 are established markers of well and poorly differentiated areas, respectively (19,30). Regardless of their apparent differentiation state, cancer cells within acinar-structured Sik-targeted and Lkb1-deficient tumors exhibited heterogeneous expression of Nkx2–1 and Hmga2 (Supplementary Fig. S7B). Thus, in addition to enabling the emergence of acinar adenocarcinoma, this uncoupling of differentiation markers is a shared feature of Lkb1-deficient and Sik-targeted tumors.
Lkb1 loss has been shown to broaden the histological spectrum of lung tumors within mouse models, enabling the emergence of tumors with squamous, large cell, and mucinous differentiation (7,28,29,31). Interestingly, LKB1 mutations are frequent in human mucinous lung adenocarcinoma and gene expression signatures of mucinous adenocarcinoma are enriched in multiple cohorts of LKB1 mutant human lung adenocarcinomas (Supplementary Table S2, S3)(20,32–37). Patients with Peutz-Jeghers syndrome have also been documented to develop mucinous lung adenocarcinoma (38,39). KT;H11LSL-Cas9 and KPT;H11LSL-Cas9 mice transduced with Sik-targeting lentiviral vectors frequently developed small regions of mucinous lung adenocarcinoma (Fig. 4H,I and Supplementary Fig. S7C–E). The abundant intracellular mucin in these regions stained strongly with alcian blue and phenocopied mucinous regions in KT;Lkb1flox/flox and KPT;Lkb1flox/flox mice as well as in KT;H11LSL-Cas9 mice with Lenti-sgLkb1/Cre-initiated tumors (Supplementary Fig. S7D,E). In contrast, mucinous lesions were neither detected in KP, KPT, or KT mice nor within KT;H11LSL-Cas9 or KPT;H11LSL-Cas9 mice bearing tumors initiated by control vectors (Fig. 4I and Supplementary Fig. S7C). Histology and immunohistochemistry for p63 and Ck5 indicated that neither Lkb1 nor Sik deficiency resulted in squamous differentiation, which is in line with lentiviral Cre vectors preferentially generating adenocarcinomas in KrasLSL-G12D/+;Lkb1flox/flox mice (Supplementary Fig. S8A–D)(28). Together, these analyses demonstrate that targeting of the Sik family recapitulates the expanded histologic spectrum elicited by Lkb1 deficiency in lung cancer, which supports the existence of an Lkb1-Sik tumor-suppressive axis.
Sik targeting in lung tumors elicits gene expression changes that parallel Lkb1 loss
Given the identification of the Sik family of kinases as tumor-suppressive Lkb1 substrates, we investigated the impact of Lkb1 and Sik inactivation on global gene expression programs. To uncover the gene expression changes induced by either Lkb1 deficiency or Sik targeting, we isolated Tomatopositive cancer cells from autochthonous tumors and performed RNA-seq. Analysis of the reads aligning to the targeted loci confirmed the presence of indels at Sik1 within all five Sik-targeted tumors (Supplementary Fig. S9A).
Hierarchical clustering of gene expression profiles revealed clustering of Sik-targeted tumors with Lkb1-deficient tumors (Fig. 5A). Principal component analysis also uncovered partial co-clustering of Lkb1-deficient and Sik-targeted tumors (Supplementary Fig. S9B). Initial comparison of gene expression changes elicited by Lkb1 deficiency and Sik targeting demonstrated extensive overlap of differentially expressed genes, with greater than 70% of the gene expression changes in Lkb1-deficient tumors being recapitulated in the Sik-targeted tumors (Fig. 5B–D and Supplementary Fig. S9C,D). Furthermore, direct comparison of all gene expression changes relative to KT tumors indicated a highly significant correlation between Lkb1-deficient and Sik-targeted tumors (Supplementary Fig. S9E).
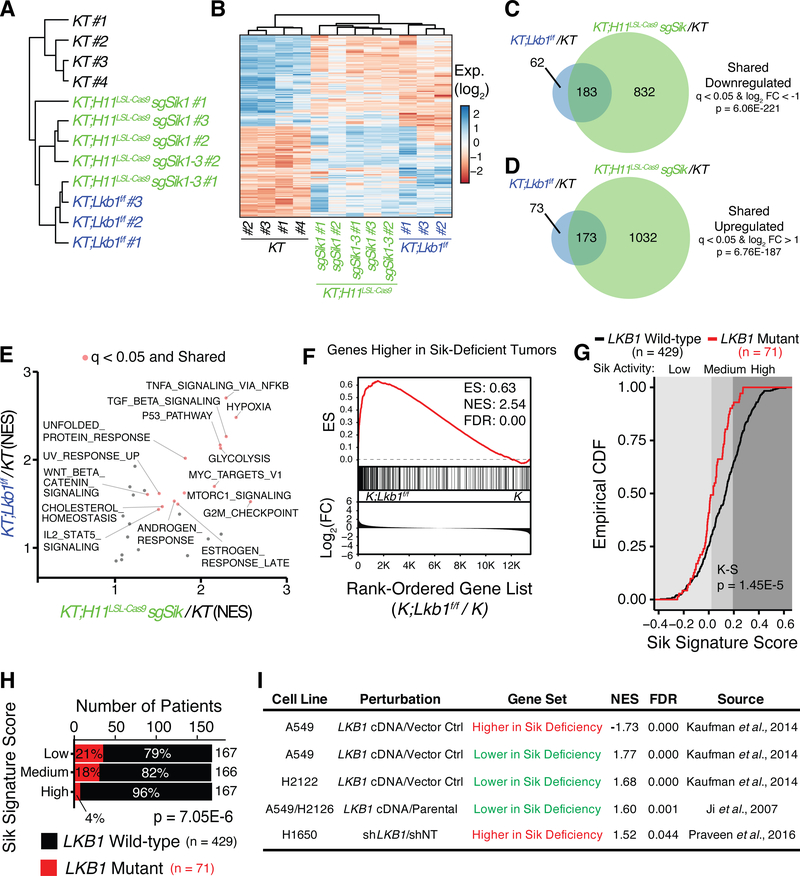
Figure 5. Sik targeting drives a transcriptional state resembling Lkb1 loss in mouse models and human lung tumors.
