Abstract
Many immunotherapeutic approaches under development rely on T-cell recognition of cancer-derived peptides bound to human leukocyte antigen molecules on the cell surface. Direct experimental demonstration that such peptides are processed and bound is currently challenging. Here we described a method that meets this challenge. The method entailed an optimized immuno-precipitation protocol coupled with two-dimensional chromatography and mass spectrometry. The ability to detect and quantify minute amounts of pre-defined antigens should be useful for both basic research in tumor immunology as well as for the development of rationally-designed cancer vaccines.
INTRODUCTION
Proteins encoded by mutant genes in cancers can be processed and presented by human leukocyte antigen (HLA) molecules on a cell surface. In some cases, this processing can result in presentation of peptides containing “Mutation-Associated Neo-antigens” (MANAs), which can be recognized by T-cells[1]. The effectiveness of immune checkpoint inhibitors, such as PD-1 antibodies, are dependent on recognition of such MANAs[2]. Many other types of immunotherapies are currently being developed to exploit MANAs with either immune modulation or with vaccines composed of the MANAs themselves[2–4]. The durable responses achieved with immunotherapies against cancer are often remarkable [5, 6].
One of the rate-limiting steps in understanding and developing MANA-based therapies is the identification of HLA-presented MANAs. Although genetic mutations within cancer cells can now be routinely identified through exome-wide sequencing, the determination of which mutations result in altered peptides that are properly processed and actually presented on the patient’s HLA molecules on the surface of cells remains difficult[7]. As a result, such determinations are rarely performed experimentally[4, 8], but rather rely on predictions of binding affinity performed in silico[4]. These in silico predictions have proved to be very helpful but are not particularly sensitive or specific[9]. Alternatively, MANAs can be determined experimentally by culturing tumor cells or antigen-presenting cells (peptide-pulsed or transfected with antigen-encoding plasmids) with autologous T cells to expand MANA-reactive T cells, with validation provided by tetramer staining or peptide-pulsing assays [10, 11]. However, these experimental methods require the presence of endogenous T cell clones recognizing the putative MANAs and are technically difficult due to the low abundance of most MANA-reactive T cell clones relative to all T-cell clonotypes. Thus, facile experimental techniques for confirming these predictions or for testing potentially important MANAs that do not score in the predictive algorithms are needed and would likely improve the odds of successful immunotherapy[7].
The most direct way to test whether an individual MANA is presented on the surface of a cell is through mass spectrometry (MS)[12]. The mutant genes and corresponding HLAs of interest can be transfected into cells, and antibodies reactive against HLA molecules can be used to immuno-precipitate the HLA-peptide complexes on the cell surface. Mass spectrometry can then, in theory, be used to determine whether the MANA is present within the immuno-precipitate but, in practice, any individual MANA represents only a tiny fraction of the immuno-precipitated complexes and their detection has proved challenging [12, 13]. Thus, commonly used MS-based techniques for detecting abundant peptides are not applicable[12]. To our knowledge, the only published MS-based technique for targeted detecting presented peptides required 2.0 to 6.7 billion cells and the efficiency of recovering MANA-related peptides was only 1%–3%[14]. To overcome this obstacle, we have developed a technique, called Mutation-Associated Neo-Antigen Selected Reaction Monitoring (MANA-SRM), that permits the direct detection and quantification of HLA-binding MANAs.
MATERIALS AND METHODS
Materials.
Cell Lines:
COS-7, CFPAC-1, Hs578.T, HL-60, HH, SW948, RD, Hs940.T, and RPMI-6666 cells were obtained from the ATCC between 2014 and 2018. COS-7, CFPAC-1 and SW948 were cultured in McCoy’s 5A Media (Thermo Fisher Scientific) with 10% FBS (HyClone) and 1% penicillin streptomycin (Thermo Fisher Scientific). Hs578.T, Hs940.T, and RD were cultured in Dulbecco’s Modified Eagle’s Media (Thermo Fisher Scientific) with 10% FBS (HyClone) and 1% penicillin streptomycin (Thermo Fisher Scientific). HL-60 was cultured in Iscove’s Modified Dulbecco’s Medium (ATCC) with 20% FBS and 1% penicillin streptomycin. HH was cultured in RPMI-1640 (ATCC) with 10% FBS and 1% penicillin streptomycin. RPMI-6666 was cultured in RPMI-1640 (ATCC) with 20% FBS and 1% penicillin streptomycin. Upon receipt of each cell line, the line was expanded and stock vials frozen. Each cell line was cultured for no longer than six months before thawing a new vial from the original stock. Cell lines were not reauthenticated but were routinely tested for mycoplasma.
