Abstract
High-grade serous ovarian cancer (HGSOC) is often sensitive to initial treatment with platinum and taxane combination chemotherapy, but most patients relapse with chemotherapy-resistant disease. To systematically identify genes modulating chemotherapy response, we performed pooled functional genomic screens in HGSOC cell lines treated with cisplatin, paclitaxel, or cisplatin plus paclitaxel. Genes in the intrinsic pathway of apoptosis were among the top candidate resistance genes in both gain- and loss-of-function screens. In an open reading frame overexpression screen, followed by a mini-pool secondary screen, anti-apoptotic genes including BCL2L1 (BCL-XL) and BCL2L2 (BCL-W) were associated with chemotherapy resistance. In a CRISPR-Cas9 knockout screen, loss of BCL2L1 decreased cell survival while loss of pro-apoptotic genes promoted resistance. To dissect the role of individual anti-apoptotic proteins in HGSOC chemotherapy response, we evaluated overexpression or inhibition of BCL-2, BCL-XL, BCL-W, and MCL1 in HGSOC cell lines. Overexpression of anti-apoptotic proteins decreased apoptosis and modestly increased cell viability upon cisplatin or paclitaxel treatment. Conversely, specific inhibitors of BCL-XL, MCL1, or BCL-XL/BCL-2, but not BCL-2 alone, enhanced cell death when combined with cisplatin or paclitaxel. Anti-apoptotic protein inhibitors also sensitized HGSOC cells to the poly (ADP-ribose) polymerase inhibitor olaparib. These unbiased screens highlight anti-apoptotic proteins as mediators of chemotherapy resistance in HGSOC, and support inhibition of BCL-XL and MCL1, alone or combined with chemotherapy or targeted agents, in treatment of primary and recurrent HGSOC.
Implications:
Anti-apoptotic proteins modulate drug resistance in ovarian cancer, and inhibitors of BCL-XL or MCL1 promote cell death in combination with chemotherapy.
Keywords: ovarian cancer, cisplatin, paclitaxel, apoptosis, resistance
Introduction
High-grade serous ovarian cancer (HGSOC) comprises ~70% of ovarian cancers and is frequently diagnosed at an advanced stage; 5-year survival is <50% (1). Most patients with HGSOC respond to surgery and combination chemotherapy with platinum and taxanes, but >80% eventually develop recurrent disease and chemotherapy resistance, for which few effective therapies exist (1). HGSOC is characterized by TP53 mutations (nearly 100%) and defects in homologous recombination DNA repair (HRR), including BRCA1/2 mutations (1). HGSOC with HRR defects are more sensitive to platinum chemotherapy and poly (ADP-ribose) polymerase (PARP) inhibitors (1).
Numerous resistance mechanisms to platinum and taxanes have been reported in ovarian cancer, although their clinical significance is often unclear. Reversion mutations in BRCA1/2 and other genes involved in HRR have been reported to confer clinical resistance to platinum and PARP inhibitors (1,2). In addition, recurrent fusions driving ABCB1 overexpression occur in platinum-resistant HGSOC (3); ABCB1 encodes MDR1 (multidrug resistance-1, P-glycoprotein) which mediates efflux of drugs including paclitaxel and some PARP inhibitors, leading to drug resistance (4).
Anti-apoptotic proteins have also been linked to chemotherapy resistance in ovarian cancer. Platinum and taxanes cause cell death primarily via the intrinsic pathway of apoptosis (5); activity of this pathway is restrained by BCL-2 family anti-apoptotic proteins (BCL-2, BCL-XL, BCL-W, MCL1, BFL1) (5). Increased BCL-XL protein expression was observed in recurrent compared to primary ovarian cancers (6) and was associated with clinical resistance to chemotherapy (7) and decreased survival (6,7). BCL-2 overexpression correlated with poor responses to primary chemotherapy and decreased survival in ovarian cancer patients (8,9), and MCL1 expression was also associated with poor prognosis (10). In ovarian cancer cell lines (including non-high-grade serous subtypes (11)), enforced overexpression of BCL-XL conferred resistance to cisplatin or paclitaxel (6,12,13), and modulating MCL1 levels altered sensitivity to chemotherapy and targeted drugs (14–18). The role of BCL-W in ovarian cancer is unknown, though in other solid cancers BCL-W protects cells from drug-induced apoptosis (19). Targeting anti-apoptotic proteins with genetic knockdown of BCL-XL or with small molecule inhibitors of BCL-2/BCL-XL or BCL-XL enhanced sensitivity to platinum or paclitaxel in ovarian cancer cell lines (7,17,20–24) and ex vivo patient samples (23,24).
Despite the clinical use of platinum and taxanes for decades, and known mechanisms of resistance including reversion of HRR gene mutations, overexpression of ABCB1, and increased levels of certain anti-apoptotic proteins, the diverse landscape of chemotherapy resistance mechanisms in HGSOC remains incompletely understood. Correspondingly, few therapies are available to effectively target resistance pathways.
To investigate mechanisms of resistance to standard-of-care chemotherapy in HGSOC, we performed a pooled, near-genome-scale open reading frame (ORF) overexpression screen and a pooled genome-scale CRISPR-Cas9 knockout screen for mediators of resistance to cisplatin, paclitaxel, or cisplatin plus paclitaxel in HGSOC cell lines. In these unbiased screens, anti-apoptotic genes were among the most significant genes promoting resistance across all drug treatments. Based on this result, we systematically investigated the role of individual BCL-2 family anti-apoptotic proteins in HGSOC chemotherapy resistance and the effects of targeting these proteins on HGSOC survival.
Materials and Methods
Additional details are available in supplementary methods.
Genome-wide overexpression screen and mini-pool secondary screen
Kuramochi and OVSAHO cell lines were infected with a library of 17,255 individually barcoded human ORF clones in lentiviral vector pLX317 under an EF1a promoter (25,26), at a multiplicity of infection of 0.5–1. Following puromycin selection, cells were collected for an early time point sample of initial library representation, then divided into drug treatment arms: DMSO, cisplatin (0.5 μM), paclitaxel (10 nM), or cisplatin + paclitaxel combination (0.5 μM + 10 nM, respectively) in quadruplicate and treated with drug every 3–7 days for 21 days. Surviving cells were collected for DNA extraction, PCR, and sequencing of barcodes. The total number of sequencing reads for each clone was determined, normalized to reads per million, and log2-transformed. The log2 fold-change (LFC) of each ORF was determined relative to the early time point, and statistical significance (q-value) of candidate resistance ORFs was assigned using differential representation methods (see detailed methods). Candidate genes meeting the indicated criteria for LFC and q-values were further validated in a mini-pool secondary screen of 376 ORFs including candidate resistance genes and neutral and candidate sensitizing genes. Cells were infected with the mini-pool library and treated with drugs for 10–14 days, and collected for PCR and sequencing to identify resistance genes based on LFC and Z-score relative to the population.
CRISPR-Cas9 knockout screen
Kuramochi and OVSAHO cells stably expressing Cas9 were infected with a library of 73,687 barcoded sgRNAs targeting 18,454 genes and 1000 non-targeting guides (26,27). Following selection and collection of cells for the early time point, cells were divided into four drug arms in quadruplicate (cisplatin (1 μM), paclitaxel (10 nM), or two combination doses (0.5 μM + 10 nM or 0.3 μM + 5 nM cisplatin + paclitaxel)), treated with drug every 3–4 days, and harvested at 14 days for PCR and sequencing of barcodes. The enrichment or depletion of each sgRNA was determined relative to the initial time point.