A. Hierarchical clustering of tumors derived from KT and KT;Lkb1flox/flox mice as well as KT;H11LSL-Cas9 mice transduced with lentiviral sgRNA vectors targeting either Sik1 alone or Sik1–3.
B. Heatmap of differentially expressed genes in KT;Lkb1flox/flox tumors relative to KT tumors depicting the conservation of differential gene expression in Sik-targeted tumors. Significant differential expression defined as an absolute log2(fold change) > 1 and q < 0.05.
C,D. Venn diagrams depicting conservation of differential gene expression relative to KT tumors between KT;Lkb1flox/flox tumors and Sik-targeted tumors initiated in KT;H11LSL-Cas9 mice. Significance cutoffs and hypergeometric test p-values are indicated.
E. Direct comparison of pathways that are higher in KT;Lkb1flox/flox tumors and Sik-targeted tumors relative to KT tumors. Each dot corresponds to a gene set derived from the mSigDB Hallmarks module. NES signifies the normalized enrichment score calculated for each gene set using GSEA. Pathways that are significantly enriched in both tumor genotypes are colored red.
F. GSEA with genes that are higher in Sik-deficient tumors on the comparison of K;Lkb1flox/flox and K tumors (7).
G. CDF plot of the distributions of signature scores for human tumors stratified by LKB1 genotype using only the downregulated subset of genes within the Sik deficiency signature. Cohort size and Kolmogorov-Smirnov test p-value are indicated (20). Background shading delineates the Sik signature score tertiles.
H. Stacked barplot summarizing the distribution of LKB1 mutant human tumors across tertiles of Sik signature scores. Chi-square test p-value is indicated.
I. GSEA using subsets of upregulated or downregulated genes within the Sik signature genes performed on external gene expression data derived from the re-expression of LKB1 in LKB1-deficient cell lines (A549, H2122, H2126) or the knocking down of LKB1 expression in an LKB1-proficient cell line (H1650)(7,14,56).
From a broader perspective, analysis of dysregulated pathways also demonstrated general agreement between the programs that are altered by Lkb1 and Sik deficiency. Consistent with the Lkb1-Sik axis constraining proliferation in lung tumors, pathways related to proliferation were upregulated in Lkb1-deficient and Sik-targeted tumors (Fig. 5E)(7,40). However, many shared upregulated pathways were unrelated to proliferation, suggesting functional overlap beyond a general increase in proliferation (Fig. 5E). Analysis of motifs enriched at the promoters of genes that were significantly upregulated in Lkb1-deficient and Sik-targeted tumors relative to KT tumors uncovered a strong enrichment for Creb/Atf family binding sites (Supplementary Fig. S9F). Several known Creb target genes were significantly upregulated in both Lkb1-deficient and Sik-targeted tumors (Supplementary Fig. S9G)(41–43). These findings align with the well-established roles for Lkb1 and Siks as negative regulators of cAMP-driven transcriptional changes (44).
To evaluate the gene expression overlap between Lkb1 and Sik deficiency using independent datasets, we generated a signature composed of genes that are differentially expressed in Sik-targeted tumors relative to control KT tumors. With this signature, we analyzed published microarray-based gene expression datasets of lung adenocarcinomas generated from different oncogenic Kras-driven genetically engineered mouse models (7,29,45). Applying GSEA, we observed a highly significant enrichment of genes that are higher in Sik-targeted tumors among those genes that are higher in Lkb1-deficient tumors relative to Lkb1-proficient tumors (Fig. 5F and Supplementary Fig. S10A). The genes that are lower in Sik-targeted tumors were also enriched among those genes that are lower in Lkb1-deficient tumors relative to other tumor genotypes (Supplementary Fig. S10A). Overall, Lkb1-deficient tumors were more similar to Sik-targeted tumors than other highly proliferative genotypes such as those with concomitant ablation of either Trp53 or p16Ink4a (Supplementary Fig. S10A). Together, these analyses indicate that the transcriptional changes resulting from Sik targeting are uniquely related to the Lkb1-deficient state.
Transcriptional state arising from Sik loss resembles that of LKB1-deficient human lung adenocarcinoma
To extend our gene expression analyses to human lung adenocarcinoma, we performed single-sample GSEA on the TCGA lung adenocarcinoma cohort to score individual patients on the basis of the genes in our Sik signature that were either higher or lower in the Sik-targeted state (20). Upon stratification of patients on the basis of LKB1 genotype, those genes that were lower in Sik-targeted tumors most appropriately changed in LKB1 mutant tumors relative to all other tumors (Fig. 5G and Supplementary Fig. S10B). Alternatively, classification of LKB1-deficient tumors on the basis of a previously published signature of LKB1 deficiency revealed that this subset of tumors exhibited significantly lower expression of Sik-dependent genes (Supplementary Fig. S10C)(46). Consistent with the derivation of our Sik signature from an oncogenic Kras-driven lung cancer model, the lower scoring of LKB1 mutant tumors relative to all others was conserved in human tumors with oncogenic KRAS alterations (Supplementary Fig. S10D).
Taking an inverse approach, stratification of the distribution of Sik activity scores across patients into tertiles revealed a significant enrichment of LKB1 mutant tumors among those with the lowest Sik signature scores (Fig. 5H). Notably, the few SIK1 mutant tumors in this cohort were binned within the two lowest Sik signature strata, in agreement with these tumors lacking functional SIK1 activity (Supplementary Fig. S10E). Our signature of Sik-dependent genes was also strongly enriched among genes that were lower in LKB1 mutant lung adenocarcinomas relative to all other tumors in three independent datasets as well as in LKB1 mutant human lung adenocarcinoma cell lines relative to LKB1-proficient cell lines (Supplementary Fig. S10F)(47–51).
