Abstract
RX-5902 is a first-in-class anti-cancer agent targeting phosphorylated-p68 and attenuating nuclear shuttling of β-catenin. The purpose of this study was to evaluate the efficacy of RX-5902 in preclinical models of TNBC and to explore effects on β-catenin expression. A panel of 18 TNBC cell lines were exposed to RX-5902 and changes in proliferation, apoptosis, cellular ploidy and effector protein expression were assessed. Gene expression profiling was used in sensitive and resistant cell lines with pathway analysis to explore pathways associated with sensitivity to RX-5902. The activity of RX-5902 was confirmed in vivo in cell line and patient-derived tumor xenograft (PDX) models. RX-5902 demonstrated potent antiproliferative activity in vitro against TNBC cell lines with an average IC50 of 56 nM in sensitive cell lines. RX-5902 treatment resulted in the induction of apoptosis, G2/M cell cycle arrest and aneuploidy in a subset of cell lines. RX-5902 was active in vivo against TNBC PDX models and treatment resulted in a decrease in nuclear β-catenin. RX-5902 exhibited dose-proportional pharmacokinetics and plasma and tumor tissue in nude mice. Pathway analysis demonstrated an increase in the EMT, TGF beta, and Wnt/β-catenin pathways associated with sensitivity to RX-5902. RX-5902 is active against in vitro and in vivo preclinical models of TNBC. Target engagement was confirmed with decreases in nuclear β-catenin and MCL-1 observed, confirming the proposed mechanism of action. This study supports the continued investigation of RX-5902 in TNBC and combinations with immunotherapy.
Keywords: RX-5902, triple-negative breast cancer, β-catenin, Phosphorylated-p68, MCL-1
Introduction
Triple-negative breast cancer (TNBC) is an aggressive breast cancer subtype that is defined by a lack of expression of the estrogen and progesterone receptors and human epidermal growth factor receptor 2 (HER2) overexpression [1-5]. TNBC comprises 10–20% of all breast cancers and is associated with a more aggressive disease course and early development of resistance to chemotherapy [1, 2, 6]. TNBC is more common in young women, African Americans, and women carrying deleterious mutations in BRCA1 [5, 7]. While TNBC can be subdivided into molecular subtypes using gene expression profiling, approved therapies targeting these subtypes remain elusive and there is still an urgent need for effective targeted anti-cancer therapies [8-12].
p68 RNA helicase is a prototypical member of the DEAD box family of RNA helicases with roles in pre-mRNA, rRNA and miRNA processing, ribosome biogenesis and cellular proliferation [13-16]. p68 can be phosphorylatedrylated by c-Abl in response to PDGF stimulation and phosphorylated-p68 is associated with tumorigenic transformation, cancer progression, invasion and metastasis [17-19]. Phosphorylated-p68 is aberrantly expressed in cancer cells, including TNBC, and minimally expressed in normal cells [13, 20-22]. Phosphorylated-p68 promotes epithelial-mesenchymal transformation (EMT) through β-catenin dependent ATPase activity and up-regulation of Snail 1 [17]. Based on its association with malignant transformation, cancer progression and EMT, inhibition of phosphorylated-p68 represents a novel strategy to disrupt Wnt-independent β-catenin signaling and cancer cell growth.
RX-5902 is a first-in-class, orally bioavailable phosphorylated-p68 inhibitor that has demonstrated potent in vitro inhibition of cancer cell growth and induction of apoptosis [17, 23]. RX-5902 is a quinoxalinyl-piperazine compound selected for development based on potent antiproliferative activity in vitro (IC50 of 10-20 nM), ability to induce apoptosis, and favorable pharmacokinetics in rat models [24, 25]. RX-5902 interacts with Y593 phosphorylated-p68 and inhibits β-catenin dependent ATPase activity with little effect on RNA dependent ATPase activity [23]. RX-5902 was shown to lead to a decrease in phosphorylated-p68 and expression of downstream proteins of β-catenin including cyclin D1, phosphorylated-c-Jun and c-Myc in MDA-MB-231 TNBC cells, but not in normal fetal fibroblast cells [23]. Given the high expression of phosphorylated-p68 in TNBC and preliminary efficacy observed against the MDA-MB-231 TNBC cell line in vitro, this study aimed to further investigate the anticancer activity of RX-5902 against a broad panel of TNBC cancer cell lines and cell line-based and patient-derived xenograft (PDX) models in vivo. Additionally, we investigated the effect of RX-5902 treatment on nuclear β-catenin and explored gene expression profiles associated with sensitivity to RX-5902 in vitro.
Material and Methods
Drug
RX-5902 was provided by Rexahn, Inc. (Rockville, MD). The drug was dissolved in DMSO to prepare a stock solution of 10 mM and stored at −20°C. For in vitro use, the drug was diluted with complete media to prepare the appropriate concentration. For use in vivo, RX-5902 was prepared in sterile milliQ H2O. The chemical structure of RX-5902 is 1-(3,5-Dimethoxyphenyl)-4-[(6-fluoro-2-methoxyquinoxalin-3-yl)aminocarbonyl] piperazine and the drug was synthesized at Rexahn, Inc. according to the method described by Lee et al [24]. RX-5902 is compound 25 in this reference.
Cell lines and tissue culture
Human TNBC cell lines MDA-MB-231, MDA-MB-436, MDA-MB-157, HCC1806, HCC1937, Hs578T, HCC38, BT549, HCC1187, and HCC1395 were obtained from American Type Culture Collection (ATCC, Manassas, VA). CAL-51, CAL-85-1, CAL-120, CAL-148 and HDQ-P1 were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). MDA-MB-468, BT20, and HCC70 were obtained from the University of Colorado Cancer Center (UCCC) Tissue Culture Core laboratory. The MCF-10A cell line was obtained as a kind gift from Traci Lyons, PhD and used as a control. All breast cancer cells were cultured in DMEM supplemented with 10% FBS, 1% penicillin–streptomycin, 1% MEM nonessential amino acids and 1% normocin. All cells were grown in an incubator at 37°C containing 5% CO2. Cell lines were routinely authenticated by the Barbara Davis Center for Childhood Diabetes Core and screened for mycoplasma every 3 months.
