Abstract
Background
Distinctions in the etiology of triple negative versus luminal breast cancer have become well-established using immunohistochemical surrogates (notably estrogen receptor [ER], progesterone receptor [PR], and human epidermal growth factor receptor 2 [HER2]). However, it is unclear whether established immunohistochemical subtypes are the sole or definitive means of etiologically subdividing breast cancers.
Methods
We evaluated clinical biomarkers and tumor suppressor p53 with risk factor data from cases and controls in the Carolina Breast Cancer Study, a population-based study of incident breast cancers. For each individual marker and combinations of markers, we calculated an aggregate measure to distinguish the etiologic heterogeneity of different classification schema. To compare schema, we estimated subtype-specific case-control odds ratios for individual risk factors and fit age-at-incidence curves with two-component mixture models. We also evaluated subtype concordance of metachronous contralateral breast tumors in the California Cancer Registry.
Results
ER was the biomarker that individually explained the greatest variability in risk factor profiles. However, further subdivision by p53 significantly increased the degree of etiologic heterogeneity. Age at diagnosis, nulliparity, and race were heterogeneously associated with ER/p53 subtypes. The ER-/p53+ subtype exhibited a similar risk factor profile and age-at-incidence distribution to the triple negative subtype.
Conclusions
Clinical marker-based intrinsic subtypes have established value, yet other schema may also yield important etiologic insights.
Impact
Novel environmental or genetic risk factors may be identifiable by considering different etiologic schema, including cross-classification based on ER/p53.
Keywords: breast cancer, estrogen receptor, tumor suppressor protein p53, risk factors, tumor biomarkers
Introduction
Numerous studies have evaluated subtypes of breast tumors from an etiologic perspective, with many studies suggesting strong heterogeneity in risk factor associations by ER status and according to luminal A (ER+/HER2-) versus triple negative (ER-/PR-/HER2- with or without positive basal markers) (1–6).In addition to distinct risk factor profiles in population-based studies, these clinical marker-based subtype definitions show bimodal age-at-incidence frequency in large data sources such as Surveillance Epidemiology and End Results (SEER), which has been interpreted to signify residual etiologic heterogeneity even within defined clinical marker-based subtypes (7–10). Finally, some studies evaluating marker concordance of double primary breast cancers have shown that second primaries tend to share the ER status or triple negative status of the first cancer occurrence, suggesting both cancers arise from the same etiologic milieu (11–13). However, most efforts to understand etiologic heterogeneity have focused on clinical markers without considering other markers of potential etiologic significance.
The tumor suppressor p53 is mutated in 30–50% of breast cancers and tends to have high variant allele frequencies suggestive of monoclonality (14). These findings, together with evidence that p53 may define etiologic subtypes of ovarian cancers, have led to interest in p53 as a marker for breast cancer subtypes (15, 16). A previous paper used a data driven approach to evaluate ER, PR, HER2, p53 and clinical multi-marker schemes for intrinsic subtype, and found that a four-group solution best described risk factor segregation (17). The cross-classification of ER and p53 was the optimal marker combination for describing these four etiologic subgroups. We sought to evaluate these same four markers (ER, PR, HER2, and p53) in the Carolina Breast Cancer Study, a large population-based study with rich risk factor data, to assess which marker combinations showed greatest evidence for etiologic heterogeneity.
Materials and Methods
Study Population
The Carolina Breast Cancer Study (CBCS) is a population-based study conducted in North Carolina in three phases (Phase 1: 1993–1996, Phase 2: 1996–2001 and Phase 3: 2008–2013); study details and sampling schema have been described previously (18, 19). Briefly, cases were women aged 20 – 74 years diagnosed with a first primary invasive breast cancer enrolled using rapid case ascertainment. For Phases 1 and 2, controls were identified using the Driver’s License and Medicare beneficiary lists. Phase 3 did not enroll controls. Note that as a result use of Phase 3 cases in this study is limited to our analyses of age-at-incidence curves. Black and younger women (age <50) were oversampled to allow sufficient sample sizes for subset analyses. Race was determined by self-report and categorized as white or black. Less than 2% of non-black participants self-identified as multiracial, Hispanic or other race/ethnicities and were classified as white for statistical analyses. Tumor characteristics for cases (e.g., tumor size, node status, and stage) were abstracted from medical records and pathology reports. The study was approved by the Office of Human Research Ethics/Institutional Review Board at the University of North Carolina at Chapel Hill, conducted in accordance with U.S. Common Rule, and informed consent was obtained from each participant.
Risk factor data
In-person interviews were conducted by trained nurses for both cases and controls to measure body mass index (BMI) and collect medical history, family history, and other risk factor information (4, 20). BMI was classified as pre-menopausal BMI if the participant was pre-menopausal and as post-menopausal BMI if the participant was post-menopausal. The set of individual risk factors was restricted to those used in an earlier analysis of etiologic heterogeneity in a pooled case-control study population, to facilitate comparison of the results (17). We did, however, consider including mammographic density due to its importance as a risk factor but unfortunately Breast Imaging Reporting and Data System measures were only available for a limited subset of the cases.