We also examined whether Sik-targeted tumors exhibit features that have been previously associated with LKB1 loss in human lung cancer and mouse models thereof. Specifically, we used GSEA to assess the enrichment of gene sets generated from previous gene expression profiling studies of LKB1 deficiency (Supplementary Table S4, S5)(46,48,52–54). FOX/CREB-regulated genes were significantly higher in Sik-targeted relative to KT tumors (46). Genes related to mitochondrial function as well as molecular chaperoning and stress response were also modestly enriched in Sik-targeted tumors (48). Additionally, several chemokines and cytokines have been previously shown to be altered in the Lkb1-deficient state, such as Ppbp, Il33, Cxcl5, Lgasl9, Cccl5, and Cxcl12, were also dysregulated in Sik-deficient tumors (Supplementary Table S6)(52). In line with frequent downregulation of PD-L1 expression in LKB1 mutant human lung tumors and cell lines, we also observe lower expression of Pdl1 (Cd274) in Sik-targeted tumors (48,55). More generally, Sik-targeted tumors also have significant enrichment of those genes that are upregulated in KRAS mutant, LKB1-deficient human tumors and cell lines relative to the KRAS mutant, P53-deficient state (53) (Supplementary Table S5). Collectively, these findings demonstrate extensive parallels at the molecular level between LKB1-deficient human lung tumors and Sik-targeted tumors, further supporting the operation of Siks within an Lkb1-regulated tumor-suppressive axis.
Sik-dependent genes are regulated by LKB1
Beyond examining the enrichment of genes dysregulated by Sik deficiency in LKB1-deficient tumors, we analyzed gene expression datasets derived from LKB1-deficient human lung adenocarcinoma cell lines in which LKB1 had been re-expressed or LKB1-proficient cell lines in which LKB1 had been knocked down (7,46,56). The genes that were lower in the Sik-targeted state were highly enriched among those upregulated upon re-expression of LKB1 in three LKB1-deficient cell lines across two independent studies (Fig. 5I)(7,46). The genes that were higher in the Sik-targeted state were dramatically enriched among those downregulated upon re-introduction of LKB1 into LKB1-deficient A549 cells (Fig. 5F)(46). Complementary to these findings, the genes that were lower in Sik-targeted tumors were strongly enriched among those upregulated upon LKB1 knockdown in LKB1-proficient lung cancer cells (Fig. 5I)(56). Together, these observations suggest that LKB1 regulates genes that are dysregulated by Sik deficiency, thus further reinforcing the functional overlap between LKB1 and SIKs.
Sik-mediated tumor suppression is partially maintained in the absence of Lkb1
Our gene expression analyses substantiated a functional association between Siks and Lkb1, however we also found that Sik-targeting resulted in more extensive remodeling of the transcriptional landscape than Lkb1 loss (Fig. 5C,D and S7C–E). To address whether Siks possess additional tumor-suppressive function beyond that controlled by Lkb1, we determined whether Sik inactivation conferred a growth advantage to Lkb1-deficient tumors. We initiated tumors in KT;Lkb1flox/flox;H11LSL-Cas9 mice with either Lenti-sgControl/Cre or Lenti-sgSik1–3/Cre (Supplementary Fig. S11A). Tumor burden, as assessed by lung weight, surface tumor number, and tumor area was not significantly different between the cohorts (Supplementary Fig. S11B–E). However, the size of tumors in the Sik-targeted setting appeared larger than those initiated with Lenti-sgControl/Cre, suggesting some Sik-mediated tumor suppression in the absence of Lkb1 (Supplementary Fig. S11F).
To quantify the effect of Sik-targeting in Lkb1-deficient tumors, we employed Tuba-seq. We initiated tumors in KT;Lkb1flox/flox;H11LSL-Cas9 mice using the same pool of dual and triple Lenti-sgRNA/Cre vectors that we initially used in KT;H11LSL-Cas9 mice (Fig. S11G). Multiple metrics of tumor size revealed that targeting of Sik1 or Sik3 individually did not result in a significant increase. However, three of the four vectors targeting Sik1 and Sik3 as well as the vector targeting all three Siks increased tumor growth (Fig. S11H–I). These findings suggest that Sik-mediated tumor suppression is not entirely inactivated by Lkb1 loss (Supplementary Fig. S12). Interestingly, consistent with previous reports, we find that Sik1 mRNA levels are generally higher in the Lkb1-deficient setting (Supplementary Table S7)(46,57). This increased Sik1 expression may cooperate with an alternate input upstream of Siks to sustain some degree of tumor suppression in the absence of Lkb1 (Supplementary Fig. S12).
DISCUSSION
Despite being one of the most frequently mutated genes in human lung adenocarcinoma and a well-established familial cancer susceptibility gene, our understanding of the mechanisms by which LKB1 constrains tumorigenesis remains limited. AMPK is often viewed as a principal downstream tumor suppressor given its ability to constrain pro-growth processes during periods of energetic deficit (9,16,17). However, rather than driving proliferation, the ablation of Ampk renders cells more sensitive to metabolic perturbation and severely impairs subcutaneous tumor growth (17). Furthermore, Ampk has been shown to be required for oncogenic Kras-driven tumor growth in the lung (18). Given the growing complexity surrounding Ampk function during tumorigenesis, there has emerged a pressing need to determine which LKB1 substrates actually constrain tumor growth in vivo.
To identify Lkb1 substrates that suppress tumor growth in vivo, we employed CRISPR/Cas9-mediated genome editing in a genetically engineered mouse model of lung adenocarcinoma. By simultaneously targeting multiple paralogs across Lkb1 substrate families, we identified the salt-inducible kinases as potent suppressors of lung tumor growth. Beyond driving tumor growth, we identified extensive parallels between Lkb1 and Sik loss at the histological and gene expression levels, suggesting that Siks compose a critical tumor-suppressive arm downstream of Lkb1.
While Sik inactivation drives lung tumor growth in vivo, members of this family are rarely inactivated in human cancers (Supplementary Fig. S1C and data not shown). The functional redundancy between SIK1 and SIK3 may render them less favorable targets of direct genetic inactivation relative to LKB1. Consistent with this notion, obligate components of the LKB1 activation complex, namely STRAD and MO25, also exist as multiple paralogs and are only rarely mutated in human cancer (Supplementary Fig. S1C)(9). Beyond genetic inactivation, Sik activity can also be suppressed at the post-translational level (58). Active cAMP signaling can drive Pka-mediated inactivation of Siks, and this mechanism of Sik inactivation has been recently shown to promote the growth of pancreatic organoids in vitro and after subcutaneous transplantation (59).