Cell viability assay
Cellular proliferation was assessed using the Cell Titer-Glo Luminescent Cell Viability Assay (Promega Corporation, Madison, WI, USA). This assay estimates the number of viable cancer cells present in culture by quantification of the ATP present in cells as a signal of metabolic activity. Cells were harvested in the logarithmic growth phase and then plated in 96 well flat-bottomed plates with lids for each experiment. The next day, cells were treated with increasing doses of RX-5902 (0-10 μmol/l) for 72 hours. Next, 100 μl of Cell Titer-Glo reagent was added followed by a 10 min incubation. The fluorescent intensity of each plate was then assessed using a plate reader (Biotek Synergy 2, Winooski, VT, USA). The raw luminescent data was used to determine IC50 values from at least 3 independent experiments for each cell line.
Long-term live cell microscopy and caspase 3/7 analysis
Apoptosis was assessed using the Incucyte Caspase 3/7 Green apoptosis assay (Essen Bioscience) which measures activated caspase-3/7 to quantify apoptosis over time. MDA-MB-231, HCC 1806, MDA-MB-436 and CAL-120 cell lines were selected for use in this experiment. Depending on the growth kinetics of the cell lines, 1500–5000 cells were plated in 96 well black-walled plates and allowed to adhere overnight. Cells were then exposed to increasing concentrations of RX-5902 (0-1μmol/l) for 72 hours. Caspase 3/7 Green reagent 1:1000 was added to each well on the first day of treatment. Live cell imaging was used to monitor the activation of caspase-3/7 for quantification using the IncuCyte basic analyzer (Essen BioScience Inc, USA). Experiments were performed in triplicate.
Flow-cytometric analysis of cell-cycle distribution
MDA-MB-231, HCC-1806, MDA-MB-436 and Cal-120 cell lines were seeded in 6 well plates (2×105 per well) for 24 hours at 37 °C and then exposed to RX-5902 (0, 20 and 100nM) for 24 hours. Analysis was performed using Krishan’s stain, as previously described [26]. Experiments were performed in at least triplicate.
Immunoblotting
MDA-MB-231, MCF-10A, Cal-51, HCC 1806, MDA-MB-468, CAL-120 and MDA-MB-436 cells were seeded in 6 well plates and allowed to adhere for 24 hours. Cells were then treated with increasing concentrations of RX-5902 (0, 20, 100 or 200 nM) for 24 to 48 hours. Cells were harvested by scraping or with trypsin/EDTA and then lysed in Cell Lysis Buffer (Cell Signaling Technology). Nuclear and cytoplasmic extract isolation was performed using a nuclear extract kit (Active Motif). Protein was isolated from flash frozen tumor samples from xenograft models. In brief, flash frozen samples were homogenized in Cell Lysis Buffer (Cell Signaling Technology) using the Qiagen tissue lyser and centriguged at 16,000 rpm at 4°C for 15 minutes. Fifteen to 50 μg of total protein was loaded onto a 4% to 20% BIS-TRIS 1.5 mm gradient gel (NuPage, Novex Invitrogen), electrophoresed, and transferred to nitrocellulose using the invitrogen Xcell II blotting apparatus (BioRad). Membranes were blocked in blocking buffer for 1 hour, washed × 3 for 10 min with TBS-Tween (0.1%) and incubated overnight at 4°C with primary antibodies: β-catenin (Bethyl Lab, 1:1000, rabbit), c-Myc (Cell Signaling, 1:1000, rabbit), MCL-1 (Cell Signaling, rabbit), phosphorylated-p68 (Abcam, 5μg/ml, rabbit), cyclin D1 (Cell Signaling, 1:500, rabbit), c-Myc (Cell Signaling, 1:1000, rabbit) and actin (Santa Cruz, 1:1000, mouse). The blots were washed in TBS-Tween (0.1%) × 3 and secondary anti-rabbit or anti-mouse IgG1 horseradish peroxidase-linked antibody was added at 1:15,000 (Jackson Immuno Research, West Grove, PA, USA) for 1 hour at room temperature (RT). Blots were developed using the enhanced chemiluminescence detection system 9 pierce, Rockford, IL) or the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA). Experiments were repeated at least 3 times.
In vivo cell line and patient-derived xenograft (PDX) studies
Five- to six-week-old female athymic nude (nu/nu) mice (Envigo, formally Harlan Sprague Dawley) were allowed to acclimate for a week before handling in cages housing up to 5 mice. Animals were provided with sterilized food and water ad libitum and a 12-hour light/dark cycle was utilized. MDA-MB-231 cells were harvested during a logarithmic growth phase, mixed 1:1 in serum-free DMEM and Matrigel (BD Biosciences) and 5 × 106 cells in 100 μL was injected bilaterally into the flank. PDX models were generated as previously describe [26, 27]. Mice were weighed, and tumor measurements obtained using digital calipers at least twice a week. Study Director software package (Studylog Systems) was used for data management. Tumor volume was calculated (volume = (length × width2) × 0.52) and mice were randomized to study groups when the mean tumor volume reached ~150 mm3. For the first MDA-MB-231 xenograft model, animals were randomized to treatment with vehicle, nab-paclitaxel 5 mg/kg IV twice a week × 3, RX-5902 160 mg/kg via oral gavage once weekly × 3, RX-5902 320 mg/kg once weekly × 3 or RX-5902 600 mg/kg once weekly × 3. Based on clinical tolerability of the compound in the first-in-human phase I study, an additional experiment was performed. Animals were randomized to treatment with vehicle, RX-5902 15 mg/kg, 30 mg/kg or 60 mg/kg administered by oral gavage × 5 days followed by 2 days off. For the PDX models, mice were randomized to treatment with vehicle or RX-5902 60mg/kg via oral gavage with once daily dosing 5 days on 2 days off. At the end of the study, mice were euthanized, and tumor samples were collected. Tumor growth inhibition (TGI) was calculated according to the formula: TGI = 100 × (Vt final -Vt initial)/(Vvc final -Vvc intial).