Tumor markers and intrinsic subtypes
ER, PR, HER2, and p53 status for cases were determined from formalin-fixed paraffin-embedded tumor tissue blocks, which were previously sectioned and stained for a panel of immunohistochemical (IHC) markers at the Immunohistochemistry Core Laboratory at the University of North Carolina, Chapel Hill; details have been described elsewhere (21–23). ER, PR, and p53 were considered positive if percent positive cells was ≥ 10%, HER2 positive was defined as IHC 3+ (24). IHC “intrinsic” subtype was defined as luminal A (ER+ or PR+ and HER2-), luminal B (ER+ or PR+ and HER2+), HER2-type (ER- and PR- and HER2+), or triple negative (ER- and PR- and HER2-).
Statistical analyses
Analysis of etiologic heterogeneity
Our approach relies on a scalar measure, denoted D, that captures the extent of etiologic heterogeneity in a set of subtypes. This method has been described in detail previously and has been used in applications to kidney cancer, breast cancer, and melanoma (17, 25–27). Briefly, a multivariable polytomous logistic regression model is fit with a set of subtypes as the outcome and all available established risk factors for disease as predictors. Then the scalar measure D is calculated based on coefficients of variation and covariation of risk predictions from this model, where a larger D indicates a higher level of etiologic heterogeneity. In essence, D captures the extent to which the subtypes differ with respect to the profiles of risk factors. We calculate this measure of etiologic heterogeneity for candidate subtypes based on individual IHC tumor markers ER, PR, HER2 and p53, as well as candidate subtypes based on combinations of these tumor markers, and seek to identify the subtype solution that maximizes D. This analysis included cases and controls from phases 1 and 2 of CBCS. To test whether one (or more) tumor markers define a statistically significant increase in etiologic heterogeneity, the baseline subtypes were fixed and cases were randomly allocated to the additional marker(s) in proportion to their relative frequencies. The corresponding D value was calculated based on this random allocation. This process was repeated 10,000 times to obtain a null reference distribution. The resulting p value is the proportion of these randomly simulated values of D that exceed the observed value. Formal statistical comparisons of subtypes of such configurations with comparators such as the intrinsic subtypes are not possible since these models are not nested.
Case-control comparison
Multivariable binary logistic regression models were used to compute case-control odds ratios to compare risk factor associations by subtype, including an offset term to account for CBCS sampling schema, allowing estimate comparison with other population-based studies. The offset term represents the age- and race-based sampling probabilities for women enrolled in CBCS and is defined as the natural log of the ratio of the sampling probability for a case in the specific age-race stratum to the sampling probability for a control in the same age-race stratum. All risk factors of interest were included as predictors and the four ER/p53 subtypes were modeled as the output, with adjustment for CBCS study phase 1 versus phase 2. The same analyses were performed for the intrinsic subtypes and for subtypes defined solely by the individual IHC markers and combinations of markers. Regression parameters for each risk factor were exponentiated to obtain odds ratios as a measure of effect size. For both subtype schemes, multivariable polytomous logistic regression models without an offset term were used to calculate a p value for heterogeneity to test the null hypothesis that each risk factor has the same effect across all subtypes.
Age-at-incidence curves
Bimodality in age at incidence has been used as a proxy for the hypothesis that cases comprise a mixture of etiologically distinct subtypes (7). Two-component statistical mixture models were used to estimate the mixing proportion of early-onset and late-onset peaks within each of the ER/p53 subtypes and intrinsic subtypes for cases from all three phases of CBCS, as previously described (7, 28). We tested the performance of a single-density model versus the two-component mixture model within each subtype. Single-density and two-component mixture models were each evaluated using normal density and semi-nonparametric density parameters (adding polynomial component to allow for skewness and heavy tails in the distributions), producing a total of four models for comparison within each subtype. Models were compared using Akaike information criterion (AIC) values, with smaller AIC values indicating a better fit. We identified the top-ranking single-density model and the top-ranking two-component mixture model, and then compared the goodness of fit between these two models using the difference in their AIC values (ΔAIC), with ΔAIC >10 indicating a substantial difference in the goodness of fit between the two models. The smooth density curve estimated from the best model is plotted for each of the subtypes along with the empirical age-at-diagnosis distribution (i.e. histogram).
Double primary data
Etiologic heterogeneity between subtypes can also be detected by examining the extent to which the subtypes are similar in pairs of independent tumors from the same patient (29). The concordance odds ratio is a suitable measure of the strength of association, with higher odds ratios demonstrating higher etiologic heterogeneity. We used data from cases of metachronous contralateral breast cancer reported to the California Cancer Registry between January 1999 and December 2004, originally reported in Brown et al. and evaluated for etiologic heterogeneity by Begg et al (29, 30). This analysis was limited to the intrinsic subtypes, which were defined using IHC ER, PR, and HER2 status as above.
Statistical analysis for age-at-incidence curves was conducted in SAS version 9.4 (SAS Institute, Cary, NC). All other statistical analyses were conducted in R software version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Demographic and tumor characteristics of cases and controls from Carolina Breast Cancer Study Phases 1 and 2 can be found in Supplemental Table 1 and baseline risk factor prevalence among cases and controls is shown in Supplemental Table 2. Cases had relatively higher prevalence of younger age at menarche, nulliparity, younger age at first live birth, and never breastfeeding. We evaluated these risk factors, along with BMI, oral contraceptive use, and menopausal status.