Given the position of Lkb1 upstream of multiple potentially tumor-suppressive substrates, the direct genetic inactivation of Lkb1 likely leads to a greater loss of tumor suppressor activity than the inactivation of any of its substrates individually. Notably, several Lkb1 substrates have been implicated in lung carcinogenesis in other experimental contexts (Fig. 1D–F)(60,61). It is likely that different substrates play important roles during distinct phases of carcinogenesis, thus Lkb1 inactivation may drive neoplastic cells past multiple barriers during tumor growth and progression in vivo. Finally, other Lkb1-regulated substrates outside of the canonical targets investigated here could also possess tumor-suppressive properties (62).
Beyond defining the compendium of Lkb1 substrates, it remains to be fully understood how each of its substrates is regulated and under what circumstances additional pathways contribute to their regulation. Interestingly, our data are consistent with there being residual Sik-mediated tumor suppression in the absence of Lkb1, suggesting more complex regulation of Sik function (Supplementary Fig. S12). In line with our findings, alternate activation signals upstream of Lkb1 substrates have been described. For instance, AMPK retains some activity in the absence of LKB1 via activation by CAMKK2 under certain conditions. Furthermore, the existence of three kinases upstream of the AMPK ortholog, SNF1, in yeast reinforces the potential for multiple regulators of Siks and other AMPK-related kinases in different settings, including cancer (63,64).
Another outstanding question concerns what properties drive the difference in tumor-suppressive capacity among the three Sik paralogs. Our data suggest that Sik1 and Sik3, but not Sik2 contribute to Lkb1-mediated tumor growth suppression. The three SIK paralogs are structurally quite similar, harboring a highly conserved kinase domain and exhibiting greater divergence at C-terminal regions (58). In contrast to SIK2 and SIK3 which are expressed nearly ubiquitously at a relatively constant level, SIK1 expression is variable and is influenced by processes such as high salt intake, hormonal signaling, cellular depolarization, and circadian rhythms (58). Furthermore, due to the limited characterization of the Sik family, it remains poorly understood as to what extent there exists paralog-specific substrate preferences.
In line with the findings reported by Hollstein et al. (in this issue), our results highlight the tumor-suppressive function of a subset of the Sik family of kinases in lung adenocarcinoma (65). Whether these Lkb1 substrates suppress the growth of diverse tumors in Peutz-Jeghers syndrome patients and/or other sporadic cancer types should be investigated. Furthermore, more detailed investigation of the molecular context in which the Lkb1-Sik axis of tumor suppression operates may reveal new strategies to pharmacologically counteract or exploit molecular changes that arise from inactivation of this pathway. Beyond its role in cancer, Lkb1 also influences a broad range of normal processes (10–15,66,67). Future combinatorial inactivation of Siks in these biological contexts may uncover a more extensive degree of functional overlap between Lkb1 and Siks.
METHODS
Generation and validation of Lentiviral sgRNA/Cre vectors
Lenti-sgRNA/Cre vectors encoding individual sgRNAs were generated as previously described (68). Three to seven sgRNAs were screened for each of the ten targeted genes. Cutting efficiency was assessed in LSL-YFP;Cas9 reporter cells as described previously (26) (Supplementary Table S1). The primers employed for targeted sequencing of targeted loci are listed in Supplementary Table S1. The final sgRNA sequences cloned into viral vectors are listed in Supplemental Fig. S2A. The Neo2, Lkb1, and non-targeting (NT) sgRNA sequences have been previously described (26,68).
Multi-sgRNA vectors were generated by inserting additional U6-sgRNA cassettes at the Sbf1/PmeI and PacI/NotI cloning sites flanking the original U6-sgRNA cassettes of existing single-sgRNA vectors. The final vectors and the strategies used to construct them are listed in Supplementary Table S1.
For the multi-sgRNA vectors employed for Tuba-seq, individual sgRNAs were instead initially cloned downstream of the U6 promoters in pMJ114 (bovine U6), pMJ179 (mouse U6), pMJ117 (human U6) (gifts from Jonathan Weissman: (Addgene plasmid # 85995, 85996, 85997; http://n2t.net/addgene:85995, http://n2t.net/addgene:85996, http://n2t.net/addgene:85997; RRIDs:Addgene_85995, Addgene_85996, Addgene_85997) by site-directed mutagenesis (69). Prior to sgRNA cloning, the loxP site-containing region of the mouse U6 promoter of pMJ179 was reverted to its wild-type sequence. For triple-sgRNA vectors, the BamHI site within the homology region between the bU6 and mU6 modules was destroyed to prevent interference with downstream cloning of sgID-BC cassette. The U6-sgRNA cassettes to be placed in tandem were then PCR-amplified to append homology arms for Gibson assembly, and then they were inserted in tandem at the site of the original U6-sgRNA cassette within our previously described Lenti-sgRNA/Cre vector (backbone previously linearized by PCR) (68). Primer sets employed for cloning individual sgRNAs, linearizing the destination vector, and amplifying the U6-sgRNA cassettes for use in Gibson assembly are summarized in Supplementary Table S1.
The assembled multi-sgRNA vectors were then diversified via addition of sgID-BC cassettes as described previously (26). In brief, unique sgID-BC inserts flanked by BamHI and BspEI sites were produced via PCR with Lenti-sgRNA/Cre as a template using unique forward primers encoding the sgID-BC region and a universal reverse primer. The sgID-BC amplicons were then digested with BamHI and BspEI and ligated into the Lenti-sgRNA/Cre backbones that been previously linearized using BamHI and XmaI. The resultant colonies were then pooled for each vector prior to plasmid DNA extraction.
Lenti-sgRNA/Cre virus was produced as previously described by co-transfection with packaging vectors (delta8.2 and VSV-G) into 293T cells using PEI (26). Viral supernatant was collected at 48 and 72 hours after transfection, concentrated by ultra-centrifugation at 2.50E+04 RPM for 90 minutes, resuspended in PBS, and titered as described previously (26). Vectors used in pooled experiments were packaged individually to prevent the formation of chimeric species (70).
The positional control vector with sgLkb12nd driven by mU6 in the triple sgRNA configuration (sgNeo1-sgLkb12nd-sgNT) was excluded from analyses as it was later found to be recombined, impairing sgLkb12nd expression. However, the positional control vector with sgLkb11st driven by mU6 in the triple sgRNA configuration (sgNeo1-sgLkb11st-sgNT) functioned as expected.