Plasma and tumor samples were collected for pharmacokinetic analysis from nude mice with MDA-MB-231 implanted tumors treated with a single dose of RX-5902 15 mg/kg, 30 mg/kg or 60 mg/kg. Samples were stored at −70°C until analysis and PK analysis was performed at QPS (Newark, DE). Compound concentrations in plasma and homogenized tumor tissue samples were determined by LC/MS/MS analysis (Triple Quadrupole mass spectrometer, Applied Biosystem, USA). RX-5902 powder compound was subsequently used to prepare standard and quality control (QC) solutions for this study. Verapamil was obtained from Sigma Aldrich and was used over the course of the study to prepare internal standard (IS) solutions for the analysis of RX-5902. For mouse plasma, quadratic regression analysis calculations were performed with 1/x2 weighting using Watson™ LIMS v.7.4.1. For mouse tumor tissue homogenate, linear regression analysis calculations were performed with 1/x2 weighting using Watson™ LIMS v.7.4.1. All statistics (e.g., Mean, S.D., %CV, %RE) found in the data were calculated by Watson™ LIMS or based on the “precision as displayed” option of Microsoft® Excel.
All animal experiments were performed in accordance with the current NIH guidelines for the care and use of laboratory animals with approval by the University of Colorado Institutional Animal Care and Use Committee prior to initiation of experiments. Experiments took place in a facility accredited by the American Association for Accreditation of Laboratory Animal Care
Gene expression profiles of breast cancer cell lines
We obtained baseline gene expression profiles from the Cancer Cell Line Encyclopedia project (NCBI Gene Expression Omnibus Accession Number: GSE36133), as previously described [28]. Data were profiled using Affymetrix HG U133 Plus 2.0 gene arrays and normalized by the Robust Multiarray Average algorithm using Affymetrix Power Tools.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) software Version 3.0 was used for gene set analysis (Broad Institute) [29]. For each analysis, gene set permutations were conducted 1,000 times. We used the Hallmark gene sets in this study and we considered gene sets with false discovery rate (FDR) < 0.05 as significant in this study.
Statistical analysis
We compared treatment groups using the ANOVA parametric analysis of the means (Prism 4.0, GraphPad). For in vivo studies, groups were compared using an unpaired parametric t test with Welch’s corrections (Prism 4.0, Graph Pad).
Results
In vitro antiproliferative activity of RX-5902 against TNBC cell lines
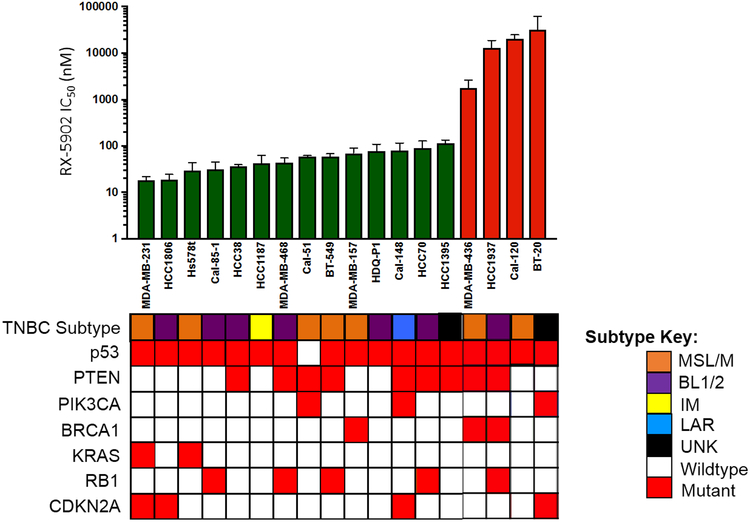
A diverse panel of 18 molecularly characterized TNBC cells lines was screened for sensitivity to RX-5902. Figure 1 depicts the in vitro response to RX-5902 with IC50 values ranging from 18 nM to > 10 μM. Using 100 nM as an IC50 value cutoff to define sensitivity to RX-5902, 14 sensitive and 4 resistant cell lines were identified. The average IC50 of the cell lines sensitive to RX-5902 treatment was 56 nM. Cell lines were sensitive to RX-5902 independent of TNBC molecular subtype or mutational profile. RX-5902 was active against cell lines with mutations in p53, RB1, CDKN2A and loss of PTEN. RX-5902 was active against cell lines of all TNBC molecular subtypes.
Figure 1: RX-5902 inhibits cellular proliferation in TNBC cell lines.
TNBC cell lines were treated with RX-5902 (0-10 μmol/l) for 72 hours and cellular proliferation was assessed using the Cell Titer-Glo Luminescent Cell Viability Assay. Using an IC50 value cutoff of 100 nM, 14 cell lines were determined to be sensitive to RX-5902 (green) and 4 resistant (red). TNBC subtypes, p53 mutation, PTEN loss, PIK3CA mutation, BRCA1 mutation, KRAS mutation, RB1 mutation and CDKN2A mutation are included based on publicly available databases. TNBC molecular subtype was obtained from Lehmann et al, using gene expression profiles [50]. MSL; mesenchymal stem-like, M; mesenchymal, BL1; basal-like 1, BL2; basal-like 2, IM; immunomodulatory, LAR; luminal androgen receptor, UNK; unknown.
RX-5902 exposure leads to G2/M cell cycle arrest and induction of apoptosis
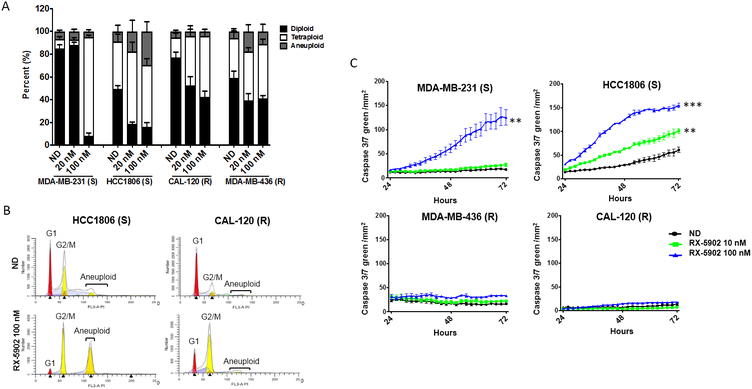
To further understand the mechanism of the observed inhibition of cellular proliferation with RX-5902 treatment in TNBC cell lines, we used flow cytometry to assess changes in cell cycle and the Incucyte Caspase 3/7 assay to assess induction of apoptosis. These experiments were performed in a subset of sensitive and resistant cell lines. As shown in Figure 2A-B, RX-5902 treatment resulted in a dose-dependent increase in tetraploid cells, consistent with induction of G2/M cell cycle arrest. This effect was more pronounced in the cell lines sensitive to the antiproliferative activity of RX-5902, but observed to some degree in all cell lines tested. These results are consistent with prior reports of RX-5902 inducing a G2/M cell cycle arrest in vitro [24].