Evaluating single markers ER, PR, HER2, and p53, we found that ER results in the highest D for discerning etiologic heterogeneity (D = 0.078; Table 1). We next considered 4-class solutions formed by cross-classifying each of the other markers with ER, and found that among 4-class solutions, the highest D resulted from the cross-classification of ER and p53 (D=0.118). The additional contribution of p53 to ER was statistically significant (p=0.002). By contrast, neither PR nor HER2 added significantly to the etiologic heterogeneity explained by ER: PR (D=0.103; p=0.190), HER2 (D=0.097, p=0.551). The 4-class IHC intrinsic subtypes (luminal A, luminal B, HER2-type, and triple negative) produced a D value substantially lower than the ER/p53 configuration (D=0.097). The extent of overlap between the ER/p53 and IHC intrinsic subtypes is displayed in Table 2. Luminal A tumors are largely p53-, though p53+ was observed in about a third of cases; luminal B tumors are evenly split between mix of p53+ and p53- cases, as are HER2 and basal-like cases.
Table 1.
D1 estimates for individual markers and subtype solutions.
| D value | |
|---|---|
| Single markers | |
| ER | 0.078 |
| PR | 0.061 |
| HER2 | 0.015 |
| p53 | 0.014 |
| Four-class solutions | |
| ER/PR | 0.103 |
| ER/HER2 | 0.097 |
| ER/p53 | 0.118 |
| Intrinsic IHC subtypes | 0.097 |
D is calculated based on a multivariable polytomous logistic regression model including age at diagnosis, age at menarche, parity, age at first live birth, months of breastfeeding, menopausal status, pre-menopausal and post-menopausal BMI, oral contraceptive use, family history of breast cancer, and race.
Table 2.
Classification of four-class subtype by immunohistochemical intrinsic subtype.
| Subtype | Luminal A N = 656 |
Luminal B N = 134 |
HER2-type N = 82 |
Triple negative N = 359 |
|---|---|---|---|---|
| ER+/p53+ | 199 (30%) | 63 (47%) | 0 (0%) | 0 (0%) |
| ER+/p53− | 383 (58%) | 55 (41%) | 0 (0%) | 0 (0%) |
| ER−/p53+ | 22 (3.4%) | 8 (6.0%) | 45 (55%) | 179 (50%) |
| ER−/p53− | 52 (7.9%) | 8 (6.0%) | 37 (45%) | 180 (50%) |
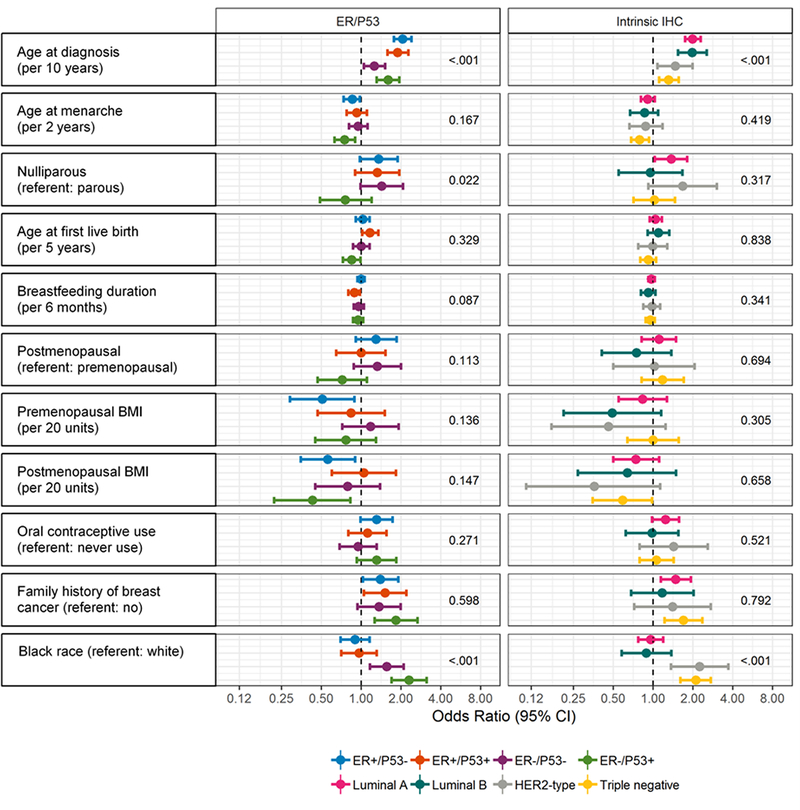
In addition to an aggregated statistical measure of heterogeneity, D, it is informative to evaluate how individual risk factor patterns differ for subtype solutions. We estimated odds ratios for each of the risk factors used in estimating D, including an offset ratio to allow for comparison of effect estimates with other cohorts (Figure 1 and Supplemental Table 3). The ER-/p53+ and triple negative subtypes showed similar risk factor profiles, with both exhibiting concordant associations with earlier age at menarche, lower post-menopausal BMI, positive family history, and black race. ER+/p53- and luminal A subtypes also showed similar risk factor profiles, though with fewer significant associations. We also formally tested for heterogeneity across subtypes within each schema (Figure 1). These analyses showed that age at diagnosis, nulliparity and race had significant heterogeneity for ER/p53 subtypes, and age at diagnosis and race had significant heterogeneity for intrinsic subtypes.