Mice and tumor initiation
KrasLSL-G12D, p53flox, Lkb1flox, H11LSL-Cas9 and Rosa26LSL-tdTomato mice have been described (68,71–74). Lung tumors were initiated by intratracheal delivery of Lenti-sgRNA/Cre vectors (75). For our initial screen for tumor-suppressive substrates, cohorts of KT and KT;H11LSL-Cas9 mice (3–5 mice/group) were transduced with 7.7 × 104 IFU/mouse of each single- and multi-sgRNA Lenti-sgRNA/Cre vector. Tumors were also initiated in KPT and KPT;H11LSL-Cas9 mice using 5 × 103 IFU of each multi-sgRNA Lenti-sgRNA/Cre vector (Fig. 2F–K and Supplementary Fig. S5A–D). For generating lung tumors for RNA-seq, tumors were also initiated in KT and KT;Lkb1flox/flox with 2.00E+05 IFU Lenti-Cre and 1.00E+03 IFU Lenti-Cre, respectively. For Tuba-seq, KT, KT;H11LSL-Cas9, and KT;Lkb1flox/flox;H11LSL-Cas9 mice were transduced with 1.2 × 105, 6 × 104, and 2.4 × 104 IFU pooled Lenti-sgRNA/Cre, respectively. Finally, for assessing Sik-mediated tumor suppression in the absence of Lkb1 in a non-multiplexed format, KT;Lkb1flox/flox;H11LSL-Cas9 mice were transduced with 6 × 104 IFU Lenti-sgRNA/Cre. The Stanford Institute of Medicine Animal Care and Use Committee approved all animal studies and procedures.
Histology and immunohistochemistry
Lung lobes were fixed in 4% formalin and paraffin embedded. H&E, alcian blue, and Masson’s trichrome staining was performed using standard methods. Total tumor burden (tumor area/total area x 100%) and individual tumor sizes were calculated using ImageJ. Immunohistochemistry was performed on 4-μm sections with the ABC Vectastain Kit (Vector Laboratories) using the Cadenza system. The following primary antibodies were used Sik1 (Santa Cruz Biotechnology: sc-83754), Sik3 (Abcam: ab110987), Nkx2–1 (Abcam: ab76013), Hmga2 (Biocheck: 59170AP), RFP/Tomato (Rockland: 600401379), Cleaved Caspase 3 (Cell Signaling Technologies: 9661S), Histone H3 phosphorylated Serine 10 (Cell Signaling Technologies: 9701S), Cytokeratin 5 (Abcam: ab52635), p63 (Cell Signaling Technologies: 13109S), and Ki-67 (BD Biosciences: 550609). Sections were developed with DAB and counterstained with hematoxylin. The fractions of H3P- and Ki-67-positive nuclei were quantified using ImageJ, and cleaved caspase 3- and TUNEL-positive cancer cells were quantified by direct counting.
Tumor dissociation and cancer cell sorting
Tumors were dissociated using collagenase IV, dispase, and trypsin at 37°C for 30 minutes as previously described (19). Cells were stained with DAPI and antibodies against CD45 (30-F11), CD31 (390), F4/80 (BM8), and Ter119 (all from BioLegend) to exclude hematopoietic and endothelial cells. FACSAria™ sorters (BD Biosciences) were used for cell sorting.
Indel analysis of ex vivo FACS-isolated neoplastic cells was performed as follows: gDNA was isolated from at least 1.00E+04 FACS-sorted Tomatopositive cancer cells using either the DNeasy Blood and Tissue kit or the AllPrep DNA/RNA/protein kit (Qiagen). PCR primers were designed to amplify sgRNA-targeted loci resulting in 350–700bp amplicons specific to each locus. Amplicons were treated with ExoSAP-IT (Thermo Fisher) and sequenced by Sanger sequencing. Cutting efficiency was determined by TIDE analysis (http://tide.deskgen.com/).
Isolation of genomic DNA from mouse lungs and preparation of Tuba-seq libraries
Genomic DNA was isolated from bulk tumor-bearing lung from each mouse following the addition of three spike-in controls (5.00E+05 cells per control) to enable absolute quantification of cell number using Tuba-seq as described previously (26). Libraries were prepared by single-step amplification the sgID-BC region from a total of 32 μg of genomic DNA per mouse across eight 100-μL reactions using the Q5 High-Fidelity 2x Master Mix (New England Biolabs, M0494X). To enable computational removal of chimeric reads that result from index hopping during ultra-deep sequencing, the sgID-BCs were amplified using defined dual-indexing primer pairs with unique i5 and i7 indices. The PCR products were subjected to double-sided purification using Agencourt AMPure XP beads (Beckman Coulter, A63881). Purified libraries were then assessed using the Agilent High Sensitivity DNA kit (Agilent Technologies, 5067–4626) on the Agilent 2100 Bioanalyzer (Agilent Technologies, G2939BA). Individual libraries were pooled in a weighted format on the basis of lung weight, and the final pool was cleaned up using Agencourt AMPure XP beads. Libraries were sequenced on an Illumina® HiSeq 2500 to generate 150-bp paired-end reads.
Tumor barcode sequencing analysis
Only those reads containing complete sgID-BC cassettes were retained, and read pairs exhibiting mismatches in this region were discarded to minimize the impact of sequencing error. Reads were then aggregated on the basis of their random barcode (clonal identifier), representing individual tumors. Barcodes containing indels were discarded to avoid potential alignment errors and miscalculation of distances between barcodes. For any given barcode pileup, smaller barcode pileups within a distance of two nucleotides were removed to minimize the impact of PCR and sequencing errors. Measures of absolute cell number for each tumor were then calculated by multiplying the read counts for each barcode pileup by the size of the spike-in controls (5× 105 cells) and subsequently dividing by the average number of reads for the three barcodes corresponding to the three spike-in controls.
Statistical analyses were performed exclusively on tumors of greater than 500 cells. Multiple metrics of tumor size distribution were examined, including various percentiles as well as the maximum-likelihood estimate of the mean assuming a log-normal distribution of tumor size (26). P-values and standard deviations were calculated by bootstrapping individual mouse tumors for 1,000 permutations. False discovery rates were calculated using the Benjamini-Hochberg procedure.