Figure 2: RX-5902 treatment results in G2/M cell cycle arrest and apoptosis in sensitive cell lines.
(A) Flow-cytometric analysis of cell-cycle distribution in two sensitive (MDA-MB-231 and HCT1806) and two resistant (MDA-MB-436 and CAL-120) cell lines exposed to RX-5902 for 24 hours. (B) Representative graphs for a sensitive (HCC1806) and a resistant (CAL-120) cell line treated with 100 nM RX-5902 or no drug control demonstrating cellular fractions in G1, G2/M and aneuploidy. (C) Indicated cell lines were treated with RX-5902 (0-100 nM) for 72 hours and apoptosis was assessed using the Incucyte Caspase 3/7 Green apoptosis assay with live cell microscopy. S; sensitive to RX-5902, R; resistant to RX-5902, ND; no drug control. *p<0.05, **p<0.01, ****p<0.0001,
RX-5902 treatment resulted in a significant increase in apoptosis in the sensitive cell lines as measured by caspase 3/7 activity (Figure 2C). As expected, no significant induction of apoptosis was observed in cell lines resistant to the antiproliferative effects of RX-5902. The observed activation of apoptosis began 24-48 hours following drug treatment and reached a peak at 72 hours. The magnitude of apoptosis was greatest with a dose of RX-5902 100 nM as compared to 10 nM.
Effect of RX-5902 on nuclear β-catenin and MCL-1
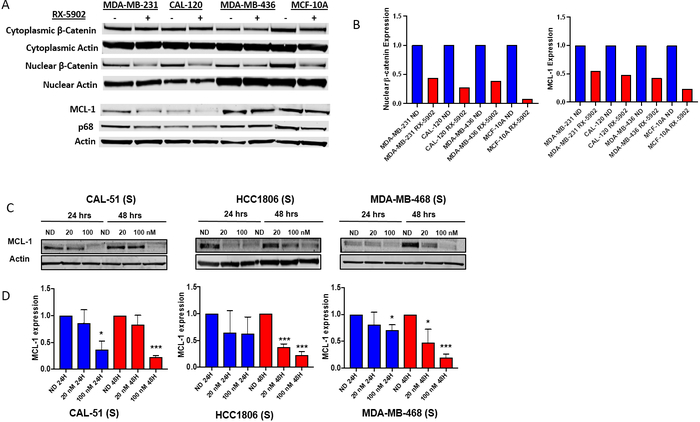
In order to further evaluate the ability of RX-5902 to disrupt the phosphorylated-p68-β-catenin interaction, we investigated the effect of RX-5902 treatment on nuclear β-catenin and cytoplasmic β-catenin in a sensitive TNBC cell line (MDA-MB-231), resistant TNBC cell lines (CAL-120 and MDA-MB-436) and a non-tumorigenic epithelial cell line (MCF-10A). It has been reported that Y593 phosphorylated-p68 binds β-catenin and induces its nuclear translocation through disrupting the interaction with cytoplasmic axin [17]. As depicted in Figure 3A, RX-5902 treatment resulted in a dose-dependent decrease in nuclear β-catenin at 24 hours in the TNBC cell lines and MCF-10 cells. RX-5902 did not significantly affect cytoplasmic β-catenin. This data support the ability of RX-5902 to inhibit the interaction of Y593 phosphorylated-p68 and β-catenin, leading to a decrease in nuclear translocation of β-catenin. This is consistent with the model of the mechanism of action of RX-5902 proposed by Kost et al based on their observations of RX-5902 interacting with Y593 phosphorylated-p68, inhibiting β-catenin dependent ATPase activity, and leading to a decrease in proteins downstream of β-catenin, including cyclin D and c-Myc in the MDA-MB-231 cell line [23].
Figure 3: Effect of RX-5902 on nuclear β-catenin and downstream effector proteins.
(A) MDA-MB-231, CAL-120, MDA-MB-436 and MCF-10A cells were treated with RX-5902 100 nM for 24 hours and immunoblotting was performed for MCL-1, phosphorylated p68, p68 and actin. Additionally, nuclear and cytoplasmic extract isolation was performed. Immunoblotting was performed for β-catenin and actin. (B) Quantification of immunoblots for nuclear β-catenin and MCL-1 normalized to actin and no drug control. (C) TNBC cell lines sensitive to RX-5902 (Cal-51, HCC-1806 and MDA-MB-468) were treated with RX-5902 and immunoblotting was performed for MCL-1 and actin. (D) Quantification of immunoblots for MCL-1. *p<0.05, ****p<0.0001.
Next, we investigated the ability of RX-5902 to modulate MCL-1 expression as a potential mechanism of the induction of apoptosis observed with drug treatment in our TNBC cell line panel. Nuclear β-catenin can bind to HIF-1α and promote transcription of the anti-apoptotic protein, MCL-1 [30]. We hypothesized that the decrease in nuclear β-catenin following RX-5902 treatment could result in a decrease in MCL-1 expression that would correlate with apoptosis observed following treatment in sensitive cell lines. As depicted in Figure 3, treatment with RX-5902 resulted in a dose-dependent, statistically significant decrease in MCL-1 in cell lines sensitive to the antiproliferative effects of RX-5902 in vitro. This decrease was observed at 24 and 48 hours post treatment and was consistent across cell lines tested. These data support a role for MCL-1 as a potential mediator of apoptotic cell death in response to RX-5902 treatment.