Figure 1:
Case-control odds ratios for ER/p53 and immunohistochemical intrinsic subtypes with p heterogeneity values. Odds ratios (dot) with 95% confidence intervals (whiskers) are plotted on log scale and estimate the association of each risk factor with each subtype. P heterogeneity values test the null hypothesis that each risk factor has the same effect across all subtypes.
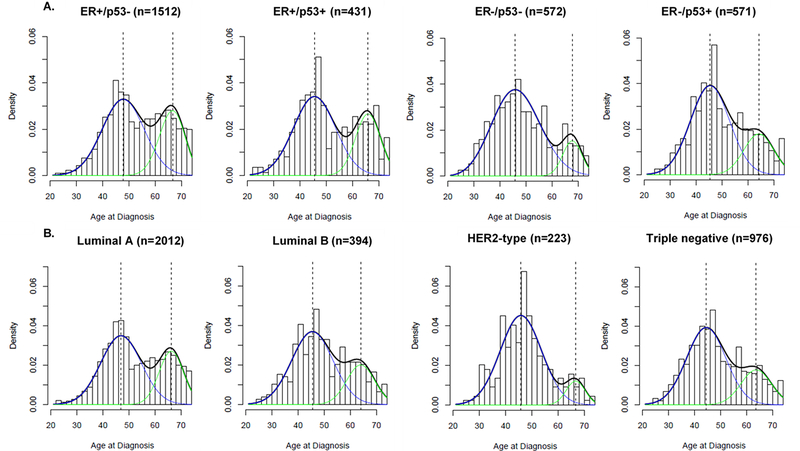
To evaluate another metric of etiologic heterogeneity, we plotted age at diagnosis distributions and applied two-component mixture models to the ER/p53 schema.Similar to the intrinsic subtypes, ER/p53 defined groups were also best fit with a bimodal model indicative of residual etiologic heterogeneity within this classification schema. Figures 2A and 2B show smoothed density plots for age at diagnosis overlaid with two-component mixture models to assess the extent of bimodality in age at diagnosis for the ER/p53 and intrinsic subtype schema, respectively. Statistical models comparing single density and two-component mixture models using AIC values are presented in Table 3. In line with previous results, we found that although some subtypes were enriched for either early or late age at onset, neither the intrinsic subtypes nor the ER/p53 subtypes captured a truly unimodal population. ER+/p53- and ER+/p53+ showed a relatively higher probability of late-onset disease, similar to the luminal subtypes, while ER-/p53- and ER-/p53+ were more enriched for early-onset disease, similar to HER2-type and triple negative subtypes.
Figure 2:
Smoothed age at diagnosis frequency distributions by ER/p53 (A.) and intrinsic subtype (B.) with two-component statistical mixture models. Smoothed density curve is plotted in black, early-onset density is plotted in blue and late-onset density is plotted in green, with dotted line representing median age at diagnosis for early- and late-onset density curves. Bar plot shows empirical distribution of age at diagnosis. Triple negative subtype and ER-/p53+ are more enriched for early-onset breast cancers, while all other subtypes more closely resemble bimodal distribution of age at diagnosis.
Table 3:
Estimates for early-onset and late-onset modes and mixing proportions by subtype.
| Total cases, n (%) | Median age at diagnosis (years) | Model fit (AIC) | Modeb (years) | Mixing proportionb | |||||
|---|---|---|---|---|---|---|---|---|---|
| AICsingle density | AICtwo-component mixture | ΔAICa (AICsingle - AICmixture) | Early onset | Late onset | Early onset | Late onset | |||
| ER/p53 subtype | |||||||||
| ER+/p53− | 1512 | 53 | 11593.4 | 11430.1 | 163.3 | 48 | 67 | 0.66 | 0.34 |
| ER+/p53+ | 431 | 49 | 3357.7 | 3299.8 | 57.9 | 46 | 66 | 0.68 | 0.32 |
| ER−/p53− | 572 | 48 | 4388.9 | 4349.6 | 39.4 | 46 | 68 | 0.86 | 0.14 |
| ER−/p53+ | 571 | 48 | 4354.5 | 4316.5 | 38.1 | 45 | 64 | 0.74 | 0.26 |
| Intrinsic subtype | |||||||||
| Luminal A | 2012 | 51 | 15499.7 | 15226.8 | 272.9 | 47 | 66 | 0.68 | 0.32 |
| Luminal B | 394 | 49 | 3012.3 | 2990.2 | 22.2 | 45 | 64 | 0.73 | 0.27 |
| HER2-Type | 223 | 48 | 1662.0 | 1649.3 | 12.8 | 46 | 67 | 0.89 | 0.11 |
| Triple negative | 975 | 48 | 7451.5 | 7374.0 | 77.5 | 44 | 64 | 0.74 | 0.26 |
Positive values favor the two-component mixture model and negative values favor the single density model, with ΔAIC >2 indicating little support for the lower-ranking model and ΔAIC >10 indicating essentially no support for the lower-ranking model.