RNA-seq on sorted cancer cells
Total RNA was prepared from FACS-sorted neoplastic cells ranging from 2.5 × 104 to 6 × 104 cells. RNA quality was assessed using the RNA6000 PicoAssay kit on the Agilent Bioanalyzer 2100 (Agilent). RNA used for RNA-seq had a mean RIN of 8.6. Total RNA (30 ng/sample) was used for cDNA synthesis using the Ovation® RNA-Seq system (NuGEN Technologies, Inc.; San Carlos, CA, USA). Two micrograms of NuGEN-amplified double-stranded cDNA was sheared using a Covaris sonicator to an average length of 400 bp and subjected to library preparation using the Illumina TruSeq™ DNA sample preparation kit according to manufacturer’s protocol and sequenced (2×100 bp) on an Illumina HiSeq 4000.
Analysis of mouse model-derived ex vivo RNA-seq datasets
Paired-end RNA-seq reads were aligned to the mm10 mouse genome using STAR under standard input parameters (76). To examine for indels at targeted loci in KT;H11LSL-Cas9 Lenti-sgSik/Cre tumors, aligned reads were visualized in IGV (77). The number of reads uniquely aligned to exons of individual genes was determined using “intersection-nonempty” mode with HTSeq against the UCSC KnownGene (mm10) transcriptome (78). The differentially expressed genes between different tumor genotypes were called by DESeq2 using the HTSeq-derived counts as input (79). The significance of overlap in terms of differentially expressed genes was computed using the hypergeometric test function in base R. The DESeq2-calculated fold changes were used to generate ranked gene lists for input into GSEA (80). The differentially expressed genes with absolute log2 fold changes greater than 1 and a false discovery rate less than 0.05 were compiled into gene lists that were input into HOMER and analyzed with the findmotifs.pl to perform motif enrichment (window of −400 bp to +100 bp from the TSS and a motif length up to 12 bp)(81). The differentially expressed genes with absolute log2 fold changes greater than 2 and a false discovery rate less than 0.05 in the comparison of KT;H11LSL-Cas9 Lenti-sgSik/Cre and KT tumors were compiled into a signature reflecting the Sik-deficient state that was utilized in the analysis of external gene expression datasets. For direct comparisons of differentially expressed genes with human tumors, a differential expression cutoff of absolute log2 fold changes greater than 1 and a false discovery rate less than 0.05 was employed.
Analysis of previously published microarray-based gene expression datasets
Gene expression data derived from tumors in humans and genetically engineered mice as well as human cancer cell lines under accession numbers GSE6135, GSE51266, GSE69747, GSE32863, GSE26939, GSE21581, GSE69552, GSE72094, GSE61913, GSE63882 were acquired from NCBI Gene Expression Omnibus using the GEOquery package (7,29,45–51,56). Differential expression was computed using limma, and the resulting log2 fold changes were used to generate ranked gene lists for input into GSEA (80,82). For direct comparisons of differentially expressed genes, a differential expression cutoff of absolute log2 fold changes greater than 1 and a false discovery rate less than 0.05 was employed.
Analysis of human lung adenocarcinoma RNA-seq-based gene expression datasets
Scaled estimates of gene expression, as well as mutation and copy number data, from The Cancer Genome Atlas lung adenocarcinoma cohort were obtained from the GDC data portal (gdc-portal.nci.nih.gov)(20). Only those patients for which gene expression data were available and LKB1 genotype could be determined were included in subsequent analyses. Patient tumors harboring homozygous deletions of or non-“Putative Passenger” alterations in LKB1 were regarded as LKB1 mutant. Patient tumors harboring amplifications of or non-“Putative Passenger” alterations in KRAS were regarded as KRAS mutant. Single-sample GSEA of GSVA was employed to calculate scores for individual patient samples based on the collective expression levels of genes within the ‘lower’ or ‘higher’ subsets of the Sik deficiency signature (83). Analysis with the Kolmogorov-Smirnov test function in base R was used to determine significant changes signature score distributions. Analysis with the Chi-Squared test function in base R was used to determine significant changes in the representation of LKB1 mutants across strata defined by Sik signature scores.
Data Availability
Next-generation sequencing data for the Tuba-Seq and RNA-Seq experiments have been deposited at Gene Expression Omnibus under accession number GSE133896.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Uncovering the effectors of frequently altered tumor suppressor genes is critical for understanding the fundamental driving forces of cancer growth. Our identification of the Sik family of kinases as effectors of Lkb1-mediated tumor suppression will refocus future mechanistic studies and may lead to new avenues for genotype-specific therapeutic interventions.
ACKNOWLEDGEMENTS
We thank the Stanford Shared FACS facility for technical support; A. Orantes for administrative support; P. Hollstein and R. Shaw for discussions; D. Petrov, D. Feldser, L. Attardi, S.H. Chiou, and members of the Winslow laboratory for helpful comments.
Financial Support
C.W.M. and I.P.W were supported by the NSF Graduate Research Fellowship Program. C.W.M. was additionally supported by an Anne T. and Robert M. Bass Stanford Graduate Fellowship. I.P.W was additionally supported by NIH F31-CA210627 and NIH T32-HG000044. J.J.B was supported by NIH F32-CA189659. C.L. is the Connie and Bob Lurie Fellow of the Damon Runyon Cancer Research Foundation (DRG-2331). R.T. was supported by a Stanford University School of Medicine Dean’s Postdoctoral Fellowship and a TRDRP Postdoctoral fellowship (27FT-0044). This work was supported by NIH R01-CA175336 (to M.M.W), NIH R01-CA207133 (to M.M.W) and NIH R01-CA230919 (to M.M.W) and in part by the Stanford Cancer Institute support grant (NIH P30-CA124435). RNA sequencing data was generated on an Illumina HiSeq 4000 that was purchased with funds from the NIH under award number S10OD018220.
Footnotes
Disclosure of Potential Conflicts of Interest
Stanford University has filed a patent on related work on which I.P.W. and M.M.W. are co- inventors. I.P.W. and M.M.W. are co-founders of and hold equity in D2G Oncology, Inc. The authors declare no additional competing interests.