Cell lines, drug treatments, and viability and apoptosis experiments
Lentiviral supernatant was produced by transient transfection of 293T cells with viral constructs and FuGene6 reagent. Cells were transduced with viral supernatant using polybrene and spin-infection by centrifugation, followed by selection with puromycin (2 μg/mL) for 4–5 days. Protein overexpression was confirmed by Western blot using standard methods and protein-specific primary antibodies. For drug experiments, drug stocks were diluted in media and added to cells manually or by HP/Tecan d300e digital dispenser. Drugs included cisplatin solution 1 mg/mL in 0.9% NaCl; and 10 mM solutions in DMSO of paclitaxel, PARP inhibitor olaparib, or anti-apoptotic protein inhibitors ABT-199/venetoclax, ABT-263/navitoclax, WEHI-539 hydrochloride, S63845, and A1331852. Cells were treated with a single dose of drug unless otherwise indicated, at physiologically relevant concentrations (28). For viability assays, cells were seeded at 10–20% confluence and 24 hours later drugs or vehicle were added at multiple doses in duplicate or triplicate; viability was assessed at 5 days using the CellTiterGlo luminescent reagent. For apoptosis assays, cells were treated with drugs 24 hours after seeding, and 24–72 hours later caspase 3/7 activity was evaluated by Caspase-Glo luminescent reagent. For colony formation assays, cells were seeded at low density and treated with vehicle or drug in triplicate once weekly for 14–21 days, fixed with 4% formaldehyde, and stained with 0.5% crystal violet. For population doubling assays, cells were seeded in duplicate, and counted and treated with drugs every 3–7 days for 14–21 days. Cell lines were genotyped and routinely tested for mycoplasma. Data were analyzed using GraphPad Prism.
BH3 profiling
BH3 profiling was performed as previously described using flow cytometry (29). Briefly, permeabilized cells were incubated with BH3 domain pro-apoptotic peptides for 60 minutes and fixed in 4–8% formaldehyde followed by neutralization. Cells were stained with AlexaFluor 647-conjugated anti-cytochrome c antibody and a viability marker and analyzed by flow cytometry. For dynamic BH3 profiling, the same procedure was performed but was preceded by incubation of the cells with drug for 24 hours prior to collection.
Genomic analysis
HGSOC genomic data were obtained from the Cancer Genome Atlas (Pan-Cancer) (30) (TCGA), Australian Ovarian Cancer Study Group (3), and Cancer Cell Line Encyclopedia (31). TCGA genomic data were analyzed with R packages or the cBioPortal. Copy-number alterations were analyzed by GISTIC 2.0. Statistical differences in parameters between groups were analyzed by t-test. Survival curves were generated by Kaplan-Meier analysis and significance assessed by log-rank test.
Results
Overexpression screen for mediators of cisplatin and paclitaxel response in HGSOC
To identify genes whose overexpression impacts response to cisplatin and paclitaxel treatment in ovarian cancer cell lines, we performed an unbiased, near-genome-scale, pooled ORF overexpression screen using a barcoded lentiviral library of 17,255 human cDNAs (25) (Fig. 1A). We selected Kuramochi and OVSAHO cell lines due to genomic similarity to HGSOC (11) and relative sensitivity to cisplatin and paclitaxel (Fig. S1A). Kuramochi has a BRCA2 mutation and BRCA1 copy loss, and OVSAHO has BRCA2 copy loss (11,31); both are deficient in HRR (32).
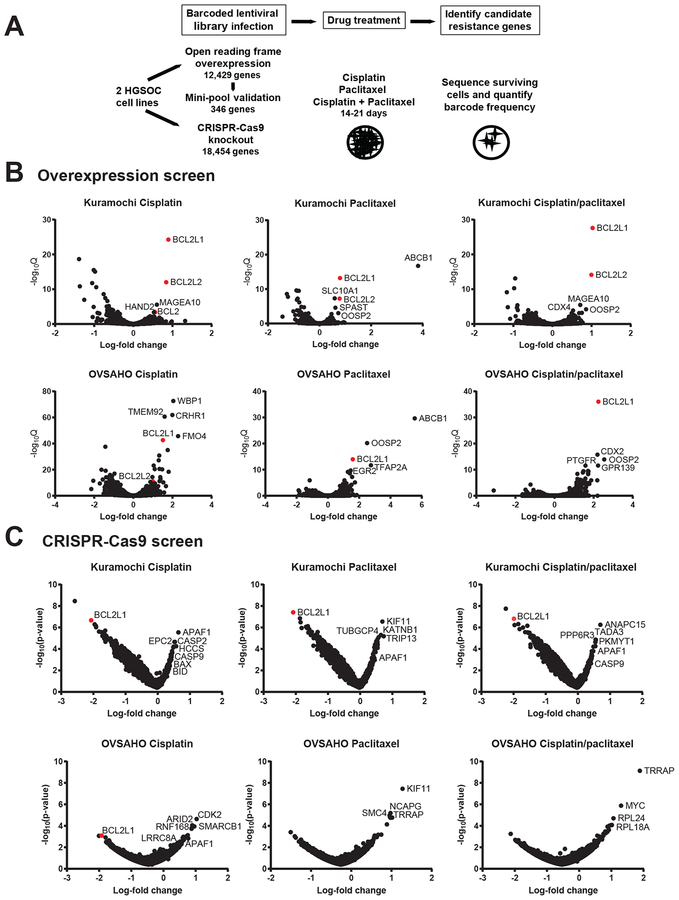
Figure 1. Overexpression and CRISPR-Cas9 screens for mediators of ovarian cancer chemotherapy resistance.
A. Schematic of primary pooled open reading frame (ORF) screen; secondary mini-pool ORF screen; and primary CRISPR-Cas9 screen for genes mediating cisplatin and paclitaxel resistance.
B. Overexpression screen results. Average log2-fold change (x-axis) compared to the early time point, versus -log10 q-value (y-axis) for all ORFs for Kuramochi and OVSAHO cell lines for each indicated drug treatment. Negative average log2-fold change indicates depletion of cells with the ORF, whereas positive average log2-fold change indicates enrichment of cells with the ORF, compared to the early time point. Candidate resistance genes are have positive log2-fold change. Anti-apoptotic genes are highlighted in red.
C. CRISPR-Cas9 screen results. Average log2-fold change (x-axis) of the guide RNAs representing each gene compared to the early time point, versus -log10 p-value (y-axis) representing statistical significance relative to the entire pool. Negative average log2-fold change indicates depletion of cells with the sgRNA, whereas positive average log2-fold change indicates enrichment of cells with the sgRNA, compared to the early time point. Anti-apoptotic genes are highlighted in red.
After lentiviral infection and selection titrated to introduce a single barcoded cDNA to each cell, the pooled cells were cultured with DMSO, cisplatin (0.5 μM), paclitaxel (10 nM), or cisplatin plus paclitaxel (0.5 μM + 10 nM) for 21 days (Fig. S1B). We collected the surviving cells post-treatment and performed sequencing of barcodes to determine representation of each cDNA clone in the post-treatment condition compared to the pre-treatment pool (Table S1). We evaluated the statistical significance of the resistance conferred by each ORF by comparing the relative representation of each ORF clone at the pre- and post-treatment time points to generate a log-fold change value and q-value (Supplementary methods; Fig. 1B, Table S2). Genes whose ORF-specific barcode showed higher abundance following drug treatment, compared to pre-treatment, were nominated as candidate resistance genes.
Among individual ORFs scoring highly in single or multiple drug treatments, ABCB1 was an outlier for paclitaxel treatment but not cisplatin treatment (Fig. 1B), consistent with the known role of MDR1 in paclitaxel resistance (4). Anti-apoptotic genes BCL2L1 (BCL-XL) and BCL2L2 (BCL-W) scored consistently across all three drug arms in both cell lines (Fig. 1B, S1C). BCL2L1 and BCL2L2 also showed a stronger positive enrichment following drug treatment compared to DMSO controls, suggesting a drug-specific survival advantage (Table S2). Additional high-scoring genes (Fig. 1B, S1C) were identified which have not been previously associated with drug resistance, e.g. OOSP2 (PLAC1L), encoding oocyte-specific protein 2.