Antibodies:
Purified anti-human HLA-A, B, C antibody (clone W6/32) was purchased from Bio X Cell. Anti-HLA-DR antibody (L243) was purchased from Abcam.
Plasmids:
Plasmids encoding K-RAS (wild-type, G12C, G12D, G13D, Q61H, Q61L, and Q61R), IDH2 (wild-type and R140Q mutant), and HLA class I alleles (A1, A2, A3, B7) followed by a T2A sequence and the GFP gene were synthesized and cloned into pcDNA3.1 by GeneArt (ThermoFisher Scientific). An exemplary plasmid map showing a gene of interest (HLA or Oncogene) followed by the T2A and GFP as well as the sequence inserts used are provided (Supplementary Fig. S1, Supplementary Table S1). All other reagents were purchased from Sigma-Aldrich unless otherwise indicated.
Preparation of Solutions.
Lysis buffers.
Lysis buffer I (10 mL) contained 6.87 mL RIPA buffer (68.7 μL Nonidet P-40, 687 μL 10% wt/vol sodium deoxycholate, 68.7 μL 10% wt/vol SDS) (Invitrogen), 206.1 μL 5 M NaCl, 68.7 μL 1 M sodium phosphate (pH 7.2), 1 mL water, one Complete EDTA-free Protease Inhibitor Mixture tablet (Roche), 1 mL 0.5 M NaF, 10 μL 80 mM b-glycerophosphate, 1 mL 20 mM Na pyrophosphate, 10 μL 300 mM Na orthovanadate, 10 μL 1 M DTT, and 100 μL 100 mM PMSF. Lysis buffer II (10 mL) was a buffer reported to extract and enrich membrane proteins, and it was prepared in PBS containing 0.25% sodium deoxycholate, 0.2 mM iodoacetamide, 1 mM EDTA, 1:200 Protease Inhibitors Cocktail (Sigma-Aldrich), 1 mM PMSF, 1% octyl-β-D glucopyranoside (Sigma-Aldrich)[15]. Lysis buffer III (10 mL) was a buffer reported to specifically enrich HLA-binding peptides, and it was composed of 0.5% IGEPAL 630, 150 mM NaCl, 50 mM Tris [pH 8.0])[16].
Modified RIPA buffer.
Modified RIPA buffer (10 mL) contained 300 μL 5 M NaCl, 500 μL 1 M Tris (pH 7.4), 100 μL Nonidet P-40, 250 μL 10% (wt/vol) sodium deoxycholate, 20 μL 0.5 M EDTA, and 8.83 mL water.
IP buffers.
IP system I is composed of Lysis buffer I and modified RIPA buffer; IP system II and III are composed of Lysis buffer II and III respectively as described[15, 16].
Antibody conjugation reaction buffer.
Antibody conjugation reaction buffer (ACRB, 0.2 M triethanolamine [pH 8.2] and 25 mM dimethyl pimelimidate dihydrochloride) was prepared fresh before each use.
Neo-antigen elution buffer.
Neo-antigen elution buffer (NEB) contained 50 mM ammonium bicarbonate and 50% acetic acid (pH 2.0).
bRPLC solvents.
bRPLC Solvent A contained 15 mM triethylammonium bicarbonate; bRPLC Solvent B contained 90% (vol/vol) acetonitrile, 15 mM triethylammonium bicarbonate.
Mass spectrometry solvents.
Solvent A contained 3% (vol/vol) acetonitrile, 0.1% formic acid; Solvent B contained 90% (vol/vol) acetonitrile, 0.1% formic acid.
Cell Transfection.
COS-7 cells seeded into 24.5 x 24.5 cm2 plates were transfected with plasmids containing the genes indicated above at 95% confluency using Lipofectamine 3000 Reagent (Thermo Fisher Scientific). For each plate, 125 μg of plasmid (50 μg of HLA plasmid and 75 μg of mutant or wildtype protein plasmid) was mixed with 200 uL of Lipofectamine P3000 in 6mL of Opti-MEM (Thermo Fisher Scientific). In a separate tube, 200 μL of Lipofectamine 3000 Reagent was mixed with 6mL of Opti-MEM. The contents of the two tubes were mixed and allowed to complex for 10 minutes. Medium bathing cells were removed and 50 mL of fresh complete medium was added followed by the Lipofectmine-DNA mixture. Cells were harvested 48-72 h post-transfection. The transfection efficiencies of COS-7 cells were measured to be 93.5 ± 4.9%, in multiple assays as assessed by GFP+ fraction on flow cytometry (BD LSRII). Representative data for assessing transfection efficiencies are displayed in Supplementary Fig. S2.