In vivo activity of RX-5902 in cell line and PDX TNBC models
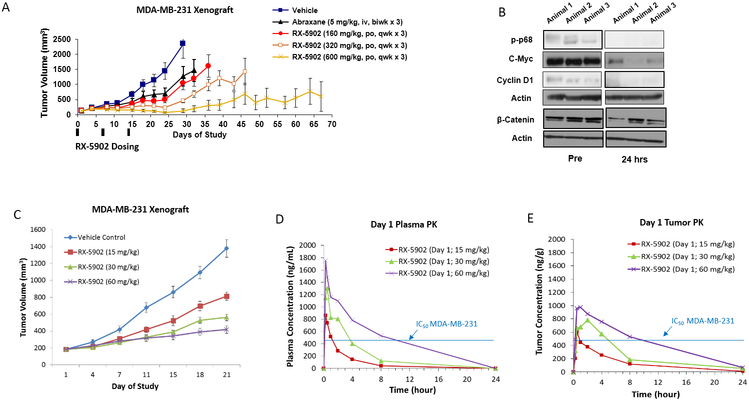
To confirm the in vitro antiproliferative activity of RX-5902 in vivo, we investigated the effects of RX-5902 treatment in a MDA-MD-231 xenograft model and 3 TNBC PDX models. As shown in Figure 4A and Supplementary Figure 1, RX-5902 treatment once weekly × 3 weeks resulted in significant dose-dependent tumor growth inhibition in the MDA-MB-231 model (RX-5902 160mg/kg TGI 55.7%, RX-5902 320mg/kg TGI 80.29% and RX-5902 600mg/kg TGI 94.58%). RX-5902 was more efficacious than the chemotherapy control arm of nab-paclitaxel (TGI 45%). In order to confirm target engagement in vivo, tumors were excised following treatment for 24 hours in a subset of animals and western blots were performed for phosphorylated-p68, c-Myc and cyclin D1. Consistent with in vitro data from Kost et al, treatment with RX-5902 resulted in a decrease in phosphorylated-p68 and the β-catenin-downstream proteins c-Myc and cyclin D1 (Figure 4B)[23].
Figure 4: RX-5902 has anti-tumor activity against a MDA-MB-231 xenograft model, dose-proportional pharmacokinetics and treatment results in decreased phosphorylated-p68, c-myc and β-catenin.
(A) Effect of RX-5902 administered once weekly in the MDA-MB-231 xenograft model. Animals were randomized to treatment with vehicle, nab-paclitaxel 5 mg/kg IV twice a week × 3, RX-5902 160 mg/kg via oral gavage once weekly × 3, RX-5902 320 mg/kg once weekly × 3 or RX-5902 600 mg/kg once weekly × 3. Nab-paclitaxel was used as a chemotherapy control. Tumor growth inhibition (TGI) RX-5902 160 mg/kg 55.7%, 320 mg/kg 80.29% and 600 mg/kg 94.58%, p < 0.001. TGI nab-paclitaxel 45%. B) Immunoblots performed using protein isolated from tumor tissue obtained pre and 24h post RX-5902. Three mice were included in each group. Immunoblotting was performed for p-p68, C-myc, Cyclin D1, β-catenin and actin. C) Based on observed toxicity in the ongoing Phase I dose escalation study, the effect of lower dose RX-5902 5 days on 2 days off was investigated in the MDA-MB-231 xenograft model. TGI RX-5902 15 mg/kg 45%, 30 mg/kg 58% and 60 mg/kg 72%, p < 0.001. D) Plasma concentration-time curve after a single dose of RX-5902 15 mg/kg, 30 mg/kg or 60 mg/kg in thenMDA-MB-231 xenograft model in nude mice. E) Tumor concentration-time curve after a single dose of RX-5902 15 mg/kg, 30 mg/kg or 60 mg/kg in the MDA-MB-231 xenograft model in nude mice.
Based on the clinical tolerability of RX-5902 in an ongoing first-in-human dose escalation study, we investigated lower doses of RX-5902 administered 5 days on 2 days off. As shown in Figure 4C, RX-5902 treatment administered in this schedule resulted in significant dose-dependent tumor growth inhibition. Pharmacokinetic analysis for RX-5902 was performed in plasma and tumor samples in the MDA-MB-231 xenograft model. The pharmacokinetics were dose-proportional and with a dose of 60 mg/kg, the concentration of RX-5902 remained above the IC50 of the MDA-MB-231 cell line in vitro for more than 8 hours. This dose was selected for the TNBC PDX models.
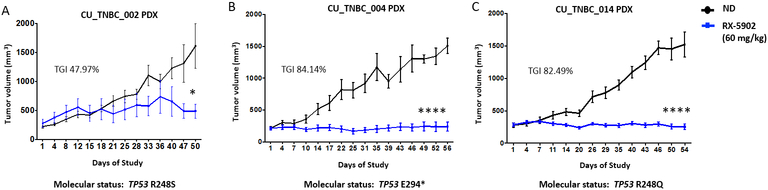
To further confirm the in vivo activity of RX-5902 in a biologically relevant model, we explored the activity of RX-5902 in 3 TNBC PDX models. RX-5902 was active in vivo in these models where oral once daily dosing 60 mg/kg 5 days on 2 days off resulted in statitstically significant tumor growth inhibition (Figure 5A, B and C) (CU_TNBC_002, TGI 47.97%, p<0.05, CU_TNBC_004, TGI: 84.14%, p<0.0001, and CU-TNBC_014, TGI 82.49%, p<0.0001). This schedule was selected based on dose and schedule finding in the first-in-human phase I clinical trial of RX-5902 [31]. Treatment with RX-5902 was well-tolerated in the mice with no significant change in body weight or outward signs of toxicity.
Figure 5: In vivo antitumor activity of RX-5902 in TNBC PDX models.
The effect of RX-5902 was evaluated against 3 TNBC PDX models. Mice were randomized to treatment with vehicle or RX-5902 60mg/kg once daily 5 days on 2 days off. This dose was selected based on the PK data in Figure 4D-E. (A) CU_TNBC_002 (TGI 47.97%, p<0.05), (B) CU_TNBC_004 (TGI: 84.14%, p<0.0001) and (C) CU-TNBC_014 (TGI 82.49%, p<0.0001).