We next aimed to test whether individual overexpression of top resistance genes from the pooled ORF screen conferred chemotherapy resistance in orthogonal assays, and whether inhibiting resistance genes could mitigate chemotherapy resistance. As proof-of-principle, we evaluated the known paclitaxel resistance gene ABCB1, which is a clinically validated resistance mechanism (3) and the top-scoring ORF in our paclitaxel screen. HGSOC cell lines infected with the ABCB1 ORF and overexpressing MDR1 (Fig. S2A) showed increased viability following paclitaxel treatment (Fig. S2B, C) and increased cell proliferation in the presence of paclitaxel in a population doubling assay (Fig. S2D) and a colony formation assay (Fig. S2E). However, treatment of ABCB1-overexpressing ovarian cancer cells with MDR1 inhibitor elacridar (1 μM) re-sensitized the HGSOC cells to paclitaxel (Fig. S2E, F). Although ABCB1-mediated drug resistance has been difficult to exploit with clinical therapeutics (4), this example suggests that our unbiased overexpression screen could detect clinically relevant, targetable mechanisms of chemotherapy resistance.
To validate the primary overexpression screen, we performed a secondary screen with a mini-pool of 376 ORFs consisting of top candidate resistance genes balanced by neutral genes and candidate sensitizer genes. We selected 191 candidate resistance ORFs with log-fold change (LFC) >0.5 (enrichment after drug treatment) and q-value<0.001 in any cell line plus drug condition (Table S2, S3), plus 48 ORFs that scored in multiple cell line/drug conditions (LFC>0.5 and q-value<0.1 in ≥2 screen arms) (Table S3). ABCB1 was excluded from the mini-pool, enabling more complete assessment of other paclitaxel resistance genes. A lentiviral pool of selected candidate resistance, sensitizer, and neutral ORFs was transduced into Kuramochi and OVSAHO cells. Following selection, cells were treated with DMSO, cisplatin (0.5 μM), paclitaxel (10 nM), or cisplatin/paclitaxel (0.3 μM + 5 nM) for 10–14 days (Fig. S3A). The surviving cells were collected for barcode sequencing to identify genes enriched (positive LFC) or depleted (negative LFC) compared to the pre-treatment pool (Table S4). Genes for which barcode abundance increased following drug treatment, relative to the abundance prior to drug exposure, were considered candidate resistance genes.
In this overexpression validation screen, anti-apoptotic genes BCL2L1, BCL2L2, and/or BCL2 were again enriched following drug treatment compared to pre-treatment (Fig. S3B; Table S4). These three genes were among the top-ranked enriched genes by log-fold change in each drug condition. Notably, BCL2L1 and BCL2L2 were the top two enriched genes by sum of the ranks across all conditions (Fig. S3C; Table S4). Additional candidate resistance ORFs such as OOSP2 were enriched in multiple drug conditions, while many candidate sensitizing ORFs were depleted following drug treatment (Fig. S3B, C; Table S4). Together, the initial pooled ORF screen and secondary mini-pool screen identified consistent candidate genes whose overexpression increases viability of HGSOC cell lines following treatment with cisplatin and/or paclitaxel, including multiple BCL-2 family anti-apoptotic genes.
CRISPR-Cas9 screen for mediators of cisplatin and paclitaxel response in HGSOC
To complement the ORF gain-of-function screen, we performed a loss-of function screen to identify genes whose loss impacts cisplatin and paclitaxel resistance in HGSOC. We performed a genome-scale CRISPR-Cas9 knockout screen with the same HGSOC cell lines and drugs, using a pooled library of 73,687 barcoded sgRNAs targeting 18,454 genes (27). HGSOC cells stably expressing Cas9 were infected with the sgRNA lentiviral pool titrated to target one sgRNA per cell, then treated with cisplatin (1 μM), paclitaxel (10 nM), or cisplatin/paclitaxel (0.5 μM + 10 nM or 0.3 μM + 5 nM) for 14 days (Fig. S4A). Using the sequencing reads of each uniquely barcoded sgRNA, we calculated log2-fold-change in abundance for each sgRNA in the surviving cells compared to the pre-treatment pool, and the q-value for statistical enrichment or depletion compared to the population (Fig. 1C, S4B; Table S5). We identified candidate genes whose sgRNA-mediated knockout promoted cell survival (positive log-fold change) and candidate genes whose knockout correlated with cell death (negative log-fold change) (Fig. 1C). Candidate genes were also ranked with the STARS algorithm (http://portals.broadinstitute.org/gpp/public/software/stars) (Table S6).
Genes whose knockout were associated with increased cell survival, and therefore may normally mediate drug-induced killing, included pro-apoptotic caspase enzymes CASP2 (caspase-2), CASP9 (caspase-9), and APAF1 (apoptotic peptidase activating factor) (Fig. 1C, S4B; Table S5, S6). Conversely, BCL2L1 was among the top genes whose knockout promoted cell death (and thus may be associated with chemotherapy resistance when intact) (Fig. 1C, S4B; Table S5, S6). This independent, genome-scale loss-of-function screen supports the significance of the intrinsic pathway of apoptosis in HGSOC resistance to chemotherapy. Other genes for which knockout was associated with response to individual drugs included spindle assembly checkpoint genes associated with paclitaxel response (33) and cisplatin importer LRRC8A associated with cisplatin resistance (34), confirming that the screen identified genes relevant to chemotherapy response.
Taken together, our systematic gain- and loss-of-function screens revealed numerous known and novel genetic mediators of cisplatin and paclitaxel response in HGSOC cell lines, and supported
anti-apoptotic signaling as a candidate pathway that could be targeted to eliminate chemotherapy-resistant HGSOC.
Effects of anti-apoptotic protein overexpression on HGSOC response to chemotherapy
Because our overexpression and knockout screens converged upon anti-apoptotic proteins in the intrinsic pathway of apoptosis as key mediators of both platinum and taxane resistance in HGSOC, and because these proteins are targets of clinically relevant small molecule inhibitors, we decided to further analyze BCL-2 family anti-apoptotic proteins in HGSOC resistance to chemotherapy.