REFERENCES
- 1.Hanahan D, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science 2013;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winters IP, Murray CW, Winslow MM. Towards quantitative and multiplexed in vivo functional cancer genomics. Nature Reviews Genetics 2018;19(12):741–55. [DOI] [PubMed] [Google Scholar]
- 4.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz–Jeghers syndrome. Nature 1998;391(6663):184–7. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez-Cespedes M A role for LKB1 gene in human cancer beyond the Peutz–Jeghers syndrome. Oncogene 2007;26(57):7825–32. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Research 2002;62(13):3659–62. [PubMed] [Google Scholar]
- 7.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007;448(7155):807–10. [DOI] [PubMed] [Google Scholar]
- 8.Lizcano JM, Göransson O, Toth R, Deak M, Morrice NA, Boudeau J, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR‐1. The EMBO Journal 2004;23(4):833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shackelford DB, Shaw RJ. The LKB1–AMPK pathway: metabolism and growth control in tumour suppression. Nature Reviews Cancer 2009;9(8):563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amin N, Khan A, St. Johnston D, Tomlinson I, Martin S, Brenman J, et al. LKB1 regulates polarity remodeling and adherens junction formation in the Drosophila eye. Proceedings of the National Academy of Sciences 2009;106(22):8941–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foretz M, Ancellin N, Andreelli F, Saintillan Y, Grondin P, Kahn A, et al. Short-term overexpression of a constitutively active form of AMP-activated protein kinase in the liver leads to mild hypoglycemia and fatty liver. Diabetes 2005;54(5):1331–9. [DOI] [PubMed] [Google Scholar]
- 12.Kishi M, Pan YA, Crump JG, Sanes JR. Mammalian SAD kinases are required for neuronal polarization. Science 2005;307(5711):929–32. [DOI] [PubMed] [Google Scholar]
- 13.Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 2008;457(7226):210–4. [DOI] [PubMed] [Google Scholar]
- 14.Patel K, Foretz M, Marion A, Campbell DG, Gourlay R, Boudaba N, et al. The LKB1-salt-inducible kinase pathway functions as a key gluconeogenic suppressor in the liver. Nature Communications 2014;5:4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zagórska A, Deak M, Campbell DG, Banerjee S, Hirano M, Aizawa S, et al. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Science Signaling 2010;3(115):ra25. [DOI] [PubMed] [Google Scholar]
- 16.Hardie DG. AMP-activated protein kinase—an energy sensor that regulates all aspects of cell function. Genes & Development 2011;25(18):1895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang J, Mills GB. AMPK: a contextual oncogene or tumor suppressor? Cancer Research 2013;73(10):2929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichner L, Brun SN, Herzig S, Young NP, Curtis SD, Shackelford DB, et al. Genetic analysis reveals AMPK is required to support tumor growth in murine Kras-dependent lung cancer models. Cell Metabolism 2018;29(2):285–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang C-H, Greenside PG, Rogers ZN, Brady JJ, Yang D, Ma RK, et al. Molecular definition of a metastatic lung cancer state reveals a targetable CD109–Janus kinase–Stat axis. Nature Medicine 2017;23(3):291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Cancer Genome Atlas Research N, Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511(7511):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nature Reviews Cancer 2007;7(9):654–58. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed AA, Lu Z, Jennings NB, Etemadmoghadam D, Capalbo L, Jacamo RO, et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell 2010;18(2):109–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bon H, Wadhwa K, Schreiner A, Osborne M, Carroll T, Ramos-Montoya A, et al. Salt-inducible kinase 2 regulates mitotic progression and transcription in prostate cancer. Molecular Cancer Research 2015;13(4):620–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miranda F, Mannion D, Liu S, Zheng Y, Mangala LS, Redondo C, et al. Salt-inducible kinase 2 couples ovarian cancer cell metabolism with survival at the adipocyte-rich metastatic niche. Cancer Cell 2016;30(2):273–89. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H, Liu P, Wang ZC, Zou L, Santiago S, Garbitt V, et al. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Science Signaling 2009;2(80):ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers ZN, McFarland CD, Winters IP, Naranjo S, Chuang C-H, Petrov D, et al. A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nature Methods 2017;14(7):737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers ZN, McFarland CD, Winters IP, Seoane JA, Brady JJ, Yoon S, et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nature Genetics 2018;50(4):483–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert-Ross M, Konen J, Koo J, Shupe J, Robinson BS, Wiles WGIV, et al. Targeting adhesion signaling in KRAS, LKB1 mutant lung adenocarcinoma. JCI Insight 2017;2(5):e940487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagaraj AS, Lahtela J, Hemmes A, Pellinen T, Blom S, Devlin JR, et al. Cell of origin links histotype spectrum to immune microenvironment diversity in non-small-cell lung cancer driven by mutant Kras and loss of Lkb1. Cell Reports 2017;18(3):673–84. [DOI] [PubMed] [Google Scholar]
- 30.Winslow MM, Dayton TL, Verhaak RGW, Kim-Kiselak C, Snyder EL, Feldser DM, et al. Suppression of lung adenocarcinoma progression by Nkx2–1. Nature 2011;473(7345):101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Fillmore Brainson C, Koyama S, Redig AJ, Chen T, Li S, et al. Lkb1 inactivation drives lung cancer lineage switching governed by polycomb repressive complex 2. Nature Communications 2017;8:14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boland JM, Maleszewski JJ, Wampfler JA, Voss JS, Kipp BR, Yang P, et al. Pulmonary invasive mucinous adenocarcinoma and mixed invasive mucinous/nonmucinous adenocarcinoma—a clinicopathological and molecular genetic study with survival analysis. Human Pathology 2018;71:8–19. [DOI] [PubMed] [Google Scholar]
- 33.Guo M, Tomoshige K, Meister M, Muley T, Fukazawa T, Tsuchiya T, et al. Gene signature driving invasive mucinous adenocarcinoma of the lung. EMBO Molecular Medicine 2017;9(4):462–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang DH, Sholl LM, Rojas-Rudilla V, Hall DL, Shivdasani P, Garcia EP, et al. KRAS and NKX2–1 mutations in invasive mucinous adenocarcinoma of the lung. Journal of Thoracic Oncology 2016;11(4):496–503. [DOI] [PubMed] [Google Scholar]