Gene set enrichment analysis to explore pathways potentially associated with sensitivity to RX-5902
RX-5902 is known to interact with Y593 phosphorylated-p68 and lead to a decrease in β-catenin downstream effector proteins cyclin D1 and c-Myc [23]. Our data further demonstrate that RX-5902 leads to a decrease in nuclear β-catenin and MCL-1 expression which may mediate apoptosis and cell death that is observed following treatment in the majority of TNBC cell lines tested. PDGF stimulation can trigger c-Abl phosphorylatedrylation of p68, however, there are potentially other cellular pathways with uncharacterized interactions with phosphorylated-p68 that may affect sensitivity to RX-5902.
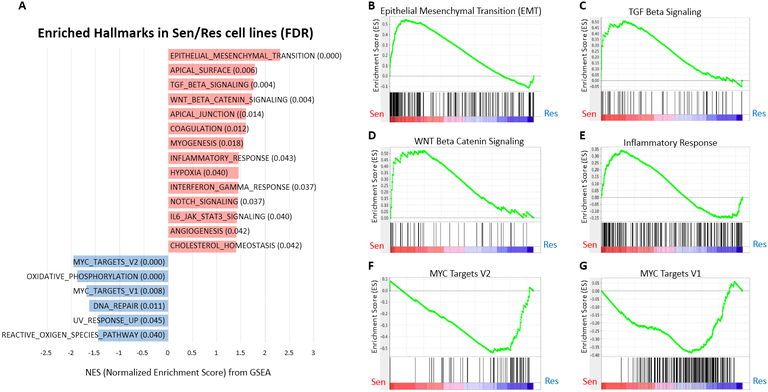
In order to evaluate baseline characteristics than may be predictive of sensitivity to RX-5902 treatment using an unbiased approach, GSEA was performed using baseline gene expression profiling in sensitive TNBC cell lines compared to resistant cell lines. As depicted in Figure 6A, we identified a number of differentially pathways enriched in sensitive vs resistance TNBC cell lines. The top differentially expressed pathways more highly expressed in sensitive cell lines included Epithelial Mesenchymal Transition (EMT, Figure 6B), TGF-β Signaling (Figure 6C), WNT/β-catenin Signaling (Figure 6D) and Inflammatory Response (Figure 6E). Pathways upregulated in resistant cell lines included MYC target V2 and V1 (Figure 6F and G). While these analysis are hypothesis generating, they support the proposed mechanism of action of RX-5902 and may lay the groundwork for predictive biomarker investigation and development.
Figure 6: Gene set enrichment analysis (GSEA) according to FDR value in RX-5902 sensitive vs. resistant cell lines.
(A) GSEA was performed using baseline gene expression profiling in 14 sensitive cell lines (IC50 ≤100 nM) and 4 resistant cell lines (IC50 >100 nM) from Figure 1. Hallmark gene sets enriched in sensitive cell lines vs. resistance cell lines (FDR < 0.05). Hallmark gene sets enriched in the sensitive and resistant cell lines are highlighted in red and blue color, respectively. Selected enrichment plots for sensitive cell lines: (B) EMT, (C) TGF beta, (D) WNT/ β -catenin signaling pathway, and (E) inflammatory response. Selected enrichment plots for resistant cell lines: (F) MYC target V2 and (G) MYC target V1.
Discussion
Although substantial progress has been made in understanding the differential molecular characteristics of TNBC, this disease remains defined by the absence of expression of the estrogen receptor, progesterone receptor and HER2 over-expression. TNBC is a highly aggressive breast cancer subtype still in need of novel, active targeted therapies. The target of RX-5902, phosphorylated-p68, is highly expressed in TNBC, therefore, we conducted these studies to characterize the antitumor activity of RX-5902 in a large panel of genomically characterized TNBC cell lines and cell line and PDX models in vivo. Furthermore, we investigated the ability of RX-5902 to affect expression of nuclear β-catenin and the anti-apoptotic protein MCL-1, and explored pathways associated with sensitivity.
Our study demonstrates that RX-5902 is active against a large number of TNBC cell lines tested, independent of TNBC genomic subtype or mutational profile. We also show that RX-5902 is active in vivo against multiple TNBC models and cell lines with gene expression profiles enriched for genes in the EMT, TGF-β, and WNT/β-catenin signaling may be more sensitive to RX-5902 whereas cell lines enriched for MYC target genes may be more resistant. RX-5902 treatment led to a G2/M cell cycle arrest and cell death was associated with robust induction of apoptosis in sensitive TNBC cell lines. We observed a treatment emergent decrease in expression of the anti-apoptotic protein MCL-1 which may mediate the induction of apoptosis following RX-5902 demonstrated in our work, as well as the work of others [23, 24]. RX-5902 is multifaceted in its ability to induce apoptosis and cell death, but also impact β-catenin signaling that may be essential to EMT and cancer metastasis.
p68 RNA helicase is well documented as mediator of cancer progression and metastasis [17, 32]. In particular, phosphorylated-p68 is aberrantly expressed in cancer cells compared to normal tissue, correlates with cancer progression, and cancer metastasis through promotion of EMT [17, 23, 33-35]. RX-5902 interferes with the interaction of Y593 phosphorylated-p68 and β-catenin, making it a unique inhibitor of Wnt-independent β-catenin signaling. EMT (epithelial-mesenchymal transition) is a cellular process that results in the loss of proteins responsible for cell junction maintenance, therefore, promoting metastasis [33, 36]. EMT is promoted through PDGF phosphorylatedrylation of p68 at the tyrosine residual (Y593) that leads to increased transcription of cyclin D1 and c-Myc, resulting in cancer cell proliferation [37]. The transcriptional activation of cyclin D1 and c-Myc is enhanced by the interaction of phosphorylated-p68 with β-catenin and the TCF/LEF transcriptional factors complex. In HT-29 colorectal cancer cell lines, phosphorylated-p68 was shown to facilitate β-catenin nuclear translocation by blocking the cytoplasmic phosphorylation of β-catenin by GSK-3β and displacing Axin [17, 37]. Through this mechanism of action, phosphorylated-p68 enables β-catenin to activate downstream genes with essential roles in cell proliferation and EMT [17, 37]. Adding to the growing body of literature describing the potential mechanism of action of RX-5902, we observed the ability of RX-5902 treatment to decrease nuclear β-catenin and confirmed the ability to decrease β-catenin dependent proteins c-Myc and cyclin D in vivo. This data support the conclusion that RX-5902 is able to disrupt the phosphorylated-p68/β-catenin interaction and inhibit β-catenin nuclear translocation, consistent with prior in vitro modeling [17, 23, 37].