Modes and mixing proportions are shown for the two-component mixture model, found to provide the best fit for all subtypes.
Finally, as a third assessment of etiologic heterogeneity, we assessed the concordance of intrinsic subtypes between first and second primary breast cancers. Data on independent contralateral primary breast cancers from the California Cancer Registry are displayed in Table 4, classified by intrinsic subtype. High odds ratios, indicative of greater etiologic heterogeneity, are observed for all subtype pairs except luminal A versus luminal B. These results suggest that there is no strong etiologic distinction between luminal A and luminal B tumors. Conversely the results suggest strong etiologic heterogeneity between the HER2-type subtype, the triple negative subtype, and a subtype that is a combination of luminal A and luminal B tumors. Unfortunately, p53 data were lacking in this study so it was not possible to compare intrinsic subtype results to ER/p53-defined results.
Table 4.
Concordance odds ratios1 (OR) of metachronous first and second primary breast cancers in the California Cancer Registry 1999–2004.
| Second cancer | |||||
|---|---|---|---|---|---|
| Luminal A | Luminal B | HER2-type | Triple Negative | ||
| First cancer | Luminal A | 208 | 28 | 15 | 32 |
| Luminal B | 40 OR 1.5 |
8 | 5 | 5 | |
| HER2-type | 12 OR 19.6 |
4 OR 6.8 |
17 | 9 | |
| Triple Negative | 23 OR 7.9 |
3 OR 14.9 |
5 OR 10.6 |
28 | |
The concordance odds ratio measures the alignment of the risks of the two subtypes under consideration in individuals at risk.29 For example, if the risk of one tumor type is directly proportional to the risk of the other tumor type, this corresponds to an odds ratio of 1, indicating no etiologic heterogeneity. Conversely, as the correlation of these risks becomes less strong the concordance odds ratio increases, reflecting increasingly divergent etiologies.
Discussion
We evaluated whether, in addition to intrinsic subtypes defined by ER, PR, and HER2, there are other biomarker-based classification schema that have potential value in defining etiologic groups. We found that subtypes formed by ER and p53 explained a higher degree of etiologic heterogeneity than the widely accepted IHC-defined intrinsic subtypes. Age at diagnosis, race, age at first birth, and postmenopausal BMI showed strong associations within ER/p53 subtypes, and age, race, and nulliparity exhibited significant heterogeneity across ER/p53 subtypes. Age-at-incidence density plots showed a more pronounced early-onset peak for ER-/p53+ cases, similar to HER2-type and triple negatives, while ER+ cases, similar to luminal subtypes, were enriched for late-onset disease.
Our findings are consistent with previous reports on the value of ER status in defining etiologic heterogeneity. ER is perhaps the most extensively studied breast cancer biomarker and a well-recognized indicator of etiology (1, 35). Our findings also match previous analyses using combined data from the Cancer and Steroid Hormone (CASH) and Womens’ Contraceptive and Reproductive Experiences (CARE) cohorts suggesting that ER/p53 cross-classification described more variation in aggregate risk factor profiles than intrinsic subtype (17). These findings may initially seem somewhat surprising given that analyses of p53 as an etiologic marker have been mixed and poorly concordant. For example, Furberg et al. found p53+ and p53- had largely overlapping risk factors profiles, consistent with findings by Ma et al., who reported that reproductive exposure associations did not vary by p53 status in the CARE study (36, 37). However, neither of these studies stratified on ER status, thus effects within ER-defined groups may have been masked. In contrast, a case-control study of environmental exposures and breast cancer risk by Gammon et al. found significant heterogeneity in risk for p53+ versus p53- cancer among current cigarette smokers, with greater heterogeneity noted for ER+ than for ER- cancers (38). The latter study is consistent with our own work, which suggests p53 alone is not a strong etiologic marker and that ER/p53 may help elucidate etiologic heterogeneity similar to findings for intrinsic subtypes.
Complementary to our findings, biological data from the Cancer Genome Atlas Project (TCGA) has also highlighted p53 mutation as a key event in certain breast cancer subtypes. Up to 50% of breast cancers harbored p53 pathway defects in recent TCGA analyses, with almost all of the basal-like breast cancers showing a mutation in p53 or another genomic defect in the pathway (39). In the Carolina Breast Cancer Study, we have observed that while p53 IHC status is not always positive in basal-like breast cancers, almost every basal-like breast cancer has a multi-gene RNA-based signature reflecting a defect in the p53 pathway (23). Thus, p53 may be a hallmark event for some intrinsic subtypes. It is also known that variant allele frequencies for p53 mutations are high (i.e. a high percentage of reads for a given tumor are p53 mutant) and p53 mutations frequently appear in both the primary tumor and metastases, suggesting that p53 mutation may be an early event that is highly advantageous for the tumor, leading to greater monoclonality (40, 41). Parallel implication of p53 as an important etiologic event both in the biological literature and in aggregation of breast cancer risk factors suggest that the combination of ER and p53 merits further investigation as an etiologic classification scheme. While the associations we found among ER/p53 subtypes mirror some of the risk factor differences that have been reported for triple negative versus luminal cancer, it is possible that some etiologic factors, such as germline variants or novel exposures, may show stronger association with ER/p53 defined subtypes than with intrinsic subtypes (1, 4, 31–34).