Generation of KRAS G12D-Knockin Cell Line.
The CFPAC-1 G12D knock-in cell line was generated by co-electroporation of a Cas9 ribonucleoprotein (RNP) obtained from Integrated DNA Technologies, Inc. (IDT) targeting the KRAS locus with a single-stranded DNA oligo repair template encoding the KRAS G12D mutation. The G12D repair template was designed with asymmetric homology arms of 72 and 40 base pairs and an altered PAM site to increase the rate of homology directed repair[17]. A co-selection strategy simultaneously targeting the ATP1A1 locus with a Cas9 RNP and repair template was employed to enrich for edited cells[18]. Alt-R® CRISPR Cas9 crRNAs (Integrated DNA Technologies) targeting KRAS exon 2 (AATGACTGAATATAAACTTG) and ATPA1 (GTTCCTCTTCTGTAGCAGCT) were duplexed individually with Alt-R® CRISPR-Cas9 tracrRNA (IDT) for 5 minutes at 95ºC. Duplexed RNA was cooled to room temperature before mixing at a 1.1:1 molar ratio with Cas9 Nuclease (IDT) for 10 minutes at room temperature. The KRAS G12D and ATP1A1 repair templates were ordered as Ultramer® DNA Oligonucleotides and re-suspended at a concentration of 100uM in TE buffer (Thermo Fisher Scientific). To electroporate, the following components were combined in a 20 μL reaction volume in Opti-MEM (Thermo Fisher Scientific): KRAS RNP (1 μM), ATP1A1 RNP (1 μM), KRAS G12D repair template (2.5 μM), ATP1A1 repair template (2.5 μM), and CFPAC-1 cells (20e6 cells/mL). The electroporation was performed in a 0.1 cm cuvette (Bio-Rad Laboratories) using an ECM 2001 (BTX). Electroporated cells were allowed to recover in culture in their normal growth medium for 3 days before selection with 0.5 μM ouabain (Sigma-Aldrich) for 10 days. Selected cells were plated by limiting dilution in their normal growth medium and incubated for 3 weeks. Individual clones were screened by isolating genomic DNA using the Quick-DNA™ 96 Kit (Zymo Research), PCR amplifying a 1 kb window surrounding the edit site 1-10ng of template DNA, and Sanger sequencing to identify and confirm the introduced G12D mutation (Supplementary Fig. S3). KRAS G12D Repair Template sequence is 5’–ATTGTTGGATCATATTCGTCCACAAAATGATTCTGAATTAGCTGTATCGTCAAGGCACTCTTGCCTACGCCATCAGCTCCAACTACGACAAGTTTATATTCAGTCATTTTCAGCAGGCCTTATAATA–3’
Cell Lysis and Protein Quantification.
Cultured cells (transfected or untransfected) were grown to near confluency in 24.5x24.5 cm2 plates. Cultured cells were washed with PBS two times, followed by another wash with pre-chilled PBS at 4 °C containing 1X cOmplete™ Protease Inhibitor (Sigma-Aldrich). Cells were scraped directly and collected in a falcon tube. The tube was centrifuged at 1,000 x g for 5 min and the supernatant was removed. Cell pellets were snap frozen in liquid nitrogen and stored at −80°C for future experiments.
Lysis buffer was added to the cell pellets at the ratio of 50 million cells per 1 mL lysis buffer. Cells were lysed by incubation in lysis buffer for 30 min on ice, with vortexing every 10 min. The lysates were clarified by centrifugation at 12,000 × g for 30 min at 4 °C. Lysates were stored at −80°C at a concentration of 50 mg of total protein per tube. A bicinchoninic acid (BCA) assay kit (Thermo-Scientific) was used to quantify protein concentrations on a BioTek Synergy H1 plate reader (BioTek).
Immobilization of Antibody on Magnetic Beads.