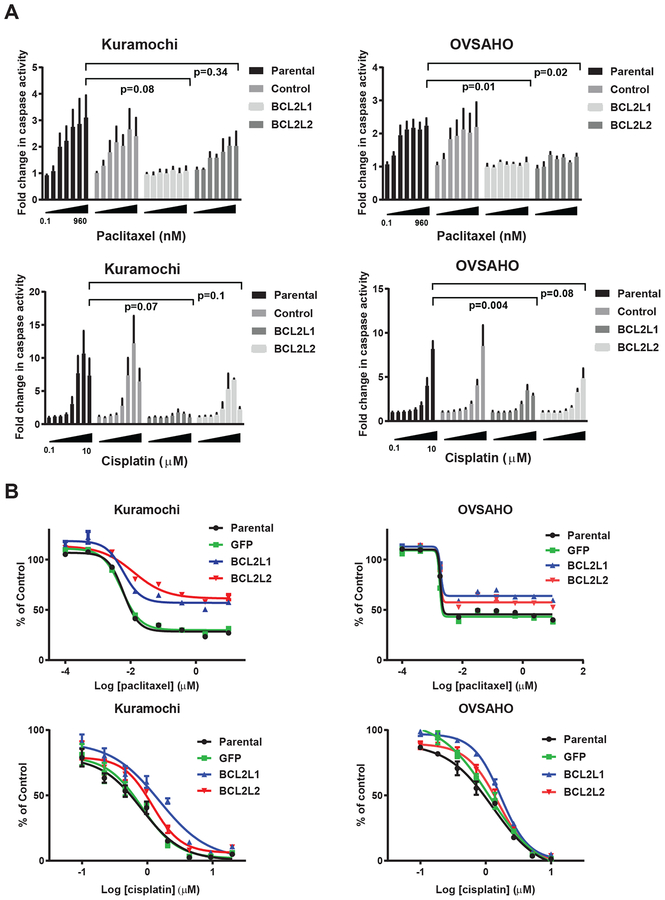
First, to evaluate the role of individual anti-apoptotic proteins in driving HGSOC chemotherapy resistance, we tested the effects of overexpression of anti-apoptotic genes in HGSOC cell lines. We first focused upon BCL2L1 and BCL2L2 as the top-ranked resistance candidates (Fig. S3C). BCL2L1 encodes a primary anti-apoptotic isoform, BCL-XL, and a minor pro-apoptotic isoform, BCL-XS. BCL2L2 encodes BCL-W, which was not previously implicated in HGSOC drug resistance. We lentivirally infected HGSOC cell lines with ORF constructs encoding BCL2L1 and BCL2L2 and confirmed protein overexpression (Fig. S5A). Cells overexpressing BCL2L1 or BCL2L2 showed decreased apoptosis by caspase 3/7 activation at 48 hours after cisplatin or paclitaxel treatment, though this difference was not statistically significant for BCL2L2 in Kuramochi cells (Fig. 2A). Overexpression of BCL2L1 or BCL2L2 increased cell viability at five days after paclitaxel treatment, and modestly increased viability after cisplatin treatment, in Kuramochi, OVSAHO, and other HGSOC cell lines (Fig. 2B, S5B).
Figure 2. Effects of overexpression of BCL2L1 and BCL2L2 on apoptosis and viability following cisplatin and paclitaxel treatment.
A. CaspaseGlo apoptosis assay for caspase 3/7 activity in parental cells or cells overexpressing BCL2L1 (BCL-XL) or BCL2L2 (BCL-W) treated with vehicle, paclitaxel, or cisplatin for 48 hours. Luminescence units were normalized to vehicle controls and expressed as fold change in caspase activity. Mean +/− SEM of 3 biological replicates. Values for parental cells were compared to cells overexpressing BCL2L1 or BCL2L2 at the highest drug dose within each figure by two-sample t-test.
B. Dose-response curves for Kuramochi and OVSAHO parental cells or cells overexpressing BCL2L1 (BCL-XL) or BCL2L2 (BCL-W), treated with paclitaxel (top) or cisplatin (bottom) once at the indicated doses (x-axis, log10 μM) followed by measurement of viability 5 days later by a CellTiterGlo ATP luminescent assay, normalized to vehicle controls (y-axis). Mean +/− standard deviation of 3 replicates; representative of at least 2 experiments. Two-tailed t-test of area under the curve (AUC) for paclitaxel: BCL-XL vs parental: Kuramochi p<0.0001, OVSAHO p<0.0001; BCL-W vs. parental: Kuramochi p<0.0001; OVSAHO p=0.0011. Two-tailed t-test of AUC for cisplatin: BCL-XL vs parental: Kuramochi p=0.0005, OVSAHO p=0.0001; BCL-W vs. parental: Kuramochi p=0.0031, OVSAHO p=0.0027.
We next applied BH3 profiling as a functional assay of mitochondrial priming for apoptosis, in which cytochrome c release from mitochondria is measured following stimulation with BH3-domain pro-apoptotic peptides (29,35). In prior BH3 profiling studies of primary ovarian cancers, greater pre-treatment apoptotic priming correlated with improved chemotherapy response and progression-free survival (35), and increased priming following carboplatin treatment correlated with clinical response to carboplatin and paclitaxel (29), supporting that apoptotic priming may impact chemotherapy response in HGSOC. Overexpression of BCL2L1 markedly decreased apoptotic priming following BIM pro-apoptotic peptide treatment of Kuramochi and OVSAHO cells, and overexpression of BCL2L2 slightly diminished priming (Fig. S5C), indicating that overexpression of anti-apoptotic proteins can reduce mitochondrial priming in HGSOC.
We also tested genes encoding other key BCL-2-family anti-apoptotic proteins, BCL2 and MCL1, using N-terminal FLAG-tagged lentiviral ORF constructs to overexpress each protein in our HGSOC cell lines (Fig. S5D). BCL2 or MCL1 overexpression decreased caspase 3/7 activity after paclitaxel treatment, which was statistically significant in OVSAHO cells (Fig. S5E). With cisplatin treatment, BCL2 overexpression decreased caspase 3/7 activity, while MCL1 overexpression had minimal effect (Fig. S5E). (Fig. S5E). BCL2 or MCL1 overexpression increased viability following paclitaxel treatment, but did not markedly impact response to cisplatin (Fig. S5F). In the ORF screen, a BCL2 ORF scored highly in the primary screen and mini-pool (Fig. 1B, S3B; Table S1–4), while MCL1 did not score in the screen nor mini-pool (potentially a false-negative due to screen conditions or translation efficiency of the ORF construct, especially as MCL1 has a short half-life). The effects on cell viability did not seem to be mediated by slower cell cycle progression, as untreated cells overexpressing each anti-apoptotic protein had similar cell cycle profiles as parental cells (Fig. S6).
Collectively, these data suggest that overexpression of specific anti-apoptotic proteins confers chemotherapy resistance via decreased apoptosis. Of note, overexpression of anti-apoptotic genes occurs frequently in HGSOC tumors, including focal amplifications and copy gains of BCL2L1 and MCL1 in patient samples and cell lines (Fig. S7, S8) (36), although such events were not significantly increased in a small cohort of chemotherapy-resistant HGSOC patients (Fig. S9). Hence, overexpression of anti-apoptotic proteins – particularly BCL-XL and MCL1 – may contribute to cancer cell survival in HGSOC patients.
Effects of inhibiting individual anti-apoptotic proteins on cisplatin and paclitaxel response
Since increased levels of anti-apoptotic proteins appear to promote platinum and taxane resistance in HGSOC cell lines, we next investigated whether, conversely, inhibiting anti-apoptotic proteins increases cell death, either with or without concomitant chemotherapy. Inhibitors of anti-apoptotic proteins are in pre-clinical and clinical development, including BCL-2 inhibitor venetoclax (ABT-199); BCL-2/BCL-XL inhibitor navitoclax (ABT-263); MCL1 inhibitors (AMG176, S64315, AZD5991); and BCL-XL inhibitors (WEHI-539, A1331852) (21,37,38).
First, we examined the effects of inhibition of individual BCL-2 family anti-apoptotic proteins on survival of ovarian cancer cell lines in the absence of chemotherapy. In our panel of ovarian cancer cell lines, individual inhibitors including ABT-199, ABT-263, A1331852, and S63845 had little effect on viability at five days after treatment with concentrations less than 2 μM (Fig. S10A).