Dysregulation of the canonical Wnt/β-catenin signaling pathway is well known to be associated with cancer progression [38]. Signaling through the frizzled family of g-protein-coupled receptors leads to stabilization and accumulation of β-catenin in the cytoplasm followed by subsequent β-catenin translocation to the nucleus where it interacts with transcriptional factors involved in tumor cell progression, migration and survival [39]. Wnt/β-catenin signaling is upregulated in many cancers, including TNBC where it is associated with a poor prognosis [40]. Targeting the Wnt/β-catenin pathway is a promising anticancer strategy and multiple agents targeting Wnt ligands and/or frizzled receptors are in preclinical and early clinical development, however, significant single agent activity has not to date been observed [41-45]. Additionally, these agents may lead to a decrease in bone density and compression fracture risk [46].
Interest has heightened recently in Wnt/β-catenin modulators based on recent identification of this pathway as a driver of cancer-mediated immune evasion [47, 48]. Activation of β-catenin in dendritic cells suppresses immunity by inhibiting cross-priming through mTOR/IL-10 signaling, leading to CD4+ and CD8+ T cell tolerance [49]. β-catenin expression is inversely correlated with CD8+ T cells infiltration, supporting a hypothesis that inhibition of the β-catenin, as an immunomodulator, could be used to increase T cell infiltration and potentiate the activity of immune check point inhibition [49]. Based on ability to modulate downstream β-catenin activity in a Wnt-independent manner, RX-5902 may represent an ideal combination partner for immune check point inhibion in TNBC with a more favorable side effect profile.
RX-5902 has been evaluated in a first-in-human phase I dose escalation study () where it was found to be well-tolerated with a favorable side effect profile [31]. The recommended phase 2 dose was determined to be 250 mg of RX-5902 administered daily for 5 on 2 days off with continuous dosing. This trial includes a planned phase II TNBC expansion that is currently enrolling. Future studies are planned to evaluate the impact of RX-5902 on immune cell infiltration and activation in order to support combination studies with immunotherapy.
Supplementary Material
Acknowledgements
We wish to thank Traci Lyons, PhD for the kind gift of MCF-10A cells.
Financial support: This work was supported by the National Institutes of Health (NIH) and the National Cancer Institute (NCI) through 5P30CA046934-25 (University of Colorado Cancer Center Support Grant), 1K23CA172691-01A1 (J.R. Diamond) and CPRIT Scholar Award #RR160093 (S. G. Eckhardt). Support was also received from Rexahn Pharmaceuticals, Inc. The funding bodies played a role in study design, data collection, analysis, and interpretation, and manuscript preparation.
Footnotes
Potential Conflict of Interest: Research funding from Rexahn Pharmaceuticals, Inc. to JRD, SGE and JJT. JGF, DJK, CG, YBK, DJK and EB are employees of Rexahn Pharmaceuticals, Inc.
References
- 1.Dent R, Trudeau M, Pritchard KI et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007; 13: 4429–4434. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB et al. Molecular portraits of human breast tumours. Nature 2000; 406: 747–752. [DOI] [PubMed] [Google Scholar]
- 3.Bauer KR, Brown M, Cress RD et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 2007; 109: 1721–1728. [DOI] [PubMed] [Google Scholar]
- 4.Carey L, Winer E, Viale G et al. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 2010; 7: 683–692. [DOI] [PubMed] [Google Scholar]
- 5.Ismail-Khan R, Bui MM. A review of triple-negative breast cancer. Cancer Control 2010; 17: 173–176. [DOI] [PubMed] [Google Scholar]
- 6.Liedtke C, Mazouni C, Hess KR et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 2008; 26: 1275–1281. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan HG, Malmgren JA. Impact of triple negative phenotype on breast cancer prognosis. Breast J 2008; 14: 456–463. [DOI] [PubMed] [Google Scholar]
- 8.Jemal A, Siegel R, Ward E et al. Cancer statistics, 2009. CA Cancer J Clin 2009; 59: 225–249. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan HG, Malmgren JA. Disease-specific survival in patient-detected breast cancer. Clin Breast Cancer 2006; 7: 133–140. [DOI] [PubMed] [Google Scholar]
- 10.Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer 2009; 9 Suppl 2: S73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol 2008; 26: 2568–2581. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann BD, Jovanovic B, Chen X et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 2016; 11: e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson RJ, Hamilton SJ, MacCallum DE et al. Expression of the ‘dead box’ RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J Pathol 1998; 184: 351–359. [DOI] [PubMed] [Google Scholar]
- 14.Jalal C, Uhlmann-Schiffler H, Stahl H. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res 2007; 35: 3590–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford MJ, Anton IA, Lane DP. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature 1988; 332: 736–738. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda T, Yamagata K, Fujiyama S et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 2007; 9: 604–611. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Lin C, Liu ZR. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell 2006; 127: 139–155. [DOI] [PubMed] [Google Scholar]
- 18.Wei Y, Hu MH. [The study of P68 RNA helicase on cell transformation]. Yi Chuan Xue Bao 2001; 28: 991–996. [PubMed] [Google Scholar]
- 19.Fuller-Pace FV. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta 2013; 1829: 756–763. [DOI] [PubMed] [Google Scholar]
- 20.Wortham NC, Ahamed E, Nicol SM et al. The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERalpha-positive breast cancer. Oncogene 2009; 28: 4053–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark EL, Fuller-Pace FV, Elliott DJ, Robson CN. Coupling transcription to RNA processing via the p68 DEAD box RNA helicase androgen receptor co-activator in prostate cancer. Biochem Soc Trans 2008; 36: 546–547. [DOI] [PubMed] [Google Scholar]