A purified human HLA-A, B, C antibody (clone W6/32) (1 mg) was added to 5 mL Protein G Dynal Magnetic Beads (purchased from ThermoFisher Scientific at 30 mg/mL, without washing), and the antibody was allowed to bind to the beads on a rotator at 30 rpm at room temperature for 2 h. The antibody-bound beads were then washed with 5 mL ACRB and collected using a DynaMag™-50 magnet (ThermoFisher Scientific). The antibody was covalently cross-linked to the protein G on the beads by incubation in 5 mL ACRB on a rotator at room temperature for 30 min. The beads were washed twice with 5 mL 50 mM Tris-HCl (pH 7.5), resuspended in 5 mL 50 mM Tris-HCl (pH 7.5), and rotated at 30 rpm at room temperature for 15 min. The cross-linking reaction was quenched by incubating the beads with 50 mM Tris-HCl. The beads were resuspended in 3 mL 50 mM Tris-HCl (pH 7.5), and 2 mL glycerol was added before the beads were stored at −20 °C. The HLA-DR antibody (clone L243) was used to prepare conjugated beads for enriching MHC class II molecules following the same procedure indicated above.
Immuno-precipitation of HLA molecules.
Cell lysates containing 50 mg of total protein were thawed on ice and diluted with four volumes of modified RIPA buffer. Antibody-conjugated Dynal beads (5 mL) were added, and the suspension incubated at 4 °C overnight (minimum of 12 h). The beads were collected on a DynaMag™-50 magnet (ThermoFisher Scientific) and were washed three times with freshly prepared modified RIPA buffer. The HLA proteins and the HLA-binding peptides (including neo-antigenic peptides) were eluted by vortexing the beads at 650 rpm in 5 mL NEB buffer for 10 min at room temperature. The beads were collected using a a DynaMag™-50 magnet (ThermoFisher Scientific) and the supernatant was transferred to an Amicon® Ultra Centrifugal Filter with a 3 kDa molecular weight cut-off (Millipore-Sigma). The filter was centrifuged at 4,000g for 30 min at 25 °C, and the filtrate was collected and snap frozen using liquid nitrogen and lyophilized.
MANA-SRM sample group establishment.
Lyophilized neo-antigenic peptides were reconstituted in bRPLC Solvent A, and 3% acetonitrile was added to the reconstituted sample to aid in the resolubilization of the peptides. An HPLC fractionation was performed to separate the neo-antigenic peptides into 96 fractions based on each peptide’s hydrophobicity in a weak basic environment (pH=8.2). The bRPLC solvents were used for this HPLC separation performed as previously described [19]. Neo-antigenic peptides were then organized into 32 groups comprising three sequential fractions each, according to the MANA-SRM fraction group scheme as previously described [19]. All MANA-SRM fractions of neo-antigenic peptides were dried in a Speed-Vac at 35 °C with vacuum pressure below 5 mbar.
Dual-reduction.
Eluted and lyophilized neo-antigenic peptides were first reconstituted in bRPLC Solvent A with 3% acetonitrile to aid the resolubilization. 200 mM triethylammonium bicarbonate was added to neutralize samples and adjust the pH to ~8. DL-dithiothreitol (DTT) was then added at 10 mM to the sample followed by an incubation at 60°C for 15 min. After the bRPLC fractionation, each fraction group was dried and could be stored at −80°C until analysis. Each sample was reconstituted in 39 μl mass spectrometry solvent A without formic acid. The dissolved peptide sample was reduced again by tris(2-carboxyethyl)phosphine (TCEP) at 10 mM at low pH (pH=3) followed by an incubation at 60°C for 15 min. The reduced samples were ready for MANA-SRM mass spectrometry assays as previously described [19].
HILIC treatment of peptides.
Peptides were purified through a GlycoWorks HILIC cartridge purchased from Waters. The samples were dried in a Speed-vac at 35 °C with vacuum pressure below 5 mbar.
SRM method development and assay.
33 heavy isotope labeled neo-antigenic peptides flanking gene mutation hotspots on cancer driver genes (including K-Ras, EGFR, TP53, CTNNB1, and IDH2) were predicted by NetMHC 4.0 [20] and subsequently synthesized as AQUA™ Peptides by Sigma-Aldrich (Supplementary Table S2). Optimization of collision energies and fractionation were performed as previously described [19]. A list of the SRM transitions and fraction group IDs for all 33 peptides are shown in Supplementary Table S2 online. All transition parameters were manually examined and curated to exclude ions with excessive noise due to co-elution with impurities. Copy number of each detectable neo-antigenic peptide was calculated using heavy isotope-labeled peptides as previously described [21].
Data deposition:
The data reported in this paper have been deposited via ProteomeXchange (identifier PASS01266).