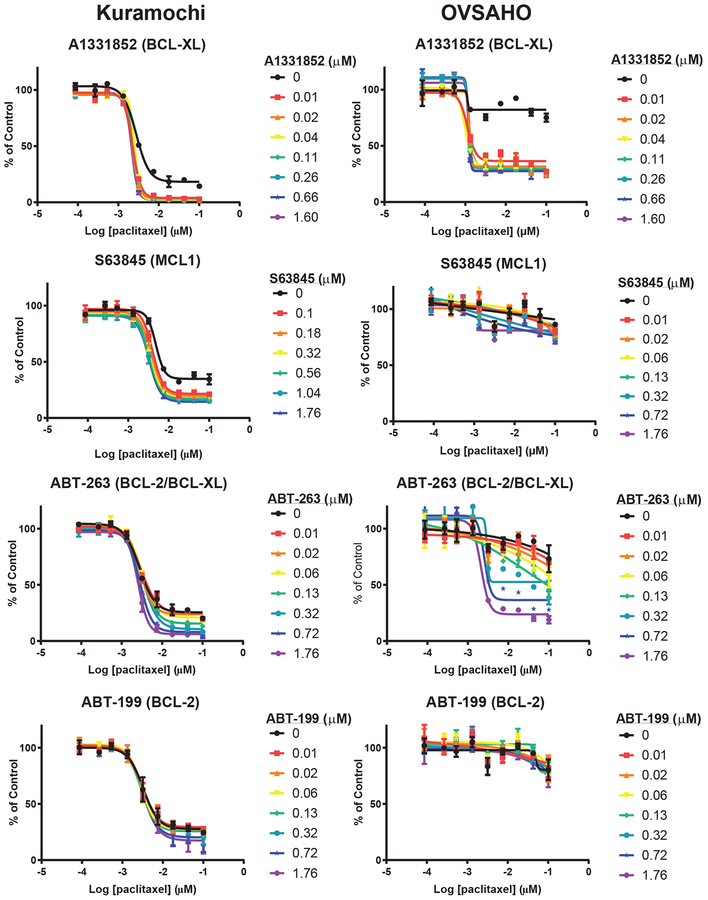
Next, we addressed whether inhibiting anti-apoptotic proteins may enhance chemotherapy-induced apoptosis. We tested whether treatment with specific anti-apoptotic protein inhibitors alters the sensitivity of Kuramochi or OVSAHO cells to chemotherapy by treating cells with a range of doses of cisplatin or paclitaxel, combined with each anti-apoptotic protein inhibitor at a series of concentrations (0–2 μM) that do not significantly affect viability as single agents (Fig. S10A). Adding BCL-XL, BCL-2/BCL-XL, or MCL1 inhibitors to cisplatin or paclitaxel treatment decreased viability of HGSOC cells, while inhibition of BCL-2 alone had little effect (Fig. 3, 4). Hence, inhibition of BCL-XL or MCL1, but not BCL-2, increased cell killing of HGSOC in combination with cisplatin or paclitaxel, and may represent an effective therapeutic strategy.
Figure 3. Effects of anti-apoptotic protein inhibitors on response to cisplatin.
Dose-response curves for Kuramochi and OVSAHO cell lines treated once with cisplatin (x-axis, log10 μM) plus increasing doses of anti-apoptotic protein inhibitors (colored lines), followed by measurement of viability normalized to vehicle controls (y-axis) at 5 days. Mean +/− standard deviation of 2 replicates; each experiment was performed at least twice.
Figure 4. Effects of anti-apoptotic protein inhibitors on response to paclitaxel.
Dose-response curves for Kuramochi and OVSAHO cell lines treated once with paclitaxel (x-axis, log10 μM) plus increasing doses of anti-apoptotic protein inhibitors (colored lines), followed by measurement of viability normalized to vehicle controls (y-axis) at 5 days. Mean +/− standard deviation of 2 replicates; each experiment was performed at least twice.
We applied an orthogonal approach, dynamic BH3 profiling, to probe the effects of combining anti-apoptotic protein inhibition by specific pro-apoptotic peptides with chemotherapy treatment (29,35). We applied BH3 profiling with promiscuous peptides that inhibit all BCL-2 family anti-apoptotic proteins (BIM, PUMA), or selective peptides that only inhibit a subset of proteins (HRK (BCL-XL), MS1 (MCL1), BAD (BCL-2, BCL-XL, BCL-W)), to interrogate anti-apoptotic dependencies in HGSOC cell lines. Kuramochi cells showed cytochrome c release upon exposure to BIM, PUMA, and BAD but not MS1, indicating mild dependence on BCL-2, BCL-XL and/or BCL-W, while OVSAHO cells had very low cytochrome c release with all peptides, suggesting inherent apoptosis resistance (Figure S10B). Treatment with carboplatin (up to 100 μg/mL for 24 hours) slightly increased BIM- and BAD-induced cytochrome c release in Kuramochi but not OVSAHO cells (Figure S10B), demonstrating that pro-apoptotic signaling is being induced by this treatment and thereby increasing dependence on pro-survival BCL-2 family proteins. We next compared these responses to those of the HGSOC cell line OVCAR3, which is primed for apoptosis. We found that cytochrome c release increased in response to BIM peptide when cells were treated with carboplatin for 24 hours (Fig. S10B). These BH3 profiles support that HGSOC cell lines can exhibit a functional apoptotic dependency upon specific anti-apoptotic proteins, and that this dependency can be further increased by carboplatin treatment in some contexts. Combination treatment strategies with chemotherapy and specific anti-apoptotic protein inhibitors can potentially exploit these induced dependencies.
To investigate factors determining response to combination treatment with chemotherapy plus anti-apoptotic protein inhibitors, we tested the impact of overexpressing each anti-apoptotic protein on the efficacy of the combinations. Upon overexpression of the target protein of each inhibitor (e.g. BCL-XL overexpression with A1331852), sensitivity to the combination treatment was maintained or enhanced (Fig. S11). However, overexpression of a non-targeted anti-apoptotic protein mitigated the sensitizing effect of the inhibitor on chemotherapy response (e.g. MCL1 overexpression reduced the efficacy of a chemotherapy plus BCL-XL inhibitor combination) (Fig. S11). These data support that baseline expression levels and/or genomic alterations in different members of the BCL-2 anti-apoptotic protein family (Fig. S7–9) may moderate the response to combination regimens with anti-apoptotic protein inhibitors, and therefore may be candidate biomarkers of effectiveness of these treatments (16,18).
Taken together, these data suggest that combining anti-apoptotic protein inhibition, particularly specific inhibitors of BCL-XL or MCL1, with platinum or taxane chemotherapy may promote apoptotic cell death in HGSOC and may represent a promising therapeutic strategy in certain patients.
Overexpression of anti-apoptotic proteins is associated with resistance to multiple drugs
Apoptosis is the final common pathway of cell death induced by multiple classes of drugs beyond platinum and taxanes. To examine correlations between anti-apoptotic gene expression and drug response, we used gene expression and drug sensitivity data for a large panel of cancer cell lines (39). Interestingly, BCL2L1 expression level was the top predictor of resistance to a variety of drugs in a panel of solid tumor cell lines (Fig. 5A). Drugs with high correlations between BCL2L1 expression and resistance included microtubule inhibitors such as paclitaxel and vincristine, DNA crosslinkers (mitomycin C), anthracyclines (doxorubicin), and inhibitors of cyclin-dependent kinases (CDKs). The correlation between BCL2L1 expression and multi-drug resistance was maintained when considering only ovarian cancer cell lines. These data suggest that BCL-XL may mediate resistance to multiple clinically relevant therapeutics in ovarian cancer.
Figure 5. Effects of anti-apoptotic proteins on response to chemotherapeutics and PARP inhibitors.
A. Relationship between BCL2L1 expression levels and drug response in cancer cell lines. Expression of 18,120 genes was evaluated for correlation with drug response in 658 solid tumor cell lines (39); each point indicates one of the 413 drugs tested. Plot shows the rank of BCL2L1 among all genes for correlation with the response to a given drug (x-axis), and an enrichment z-score of the correlation between expression and drug response (y-axis). A higher z-score indicates a stronger correlation of the selected gene with the drug response compared to the mean correlation of all genes. BCL2L1 expression was the top predictor of compound resistance in over 40 drugs tested in this assay, including microtubule agents (paclitaxel) and anthracyclines (doxorubicin).