- 35.Imielinski M, Berger Alice H, Hammerman Peter S, Hernandez B, Pugh Trevor J, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150(6):1107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagomi T, Goto T, Hirotsu Y, Shikata D, Yokoyama Y, Higuchi R, et al. Genomic characteristics of invasive mucinous adenocarcinomas of the lung and potential therapeutic targets of B7-H3. Cancers 2018;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonzogni A, Bianchi F, Fabbri A, Cossa M, Rossi G, Cavazza A, et al. Pulmonary adenocarcinoma with mucin production modulates phenotype according to common genetic traits: a reappraisal of mucinous adenocarcinoma and colloid adenocarcinoma. The Journal of Pathology: Clinical Research 2017;3(2):139–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osoegawa A, Kometani T, Nosaki K, Ondo K, Hamatake M, Hirai F, et al. LKB1 mutations frequently detected in mucinous bronchioloalveolar carcinoma. Japanese Journal of Clinical Oncology 2011;41(9):1132–7. [DOI] [PubMed] [Google Scholar]
- 39.von Herbay A, Arens N, Friedl W, Vogt-Moykopf I, Kayser K, Müller KM, et al. Bronchioloalveolar carcinoma: a new cancer in Peutz-Jeghers syndrome. Lung Cancer 2005;47(2):283–8. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y, Xiao Q, Ma H, Li L, Liu J, Feng Y, et al. LKB1 inhibits lung cancer progression through lysyl oxidase and extracellular matrix remodeling. Proceedings of the National Academy of Sciences 2010;107(44):18892–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 2004;119(7):1041–54. [DOI] [PubMed] [Google Scholar]
- 42.Parra-Damas A, Rubió-Ferrarons L, Shen J, Saura CA. CRTC1 mediates preferential transcription at neuronal activity-regulated CRE/TATA promoters. Scientific Reports 2017;7(1):18004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naqvi S, Martin Kirsty J, Arthur J Simon C. CREB phosphorylation at Ser133 regulates transcription via distinct mechanisms downstream of cAMP and MAPK signalling. Biochemical Journal 2014;458(3):469–79. [DOI] [PubMed] [Google Scholar]
- 44.Takemori H, Okamoto M. Regulation of CREB-mediated gene expression by salt inducible kinase. The Journal of Steroid Biochemistry and Molecular Biology 2008;108(3):287–91. [DOI] [PubMed] [Google Scholar]
- 45.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell 2010;17(6):547–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaufman JM, Amann JM, Park K, Arasada RR, Li H, Shyr Y, et al. LKB1 loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT. Journal of Thoracic Oncology 2014;9(6):794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLOS ONE 2012;7(5):e36530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discovery 2015;5(8):860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Research 2012;22(7):1197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schliekelman MJ, Taguchi A, Zhu J, Dai X, Rodriguez J, Celiktas M, et al. Molecular portraits of epithelial, mesenchymal, and hybrid states in lung adenocarcinoma and their relevance to survival. Cancer Research 2015;75(9):1789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2015;35(24):3209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Research 2016;76(5):999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, Sundararaman SK, et al. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discovery 2018;9(1):34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, Engel BE, Welsh EA, Yoder SJ, Brantley SG, Chen D-T, et al. A sensitive NanoString-based assay to score STK11 (LKB1) pathway disruption in lung adenocarcinoma. Journal of Thoracic Oncology 2016;11(6):838–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discovery 2018;8(7):822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Praveen P, Hülsmann H, Sültmann H, Kuner R, Fröhlich H. Cross-talk between AMPK and EGFR dependent signaling in non-small cell lung cancer. Scientific Reports 2016;6:27514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Z, Li J-L, Lin S, Cao C, Gimbrone NT, Yang R, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. The Journal of Clinical Investigation 2016;126(6):2267–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wein MN, Foretz M, Fisher DE, Xavier RJ, Kronenberg HM. Salt-inducible kinases: physiology, regulation by cAMP, and therapeutic potential. Trends in Endocrinology & Metabolism 2018;29(10):723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patra KC, Kato Y, Mizukami Y, Widholz S, Boukhali M, Revenco I, et al. Mutant GNAS drives pancreatic tumourigenesis by inducing PKA-mediated SIK suppression and reprogramming lipid metabolism. Nature Cell Biology 2018;20(7):811–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodwin JM, Svensson RU, Lou HJ, Winslow MM, Turk BE, Shaw RJ. An AMPK-independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Molecular Cell 2014;55(3):436–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou X, Liu JE, Liu W, Liu CY, Liu ZY, Sun ZY. A new role of NUAK1: directly phosphorylating p53 and regulating cell proliferation. Oncogene 2011;30(26):2933–42. [DOI] [PubMed] [Google Scholar]
- 62.Jaleel M, McBride A, Lizcano JM, Deak M, Toth R, Morrice NA, et al. Identification of the sucrose non-fermenting related kinase SNRK, as a novel LKB1 substrate. FEBS Letters 2005;579(6):1417–23. [DOI] [PubMed] [Google Scholar]
- 63.Hong S-P, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proceedings of the National Academy of Sciences 2003;100(15):8839–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia D, Shaw RJ. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Molecular Cell 2017;66(6):789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hollstein P, et al. [Google Scholar]
- 66.Kuwako K-i, Okano H. Versatile roles of LKB1 kinase signaling in neural development and homeostasis. Frontiers in Molecular Neuroscience 2018;11:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saito Y, Nakada D. The role of the Lkb1/AMPK pathway in hematopoietic stem cells and leukemia. Critical Reviews in Oncogenesis 2014;19(5):383–97. [DOI] [PubMed] [Google Scholar]
- 68.Chiou S-H, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes & Development 2015;29(14):1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 2016;167(7):1867–82.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sack LM, Davoli T, Xu Q, Li MZ, Elledge SJ. Sources of error in mammalian genetic screens. G3: Genes|Genomes|Genetics 2016;6(9):2781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caswell DR, Chuang C-H, Yang D, Chiou S-H, Cheemalavagu S, Kim-Kiselak C, et al. Obligate progression precedes lung adenocarcinoma dissemination. Cancer Discovery 2014;4(7):781–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Research 2005;65(22):10280–8. [DOI] [PubMed] [Google Scholar]
- 73.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Development 2001;15(24):3243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 2010;468(7324):653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature Protocols 2009;4(7):1064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nature biotechnology 2011;29(1):24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell 2010;38(4):576–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 2015;43(7):e47–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462(7269):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing data for the Tuba-Seq and RNA-Seq experiments have been deposited at Gene Expression Omnibus under accession number GSE133896.