- 22.Causevic M, Hislop RG, Kernohan NM et al. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene 2001; 20: 7734–7743. [DOI] [PubMed] [Google Scholar]
- 23.Kost GC, Yang MY, Li L et al. A Novel Anti-Cancer Agent, 1-(3,5-Dimethoxyphenyl)-4-[(6-Fluoro-2-Methoxyquinoxalin-3-yl)Aminocarbonyl] Piperazine (RX-5902), Interferes With beta-Catenin Function Through Y593 Phospho-p68 RNA Helicase. J Cell Biochem 2015; 116: 1595–1601. [DOI] [PubMed] [Google Scholar]
- 24.Lee YB, Gong YD, Yoon H et al. Synthesis and anticancer activity of new 1-[(5 or 6-substituted 2-alkoxyquinoxalin-3-yl)aminocarbonyl]-4-(hetero)arylpiperazine derivatives. Bioorg Med Chem 2010; 18: 7966–7974. [DOI] [PubMed] [Google Scholar]
- 25.Lee YB, Gong YD, Kim DJ et al. Synthesis, anticancer activity and pharmacokinetic analysis of 1-[(substituted 2-alkoxyquinoxalin-3-yl)aminocarbonyl]-4-(hetero)arylpiperazine derivatives. Bioorg Med Chem 2012; 20: 1303–1309. [DOI] [PubMed] [Google Scholar]
- 26.Capasso A, Pitts TM, Klauck PJ et al. Dual compartmental targeting of cell cycle and angiogenic kinases in colorectal cancer models. Anticancer Drugs 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ionkina AA, Tentler JJ, Kim J et al. Efficacy and Molecular Mechanisms of Differentiated Response to the Aurora and Angiogenic Kinase Inhibitor ENMD-2076 in Preclinical Models of p53-Mutated Triple-Negative Breast Cancer. Front Oncol 2017; 7: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamond JR, Salgia R, Varella-Garcia M et al. Initial clinical sensitivity and acquired resistance to MET inhibition in MET-mutated papillary renal cell carcinoma. J Clin Oncol 2013; 31: e254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glaab E, Baudot A, Krasnogor N et al. EnrichNet: network-based gene set enrichment analysis. Bioinformatics 2012; 28: i451–i457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iqbal S, Zhang S, Driss A et al. PDGF upregulates Mcl-1 through activation of beta-catenin and HIF-1alpha-dependent signaling in human prostate cancer cells. PLoS One 2012; 7: e30764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamond J, Eckhardt G, Gluck L et al. 258PPhase 1 study of RX-5902, a novel orally bioavailable inhibitor of phosphorylated P68, which prevents β-catenin translocation in advanced solid tumors. Annals of Oncology 2017; 28: mdx365.021–mdx365.021. [Google Scholar]
- 32.Shin S, Rossow KL, Grande JP, Janknecht R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res 2007; 67: 7572–7578. [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Lin C, Liu ZR. Signaling to the DEAD box--regulation of DEAD-box p68 RNA helicase by protein phosphorylations. Cell Signal 2005; 17: 1495–1504. [DOI] [PubMed] [Google Scholar]
- 34.Dey H, Liu ZR. Phosphorylation of p68 RNA helicase by p38 MAP kinase contributes to colon cancer cells apoptosis induced by oxaliplatin. BMC Cell Biol 2012; 13: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu ZR. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5’ splice site duplex. Mol Cell Biol 2002; 22: 5443–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Lin C, Liu ZR. Phosphorylations of DEAD box p68 RNA helicase are associated with cancer development and cell proliferation. Mol Cancer Res 2005; 3: 355–363. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, Lin C, Zhao S et al. Phosphorylation of p68 RNA helicase plays a role in platelet-derived growth factor-induced cell proliferation by up-regulating cyclin D1 and c-Myc expression. J Biol Chem 2007; 282: 16811–16819. [DOI] [PubMed] [Google Scholar]
- 38.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017; 36: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev 1997; 11: 3286–3305. [DOI] [PubMed] [Google Scholar]
- 40.Shen T, Zhang K, Siegal GP, Wei S. Prognostic Value of E-Cadherin and beta-Catenin in Triple-Negative Breast Cancer. Am J Clin Pathol 2016; 146: 603–610. [DOI] [PubMed] [Google Scholar]
- 41.Ho SY, Keller TH. The use of porcupine inhibitors to target Wnt-driven cancers. Bioorg Med Chem Lett 2015; 25: 5472–5476. [DOI] [PubMed] [Google Scholar]
- 42.Bahrami A, Amerizadeh F, ShahidSales S et al. Therapeutic Potential of Targeting Wnt/beta-Catenin Pathway in Treatment of Colorectal Cancer: Rational and Progress. J Cell Biochem 2017; 118: 1979–1983. [DOI] [PubMed] [Google Scholar]
- 43.Jimeno A, Gordon M, Chugh R et al. A First-in-Human Phase I Study of the Anticancer Stem Cell Agent Ipafricept (OMP-54F28), a Decoy Receptor for Wnt Ligands, in Patients with Advanced Solid Tumors. Clin Cancer Res 2017; 23: 7490–7497. [DOI] [PubMed] [Google Scholar]
- 44.Eguchi M, Nguyen C, Lee SC, Kahn M. ICG-001, a novel small molecule regulator of TCF/beta-catenin transcription. Med Chem 2005; 1: 467–472. [DOI] [PubMed] [Google Scholar]
- 45.Huang SM, Mishina YM, Liu S et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009; 461: 614–620. [DOI] [PubMed] [Google Scholar]
- 46.Mita MM, Becerra C, Richards DA et al. Phase 1b study of WNT inhibitor vantictumab (VAN, human monoclonal antibody) with paclitaxel (P) in patients (pts) with 1st- to 3rd-line metastatic HER2-negative breast cancer (BC). Journal of Clinical Oncology 2016; 34: 2516–2516.27269942 [Google Scholar]
- 47.Fu C, Liang X, Cui W et al. beta-Catenin in dendritic cells exerts opposite functions in cross-priming and maintenance of CD8+ T cells through regulation of IL-10. Proc Natl Acad Sci U S A 2015; 112: 2823–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spranger S, Gajewski TF. A new paradigm for tumor immune escape: beta-catenin-driven immune exclusion. J Immunother Cancer 2015; 3: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaler P, Augenlicht L, Klampfer L. Activating mutations in beta-catenin in colon cancer cells alter their interaction with macrophages; the role of snail. PLoS One 2012; 7: e45462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehmann BD, Bauer JA, Chen X et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121: 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.