B. Dose-response curves for parental cells treated with PARP inhibitor olaparib (x-axis) plus increasing doses of anti-apoptotic protein inhibitors (colored lines), followed by measurement of viability normalized to vehicle controls (y-axis) at 5 days. Mean +/− standard deviation of 2 replicates; each experiment was performed at least twice.
Due to the clinical significance of PARP inhibitors in ovarian cancer, we asked whether inhibition of BCL-XL or other anti-apoptotic proteins may sensitize to these drugs. Prior studies showed that ABT-263 is synergistic with PARP inhibitor BMN-673 (talazoparib) (40) and BCL-2/BCL-XL inhibitors ABT-263 or ABT-737 also synergize with several other targeted agents in HGSOC cell lines, including CDK inhibitor dinaciclib (41), PI3K/mTOR inhibitors (16,42), and MEK inhibitors (43). We interrogated the effects of overexpression or inhibition of each BCL-2 family anti-apoptotic protein on the response to PARP inhibitor olaparib. In our HGSOC cell lines, overexpression of several anti-apoptotic proteins resulted in a small increase in viability following olaparib exposure compared to parental cells, as did ABCB1 (olaparib is an MDR1 target) (Fig. S12A). Similar to the data for cisplatin and paclitaxel, inhibitors of BCL-XL, MCL1, or BCL2/BCL-XL, but not BCL-2 alone, increased sensitivity of HGSOC cells to olaparib (Fig. 5B), though concomitant overexpression of anti-apoptotic proteins mitigated this effect (Fig. S12B). These data support that BCL-2 family anti-apoptotic proteins may mediate resistance to multiple clinically relevant drugs, and that combination strategies with BCL-XL or MCL1 inhibition may enhance drug efficacy in HGSOC.
Discussion
Chemotherapy resistance is an important determinant of poor outcomes in HGSOC, but the mediators of chemotherapy resistance are incompletely understood. Large-scale functional genomic screens are an efficient, unbiased approach to defining the landscape of drug resistance mechanisms in cancer. Several genomic screens have explored chemotherapy resistance factors in a variety of cancers, revealing candidate mediators of taxane resistance such the spindle assembly checkpoint(33), and mediators of platinum resistance such as DNA repair proteins. Screens focusing on ovarian cancer platinum and taxane resistance include an shRNA screen in BRCA2-mutant HGSOC identifying loss of CHD4 as a cisplatin resistance factor (44), a CRISPR-Cas9 screen identifying loss of DYNLL1 as a mediator of platinum and PARP inhibitor resistance in BRCA1-mutant HGSOC cell lines (45), a kinome siRNA screen identifying loss of ATR and CHK1 checkpoint proteins as cisplatin sensitizers (46), and an siRNA screen showing that microtubule regulators impact paclitaxel responses (47).
In this study, we performed parallel near-genome-scale pooled ORF and genome-wide CRISPR-Cas9 screens in BRCA2-altered HGSOC cell lines to identify genes whose overexpression or knockout enhances survival following treatment with cisplatin, paclitaxel, or cisplatin plus paclitaxel. We identified candidate genes promoting resistance to platinum and taxane chemotherapy in HGSOC. BCL-2 family anti-apoptotic proteins were among the most strongly enriched resistance genes in each screen, highlighting the critical role of this pathway in mediating HGSOC response to chemotherapy. Enforced overexpression of BCL2L1 and BCL2L2, and to a lesser extent BCL2 and MCL1, decreased cell death in HGSOC cells following cisplatin and paclitaxel treatment, suggesting that elevated levels of anti-apoptotic proteins can promote chemotherapy resistance in HGSOC. The differences in viability in short-term assays were relatively modest, possibly due to baseline or enforced expression levels of anti-apoptotic proteins, intrinsic resistance to platinum and taxanes, the balance of pro-and anti-apoptotic proteins, or the involvement of other cell death pathways. Nonetheless, the pooled screen over a longer time frame may better model survival of HGSOC cells following repeated exposure to chemotherapy than the short-term viability assay. Our study provides the first evidence for BCL2L2 (BCL-W) as a potential resistance factor in ovarian cancer. Though BCL2L2 is expressed at lower levels than BCL2L1 and MCL1 in ovarian cancers, it may affect drug responses if aberrantly overexpressed, and further study of its role in apoptosis and drug resistance may be warranted. Our study and others suggest that anti-apoptotic proteins comprise a clinically relevant chemotherapy resistance pathway that can potentially be targeted in HGSOC to mitigate resistance.
From a therapeutic perspective, a subset of ovarian cancers may be vulnerable to inhibition of individual anti-apoptotic proteins, particularly BCL-XL and, less frequently, MCL1. A phase II study of single-agent BCL-2/BCL-XL inhibitor ABT-263 (navitoclax) showed modest activity in heavily pre-treated patients with recurrent ovarian cancer (48). To more specifically target the key anti-apoptotic proteins in HGSOC, recently developed inhibitors selective for BCL-XL or MCL1 may merit pre-clinical investigation in ovarian cancer, along with identification of biomarkers to select patients likely to respond to single-agent inhibitors. For the majority of ovarian cancer patients, however, strategies combining anti-apoptotic protein inhibitors with chemotherapy or targeted agents may be the most efficacious approach to increase apoptotic cell death in ovarian cancer cells. Previous studies showed that BCL-2/BCL-XL and BCL-XL inhibitors sensitize ovarian cancer cells to platinum or taxanes (7,17,20–24) but BCL-2 inhibition does not (20). We used a panel of specific inhibitors to show that inhibition of BCL-XL, MCL1, or BCL-2/BCL-XL can increase the response of HGSOC cells to cisplatin and paclitaxel. However, BCL-2 inhibition did not have this effect, consistent with the fact that BCL2 is not highly expressed in ovarian cancer, thus BCL-2 is likely not an optimal therapeutic target for combination treatment of HGSOC.
Although we initially focused upon chemotherapy resistance, our data and others suggest that anti-apoptotic proteins may also prevent cell death due to targeted agents, and that inhibitors of anti-apoptotic proteins can enhance sensitivity to targeted drugs. In ovarian cancer, effective combination strategies have been demonstrated between BCL-2/BCL-XL inhibition and PARP inhibitors (40), CDK inhibitors (41), PI3K/mTOR inhibitors (16,42), and MEK inhibitors (43); in some cases, the combinations were effective against cell lines resistant to the targeted drugs. Our data supported that specific inhibition of BCL-XL or MCL1 enhanced response to PARP inhibitor olaparib in HGSOC cell lines, suggesting a potential therapeutic combination, if hematologic toxicities can be managed. For example, thrombocytopenia related to ABT-263 may be improved by pre-treatment with an anti-apoptotic protein inhibitor or by staggered administration of two agents (49).
Synergy between chemotherapy and anti-apoptotic protein inhibitors may be limited by overexpression of other anti-apoptotic proteins in the BCL-2 family, which can confer cross-resistance, as observed in our study and others (16,18). Combinations of two anti-apoptotic protein inhibitors may overcome the resistance conferred by elevated levels of anti-apoptotic proteins. For example, a combination of ABT-263 and an MCL1 inhibitor was effective in sensitizing HGSOC patient-derived xenograft cells to PI3K/mTOR inhibition, especially in cells with higher MCL1 expression (16), and combined siRNA to BCL-XL and MCL1 sensitized ovarian cancer cell lines to cisplatin (50). Further studies are needed to identify the best biomarkers for response to anti-apoptotic protein inhibitors and combinations, including anti-apoptotic protein levels and functional assays such as BH3 profiling.