An informative next step in assessing the validity of joint ER/p53 status as an etiologic subtype schema will be to examine tumor subtype concordance among double primaries. As has been demonstrated, double primaries provide experimental evidence for risk factor heterogeneity among subtypes (29). A major advantage of this approach is that it is risk factor agnostic, i.e. it is influenced by all true risk factors but one does not need to observe them. We do recognize that the incidence of a second primary can be influenced by treatment for the first primary, a phenomenon that is likely to bias observed odds ratios towards the null, and so in examining concordance of subtypes in double primaries we must focus on strong trends. For example, treatment of an ER positive first primary with hormone therapy could reduce the chance of observing an ER positive second primary, lowering the corresponding concordance odds ratio for ER positive tumors. Our results still demonstrate very large odds ratios between the ER positive cases and the two subtypes defined by ER negative cases. However, there is no obvious reason why the odds ratio distinguishing luminal A and luminal B tumors should be affected by this bias, a result that suggests luminal A and luminal B tumors are etiologically similar. This finding is concordant with our case-control analyses that showed that the risk factor profiles of luminal A and luminal B tumors are very similar. This finding is also congruent with our global heterogeneity analysis (using the D measure) where we observed that HER2 status, which delineates Luminal A and Luminal B IHC subtypes, did not add significantly to the etiologic heterogeneity explained by ER. It is possible that HER2 has persisted as an important clinical biomarker because it is a therapeutic target but that it is not necessarily an informative etiologic marker. ER status has been shown to be highly correlated among first and second primary breast cancers, but p53 status has yet to be examined (42–45). Given that we have identified the ER/p53 schema on the basis of risk factor variation among subtypes, it will be insightful to assess the strength of this classification scheme using this risk factor agnostic method.
Our analysis has allowed comparison of the quality and strength of evidence for etiologic heterogeneity across multiple methods. Calculation of a single heterogeneity score demonstrated that the ER/p53 schema may reveal etiologic associations not captured by intrinsic subtypes, resulting in more distinct risk factor profiles. This approach appeared to detect subtle differences between ER/p53 and intrinsic subtype schema that were not evident using the age-at-incidence approach. The age-at-incidence approach focuses largely on age as a key etiologic variable and requires rather large datasets to statistically distinguish between two-population and one-population models. The results of these analyses suggest that none of the subtypes formed either by the ER/p53 schema or by the intrinsic schema are convincingly homogeneous subtypes, suggesting that further refinement of the subtypes will ultimately be necessary. The final approach we utilized, involving second primaries, may provide the most direct evidence for etiologic heterogeneity. However, this approach is limited in that second primaries are relatively uncommon and cannot reasonably account for intervening treatment events, including anti-estrogens, which may bias the types of tumors that occur as second primaries, thereby affecting conclusions about etiologic heterogeneity. Comparison of these three approaches in one study highlights that utilization of multiple approaches may provide the greatest weight of evidence in understanding etiologic subgroups.
A strength of our results is that they are derived from population-based sources. In the case of the CBCS we oversampled young and black women, allowing us to study the influence of race and age on etiology with increased power. We also had complete data utilizing a central laboratory, with sufficient sample size for ER and p53 to allow consideration of both markers. Finally, we used several different approaches to evaluate etiologic heterogeneity. Overall, our results are well aligned with recent biological insights implicating p53 as an important etiologic marker. However, there are some limitations inherent to our study. Our use of D as a measure of intrinsic etiologic heterogeneity is limited by the fact that the analysis only takes into account the risk factors available to us. Importantly, this excluded all genetic factors in addition to mammographic density. The value and ranking of D across schema may well vary with additional risk factors. We did include the major well-recognized risk factors and found very similar ranking of biomarker schema as the previous published CASH/CARE study, suggesting the results are moderately stable given the current set of known breast cancer risk factors (17). The use of double primaries to evaluate etiologic heterogeneity has no such limitations, since the aggregation of subtypes in this context is driven by all risk factors, both known and unknown.
We acknowledge also that we focused solely on IHC-defined subtypes in this article, as opposed to, for example, creating an mRNA-defined intrinsic subtype. This was due to the fact that mRNA data were available for only a relatively small subset of our cases (408 cases). When we examined this limited subset of cases the subtypes defined by IHC and mRNA demonstrated almost identical values of the heterogeneity measure D, and both were lower than the corresponding measure for the ER/p53 schema (data not shown).
In summary, we applied multiple quantitative strategies for detecting etiologic heterogeneity and found that more than one approach shows promise for highlighting etiologic groups. Consistent with prior studies, ER/p53 subtyping was robust in capturing etiologic distinctiveness among a large population-based cohort of breast cancer cases with detailed exposure data. This classification scheme may help identify novel environmental or genetic risk factors for breast cancer.