In summary, functional genomic screens for mediators of cisplatin and paclitaxel resistance highlighted the importance of the intrinsic pathway of apoptosis in HGSOC response to chemotherapy. Systematic overexpression and inhibition of BCL-XL, BCL-W, MCL1, or BCL-2 demonstrated that overexpression of anti-apoptotic proteins can increase chemotherapy resistance, while inhibition of BCL-XL or MCL1 may increase apoptosis in HGSOC cells treated with cisplatin, paclitaxel, or PARP inhibitors. BCL-XL and MCL1 appear to be the most critical anti-apoptotic proteins in HGSOC cell lines and the genes encoding these proteins are amplified and overexpressed in a subset of HGSOC; BCL-2 or BCL-W may be contributory in some contexts. Targeting anti-apoptotic proteins, particularly BCL-XL and/or MCL1, in combination with chemotherapy or other agents may be a promising therapeutic strategy in ovarian cancer.
Supplementary Material
Acknowledgements
We acknowledge collaborators for helpful discussions: Joan Brugge, Dipanjan Chowdhury, Claudia Iavarone, Whitney Henry, Colles Price, Kentaro Ito, Vasanthi Viswanathan, Alex Spektor, Daniel Stover, Panagiotis Konstantinopoulos, Joyce Liu, Kyle Davis, Uri Ben-David, Justin Hwang; John Daley, Suzan Lazo, and the DFCI Flow Cytometry Core Facility; and members of the Garraway, Meyerson and Letai laboratories, the Broad Institute Cancer Program, and the Boston HGSOC working group.
Financial support: This work was supported by the KL2/Catalyst Medical Research Investigator Training award (an appointed KL2 award) from Harvard Catalyst (The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health Award KL2TR001100)), Rivkin Center for Ovarian Cancer Scientific Scholar Award, American Society of Clinical Oncology Young Investigator Award, Kathryn Fox Samway Foundation, and Susan F. Smith Fellowship to E.H.S.; Victorian Cancer Agency Early Career Seed Grant (ECSG15012) and National Health & Medical Research Council Project Grant (APP1124309) to E.L.C; Stand Up To Cancer (SU2C) and The V Foundation (TVF) SU2C-TVF Convergence Scholar Awards (D2015–037) and Ramon y Cajal Programme, Ministerio de Economia y Competitividad (RYC-2015–18357) to J.M.; Ruth L. Kirschstein Postdoctoral Individual National Research Service Award F32 (F32CA180733) to J.L.G.; National Cancer Institute K99 (K99CA222554) to I.K.Z.; Breast Cancer Research Foundation to U.A.M.; National Health and Medical Research Council of Australia Program Grant (APP1092856) and NHMRC Research Fellowship Grant (APP1117044) to D.D.L.B.; National Cancer Institute Clinical and Translational Science Award U54 (1U54CA224068) and R01 (1R01CA219943) to C.M.J; National Cancer Institute R35 (1R35CA197568) and an American Cancer Society Research Professorship to M.M. This work was conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Foundation in Mexico.
Conflict of Interest Disclosure Statement: J.L.G. is a consultant for GlaxoSmithKline and receives sponsored research support from GlaxoSmithKline and Eli Lilly. J.M. is a consultant for Oncoheroes Biosciences and Vivid Biosciences. U.A.M. is a consultant for Merck, Immunogen, Fujifilm, Geneos, 2× Oncology, and Mersana and receives research support from Merck. A.L. has worked as a consultant and his lab has received research support from AbbVie, Novartis, and Astra-Zeneca; he is an equity holder and co-founder of Flash Therapeutics and Vivid Bioscience. D.D.L.B. receives research support from AstraZeneca, Beigene, and Genentech Roche. L.A.G. is an employee and shareholder at Eli Lilly and Company and is a co-founder and equity holder of Tango Therapeutics; previously he was a co-founder and equity holder at Foundation Medicine. M.M. is a consultant for OrigiMed and receives research support from Bayer, for whom he has consulted in previous years, and from Ono Pharma. He was previously a consultant for and equity holder in Foundation Medicine, and was previously a consultant for and receiving research support from Novartis. He receives post-marketing royalties for patents on EGFR mutation in lung cancer, from LabCorp, and is also an inventor on other patents and patent applications.
References
- 1.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov 2015;5(11):1137–54 doi 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol 2011;29(22):3008–15 doi 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015;521(7553):489–94 doi 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 4.Ween MP, Armstrong MA, Oehler MK, Ricciardelli C. The role of ABC transporters in ovarian cancer progression and chemoresistance. Crit Rev Oncol Hematol 2015;96(2):220–56 doi 10.1016/j.critrevonc.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Sarosiek KA, Ni Chonghaile T, Letai A. Mitochondria: gatekeepers of response to chemotherapy. Trends Cell Biol 2013;23(12):612–9 doi 10.1016/j.tcb.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams J, Lucas PC, Griffith KA, Choi M, Fogoros S, Hu YY, et al. Expression of Bcl-xL in ovarian carcinoma is associated with chemoresistance and recurrent disease. Gynecol Oncol 2005;96(2):287–95 doi 10.1016/j.ygyno.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Wong M, Tan N, Zha J, Peale FV, Yue P, Fairbrother WJ, et al. Navitoclax (ABT-263) reduces Bcl-x(L)-mediated chemoresistance in ovarian cancer models. Mol Cancer Ther 2012;11(4):1026–35 doi 10.1158/1535-7163.MCT-11-0693. [DOI] [PubMed] [Google Scholar]
- 8.Mano Y, Kikuchi Y, Yamamoto K, Kita T, Hirata J, Tode T, et al. Bcl-2 as a predictor of chemosensitivity and prognosis in primary epithelial ovarian cancer. Eur J Cancer 1999;35(8):1214–9. [DOI] [PubMed] [Google Scholar]
- 9.Kassim SK, Ali HS, Sallam MM, Fayed ST, Seada LS, abd-Elkawy E, et al. Increased bcl-2 expression is associated with primary resistance to chemotherapy in human epithelial ovarian cancer. Clinical biochemistry 1999;32(5):333–8. [DOI] [PubMed] [Google Scholar]
- 10.Shigemasa K, Katoh O, Shiroyama Y, Mihara S, Mukai K, Nagai N, et al. Increased MCL-1 expression is associated with poor prognosis in ovarian carcinomas. Jpn J Cancer Res 2002;93(5):542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nature communications 2013;4:2126 doi 10.1038/ncomms3126; 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JR, Fletcher B, Page C, Hu C, Nunez G, Baker V. Bcl-xL is expressed in ovarian carcinoma and modulates chemotherapy-induced apoptosis. Gynecol Oncol 1998;70(3):398–403 doi 10.1006/gyno.1998.5125. [DOI] [PubMed] [Google Scholar]
- 13.Dodier P, Piche A. Bcl-X(L) is functionally non-equivalent for the regulation of growth and survival in human ovarian cancer cells. Gynecol Oncol 2006;100(2):254–63 doi 10.1016/j.ygyno.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 2011;471(7336):110–4 doi 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S, Zhang M, Jing Y, Yin X, Ma P, Zhang Z, et al. Deubiquitinase USP13 dictates MCL1 stability and sensitivity to BH3 mimetic inhibitors. Nat Commun 2018;9(1):215 doi 10.1038/s41467-017-02693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zervantonakis IK, Iavarone C, Chen HY, Selfors LM, Palakurthi S, Liu JF, et al. Systems analysis of apoptotic priming in ovarian cancer identifies vulnerabilities and predictors of drug response. Nat Commun 2017;8(1):365 doi 10.1038/s41467-017-00263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonin K, N’Diaye M, Lheureux S, Loussouarn C, Dutoit S, Briand M, et al. Platinum compounds sensitize ovarian carcinoma cells to ABT-737 by modulation of the Mcl-1/Noxa axis. Apoptosis: an international journal on programmed cell death 2013;18(4):492–508 doi 10.1007/s10495-012-0799-x. [DOI] [PubMed] [Google Scholar]