Supplementary Material
Acknowledgements
Supported by the National Cancer Institute of the National Institutes of Health under Award Number F30-CA236199 (H. Benefield), T32-CA0057726 (H. Benefield), CA008748 (C. Begg and E. Zabor) and CA163251 (C Begg. and E. Zabor). This research was funded in part by the University Cancer Research Fund of North Carolina and the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/NCI P50-CA58223). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. H. Benefield is a recipient of the Gertrude B. Elion Mentored Medical Student Research Award of Triangle Community Foundation. We are grateful to CBCS participants and study staff.
Financial Support: Supported by the National Cancer Institute of the National Institutes of Health under Award Number F30-CA236199 (H. Benefield), T32-CA0057726 (H. Benefield), CA008748 (C. Begg and E. Zabor) and CA163251 (C. Begg and E. Zabor). This research was funded in part by the University Cancer Research Fund of North Carolina and the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/NCI P50-CA58223). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. H. Benefield is a recipient of the Gertrude B. Elion Mentored Medical Student Research Award of Triangle Community Foundation.
Abbreviations
- ER
estrogen receptor
- PR
progesterone receptor
- HER2
human epidermal growth factor receptor 2
- SEER
Surveillance Epidemiology and End Results
- CBCS
Carolina Breast Cancer Study
- BMI
body mass index
- IHC
immunohistochemical
- AIC
Akaike information criterion
- CASH
Cancer and Steroid Hormone
- CARE
Womens’ Contraceptive and Reproductive Experiences
- TGCA
Cancer Genome Atlas Project
Footnotes
Conflicts of interest: None
Contributor Information
Halei C. Benefield, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC
Emily C. Zabor, Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York
Yue Shan, Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, NC.
Emma H. Allott, Centre for Cancer Research and Cell Biology, Queen’s University Belfast, Belfast, United Kingdom
Colin B. Begg, Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York
Melissa A. Troester, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC
References
- 1.Althuis MD, Fergenbaum JH, Garcia-Closas M, et al. : Etiology of Hormone Receptor–Defined Breast Cancer: A Systematic Review of the Literature. Cancer Epidemiol Prev Biomarkers 13, 2004 [PubMed] [Google Scholar]
- 2.Gaudet MM, Gierach GL, Carter BD, et al. : Pooled Analysis of Nine Cohorts Reveals Breast Cancer Risk Factors by Tumor Molecular Subtype. Cancer Res 78:6011–6021, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holm J, Eriksson L, Ploner A, et al. : Assessment of Breast Cancer Risk Factors Reveals Subtype Heterogeneity. Cancer Res 77:3708–3717, 2017 [DOI] [PubMed] [Google Scholar]
- 4.Millikan RC, Newman B, Tse C-K, et al. : Epidemiology of basal-like breast cancer. Breast Cancer Res Treat 109:123–139, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Troester MA, Sun X, Allott EH, et al. : Racial Differences in PAM50 Subtypes in the Carolina Breast Cancer Study. JNCI J Natl Cancer Inst 110:176–182, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XR, Sherman ME, Rimm DL, et al. : Differences in Risk Factors for Breast Cancer Molecular Subtypes in a Population-Based Study. Cancer Epide-miol Biomarkers Prev 16:439–482, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Anderson WF, Pfeiffer RM, Dores GM, et al. : Comparison of Age Distribution Patterns for Different Histopathologic Types of Breast Carcinoma. Cancer Epidemiol Biomarkers Prev 15:1899–1905, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Anderson WF, Rosenberg PS, Prat A, et al. : How many etiological subtypes of breast cancer: two, three, four, or more? J Natl Cancer Inst 106, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuno RK, Anderson WF, Yamamoto S, et al. : Early- and Late-Onset Breast Cancer Types Among Women in the United States and Japan. Cancer Epidemiol Biomarkers & Prev 16:1437–1442, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Dickens C, Pfeiffer RM, Anderson WF, et al. : Investigation of breast cancer sub-populations in black and white women in South Africa. Breast Cancer Res Treat 160:531–537, 2016 [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Thompson W, Semenciw R, et al. : Epidemiology of contralateral breast cancer. Cancer Epidemiol Biomarkers Prev 8:855–61, 1999 [PubMed] [Google Scholar]
- 12.Horn-Ross PL: MULTIPLE PRIMARY CANCERS Multiple Primary Cancers Involving the Breast. 1993 [DOI] [PubMed]
- 13.Trentham-Dietz A, Newcomb PA, Nichols HB, et al. : Breast cancer risk factors and second primary malignancies among women with breast cancer. Breast Cancer Res Treat 105:195–207, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Spurr L, Li M, Alomran N, et al. : Systematic pan-cancer analysis of somatic allele frequency. Sci Rep 8:7735, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell D, Berchuck A, Birrer M, et al. : Integrated genomic analyses of ovarian carcinoma. Nature 474:609–615, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernardini MQ, Baba T, Lee PS, et al. : Expression signatures of TP53 mutations in serous ovarian cancers. BMC Cancer 10:237, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Begg CB, Zabor EC, Bernstein JL, et al. : A conceptual and methodological framework for investigating etiologic heterogeneity. Stat Med 32:5039–52, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman B, Moorman PG, Millikan R, et al. : The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat 35:51–60, 1995 [DOI] [PubMed] [Google Scholar]