- 18.Wei G, Margolin AA, Haery L, Brown E, Cucolo L, Julian B, et al. Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell 2012;21(4):547–62 doi 10.1016/j.ccr.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S, Tang R, Poon RY. BCL-W is a regulator of microtubule inhibitor-induced mitotic cell death. Oncotarget 2016;7(25):38718–30 doi 10.18632/oncotarget.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abed MN, Abdullah MI, Richardson A. Antagonism of Bcl-XL is necessary for synergy between carboplatin and BH3 mimetics in ovarian cancer cells. Journal of ovarian research 2016;9:25 doi 10.1186/s13048-016-0234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leverson JD, Phillips DC, Mitten MJ, Boghaert ER, Diaz D, Tahir SK, et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci Transl Med 2015;7(279):279ra40 doi 10.1126/scitranslmed.aaa4642. [DOI] [PubMed] [Google Scholar]
- 22.Stamelos VA, Robinson E, Redman CW, Richardson A. Navitoclax augments the activity of carboplatin and paclitaxel combinations in ovarian cancer cells. Gynecol Oncol 2013;128(2):377–82 doi 10.1016/j.ygyno.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Witham J, Valenti MR, De-Haven-Brandon AK, Vidot S, Eccles SA, Kaye SB, et al. The Bcl-2/Bcl-XL family inhibitor ABT-737 sensitizes ovarian cancer cells to carboplatin. Clin Cancer Res 2007;13(23):7191–8 doi 10.1158/1078-0432.CCR-07-0362. [DOI] [PubMed] [Google Scholar]
- 24.Lheureux S, N’Diaye M, Blanc-Fournier C, Dugue AE, Clarisse B, Dutoit S, et al. Identification of predictive factors of response to the BH3-mimetic molecule ABT-737: an ex vivo experiment in human serous ovarian carcinoma. International journal of cancer 2015;136(5):E340–50 doi 10.1002/ijc.29104. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, et al. A public genome-scale lentiviral expression library of human ORFs. Nature methods 2011;8(8):659–61 doi 10.1038/nmeth.1638; 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piccioni F, Younger ST, Root DE. Pooled Lentiviral-Delivery Genetic Screens. Current protocols in molecular biology 2018;121:32 1 1– 1 21 doi 10.1002/cpmb.52. [DOI] [PubMed] [Google Scholar]
- 27.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34(2):184–91 doi 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zasadil LM, Andersen KA, Yeum D, Rocque GB, Wilke LG, Tevaarwerk AJ, et al. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci Transl Med 2014;6(229):229ra43 doi 10.1126/scitranslmed.3007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell 2015;160(5):977–89 doi 10.1016/j.cell.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell 2018;33(4):690–705 e9 doi 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483(7391):603–7 doi 10.1038/nature11003; 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meghani K, Fuchs W, Detappe A, Drane P, Gogola E, Rottenberg S, et al. Multifaceted Impact of MicroRNA 493–5p on Genome-Stabilizing Pathways Induces Platinum and PARP Inhibitor Resistance in BRCA2-Mutated Carcinomas. Cell Rep 2018;23(1):100–11 doi 10.1016/j.celrep.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Martinez LA, Karamysheva ZN, Warrington R, Li B, Wei S, Xie XJ, et al. Genome-wide siRNA screen reveals coupling between mitotic apoptosis and adaptation. EMBO J 2014;33(17):1960–76 doi 10.15252/embj.201487826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Planells-Cases R, Lutter D, Guyader C, Gerhards NM, Ullrich F, Elger DA, et al. Subunit composition of VRAC channels determines substrate specificity and cellular resistance to Pt-based anti-cancer drugs. EMBO J 2015;34(24):2993–3008 doi 10.15252/embj.201592409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore Vdel G, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science 2011;334(6059):1129–33 doi 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463(7283):899–905 doi 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashkenazi A, Fairbrother WJ, Leverson JD, Souers AJ. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nature reviews Drug discovery 2017;16(4):273–84 doi 10.1038/nrd.2016.253. [DOI] [PubMed] [Google Scholar]
- 38.Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016;538(7626):477–82 doi 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 39.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol 2016;12(2):109–16 doi 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama T, Kohn EC, Brill E, Lee JM. Apoptosis is augmented in high-grade serous ovarian cancer by the combined inhibition of Bcl-2/Bcl-xL and PARP. International journal of oncology 2017. doi 10.3892/ijo.2017.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Au-Yeung G, Lang F, Azar WJ, Mitchell C, Jarman KE, Lackovic K, et al. Selective Targeting of Cyclin E1-Amplified High-Grade Serous Ovarian Cancer by Cyclin-Dependent Kinase 2 and AKT Inhibition. Clin Cancer Res 2017;23(7):1862–74 doi 10.1158/1078-0432.CCR-16-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jebahi A, Villedieu M, Petigny-Lechartier C, Brotin E, Louis MH, Abeilard E, et al. PI3K/mTOR dual inhibitor NVP-BEZ235 decreases Mcl-1 expression and sensitizes ovarian carcinoma cells to Bcl-xL-targeting strategies, provided that Bim expression is induced. Cancer letters 2014;348(1–2):38–49 doi 10.1016/j.canlet.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Iavarone C, Zervantonakis IK, Selfors LM, Palakurthi S, Liu JF, Drapkin R, et al. Combined MEK and BCL-2/XL Inhibition Is Effective in High-Grade Serous Ovarian Cancer Patient-Derived Xenograft Models and BIM Levels Are Predictive of Responsiveness. Mol Cancer Ther 2019;18(3):642–55 doi 10.1158/1535-7163.MCT-18-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guillemette S, Serra RW, Peng M, Hayes JA, Konstantinopoulos PA, Green MR, et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev 2015;29(5):489–94 doi 10.1101/gad.256214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He YJ, Meghani K, Caron MC, Yang C, Ronato DA, Bian J, et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 2018;563(7732):522–6 doi 10.1038/s41586-018-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arora S, Bisanz KM, Peralta LA, Basu GD, Choudhary A, Tibes R, et al. RNAi screening of the kinome identifies modulators of cisplatin response in ovarian cancer cells. Gynecol Oncol 2010;118(3):220–7 doi 10.1016/j.ygyno.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Ahmed AA, Wang X, Lu Z, Goldsmith J, Le XF, Grandjean G, et al. Modulating microtubule stability enhances the cytotoxic response of cancer cells to Paclitaxel. Cancer Res 2011;71(17):5806–17 doi 10.1158/0008-5472.CAN-11-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brachet PE, Fabbro M, Leary A, Medioni J, Follana P, Lesoin A, et al. A GINECO phase II study of Navitoclax (ABT 263) in women with platinum resistant/refractory recurrent ovarian cancer (ROC). Annals of Oncology: Official Journal of the European Society for Medical Oncology / ESMO 2017(28 (suppl_5): v330–v354. 10.1093/annonc/mdx372). [DOI] [Google Scholar]
- 49.Roberts AW, Wilson W, Gandhi L, O’Connor OA, Rudin CM, Brown JR, et al. Ongoing phase I studies of ABT-263: Mitigating Bcl-XL induced thrombocytopenia with lead-in and continuous dosing. Journal of Clinical Oncology 2009;27(15S):3505. [Google Scholar]
- 50.Brotin E, Meryet-Figuiere M, Simonin K, Duval RE, Villedieu M, Leroy-Dudal J, et al. Bcl-XL and MCL-1 constitute pertinent targets in ovarian carcinoma and their concomitant inhibition is sufficient to induce apoptosis. International journal of cancer 2010;126(4):885–95 doi 10.1002/ijc.24787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.