- 19.Hair BY, Hayes S, Tse C-K, et al. : Racial differences in physical activity among breast cancer survivors: implications for breast cancer care. Cancer 120:2174–82, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carey LA, Perou CM, Livasy CA, et al. : Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA 295:2492, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Allott EH, Cohen SM, Geradts J, et al. : Performance of Three-Biomarker Immunohistochemistry for Intrinsic Breast Cancer Subtyping in the AMBER Consortium. Cancer Epidemiol Biomarkers Prev 25:470–8, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furberg H, Millikan RC, Geradts J, et al. : Environmental factors in relation to breast cancer characterized by p53 protein expression. Cancer Epidemiol Biomarkers Prev 11:829–35, 2002 [PubMed] [Google Scholar]
- 23.Williams LA, Butler EN, Sun X, et al. : TP53 protein levels, RNA-based pathway assessment, and race among invasive breast cancer cases. npj Breast Cancer 4:13, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff AC, Hammond MEH, Schwartz JN, et al. : American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med 131:18–43, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Begg CB, Seshan VE, Zabor EC, et al. : Genomic investigation of etiologic heterogeneity: methodologic challenges. BMC Med Res Methodol 14:138, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Begg CB, Orlow I, Zabor EC, et al. : Identifying Etiologically Distinct Sub-Types of Cancer: A Demonstration Project Involving Breast Cancer. Cancer Med 4:1432–1439, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mauguen A, Zabor EC, Thomas NE, et al. : Defining Cancer Subtypes With Distinctive Etiologic Profiles: An Application to the Epidemiology of Melanoma. J Am Stat Assoc 112:54–63, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeiffer RM, Carroll RJ, Wheeler W, et al. : Combining assays for estimating prevalence of human herpesvirus 8 infection using multivariate mixture models. Biostatistics 9:137–51, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begg CB: A strategy for distinguishing optimal cancer subtypes. Int J cancer 129:931–7, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown M, Bauer K, Pare M: Tumor marker phenotype concordance in second primary breast cancer, California, 1999–2004. Breast Cancer Res Treat 120:217–227, 2010 [DOI] [PubMed] [Google Scholar]
- 31.Chen WY, Colditz GA: Risk factors and hormone-receptor status: epidemiology, risk-prediction models and treatment implications for breast cancer. Nat Clin Pract Oncol 4:415–423, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Hwang ES, Chew T, Shiboski S, et al. : Risk Factors for Estrogen Receptor–Positive Breast Cancer. Arch Surg 140:58, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Colditz GA, Rosner BA, Chen WY, et al. : Risk factors for breast cancer according to estrogen and progesterone receptor status. J Natl Cancer Inst 96:218–28, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Kerlikowske K, Gard CC, Tice JA, et al. : Risk Factors That Increase Risk of Estrogen Receptor–Positive and –Negative Breast Cancer. J Natl Cancer Inst 109:djw276, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pike MC, Spicer DV, Dahmoush L, et al. : Estrogens, progestogens, normal breast cell proliferation, and breast cancer risk. Epidemiol Rev 15:17–35, 1993 [DOI] [PubMed] [Google Scholar]
- 36.Furberg H, Millikan RC, Geradts J, et al. : Reproductive factors in relation to breast cancer characterized by p53 protein expression (United States). Cancer Causes Control 14:609–618, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Ma H, Wang Y, Sullivan-Halley J, et al. : Prevention and Epidemiology Use of Four Biomarkers to Evaluate the Risk of Breast Cancer Subtypes in the Women’s Contraceptive and Reproductive Experiences Study, 2010 [DOI] [PMC free article] [PubMed]
- 38.Gammon MD, Hibshoosh H, Terry MB, et al. : Cigarette Smoking and Other Risk Factors in Relation to p53 Expression in Breast Cancer among Young Women 1. Cancer Epidemiol Biomarkers Prev 8:255–263, 1999 [PubMed] [Google Scholar]
- 39.Koboldt DC, Fulton RS, McLellan MD, et al. : Comprehensive molecular portraits of human breast tumours. Nature 490:61–70, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegel MB, He X, Hoadley KA, et al. : Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest 128:1371–1383, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoadley KA, Siegel MB, Kanchi KL, et al. : Tumor Evolution in Two Patients with Basal-like Breast Cancer: A Retrospective Genomics Study of Multiple Metastases. PLoS Med 13:e1002174, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kollias J, Pinder SE, Denley HE, et al. : Phenotypic similarities in bilateral breast cancer. Breast Cancer Res Treat 85:255–261, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Bachleitner-Hofmann T, Pichler-Gebhard B, Rudas M, et al. : Pattern of hormone receptor status of secondary contralateral breast cancers in patients receiving adjuvant tamoxifen. Clin Cancer Res 8:3427–32, 2002 [PubMed] [Google Scholar]
- 44.Holdaway IM, Mason BH, Bennett RC, et al. : Estrogen receptors in bilateral breast cancer. Cancer 62:109–13, 1988 [DOI] [PubMed] [Google Scholar]
- 45.Swain SM, Wilson JW, Mamounas EP, et al. : Estrogen receptor status of primary breast cancer is predictive of estrogen receptor status of contralateral breast cancer. J Natl Cancer Inst 96:516–23, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.