RESULTS
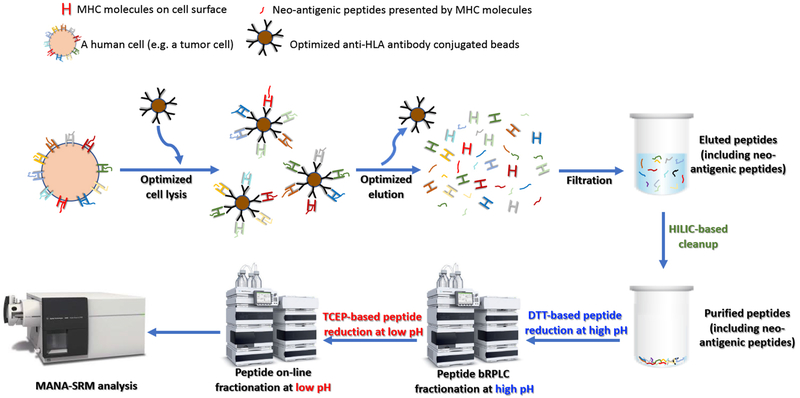
MANA-SRM entailed two steps, an antibody-based neo-antigen enrichment and a multidimensional chromatography-based MS assay (Fig. 1, Supplementary Table S2 and Materials and Methods). For the immuno-precipitation (IP) step, we first explored various ways of cross-linking anti-HLA antibodies on beads. Such cross-linking prevented co-elution of the antibodies with peptides, which otherwise would have caused a major ion-suppression because the amount of antibody was enormous relative to the amount of any individual peptide evaluated. After comparing different types and concentrations of cross-linkers, we found that 25 mM dimethyl pimelimidate dihydrochloride (DMP) maintained the antigen-binding capability of the anti-HLA antibody while remaining firmly attached to protein G magnetic beads (Supplementary Fig. S4A). We then compared various cell lysis and IP protocols to identify one with an optimum yield of HLA molecules after binding to the anti-HLA antibody-bound beads (Supplementary Fig. S4B). Finally, we tested various elution buffers at numerous concentrations and found that 20% acetic acid in 50 mM ammonium bicarbonate produced the highest HLA yield compatible with minimal release of antibody from the beads (Supplementary Fig. S4C).
Fig.1.
Schematic of MANA-SRM pipeline. Cells were lysed and HLA-binding peptides were enriched through immuno-precipitation with an antibody targeting HLA molecules. HLA molecules together with their presented peptides were eluted and dissociated. The eluate containing the neo-antigenic peptide was filtered where only short peptides (MW<3 kDa) were allowed to pass through. The filtered peptide sample was lyophilized and made ready for MANA-SRM analysis with additional improvements through HILIC-based cleanup and the D-Red strategy (see details in Materials and Methods).
Following elution, the peptides were purified from the HLA molecules by a simple filtration through a cellulose membrane. The peptides were then processed through two chromatographic steps. The first separated the peptides into 32 fractions based on hydrophobicity in a weak basic environment (pH=8.2). The fraction containing each MANA of interest was identified using a synthetic heavy isotope-labeled peptide identical in sequence to the MANA. This fraction was subjected to a second, reverse phase HPLC-based separation linked directly to the mass spectrometer. Selective reaction monitoring was used for the specific detection of each MANA after identifying the optimum collision energies, transitions, and other parameters required for SRM-based analyses.
Two further modifications of this overall approach were required for the efficient detection of MANAs. First, we noticed contaminants with characteristic 44-Da peak intervals in MS2 scans, which we interpreted to represent polyethylene glycol (PEG) derivatives (Supplementary Fig. S5). PEG-based detergents (e.g. NP-40) are commonly used in cell lysis and IP buffers [22]. We found that these contaminants could be removed by incubating the peptides with a Hydrophilic Interaction Liquid Chromatography (HILIC) matrix (Supplementary Fig. S6). Second, many potential MANAs contain unstable amino acids such as cysteine or methionine. Peptides containing such residues are typically not chosen for targeted analysis by MS[23]. Such exclusion is generally not detrimental when analyzing proteins in general, since there are usually numerous other peptides from the same protein that do not contain unstable amino acids and are preferable for targeted evaluation. However, in the case of MANAs, there is only one peptide of interest to be evaluated and no other peptides from the protein can substitute for it. We found that such unstable peptides could be stabilized through a dual-reduction strategy (Materials and Methods). The peptides were reduced first with DL-dithiothreitol (DTT) at pH 8.2 prior to bRPLC fractionation and subsequently reduced in tris(2-carboxyethyl)phosphine (TCEP) at low pH before the second chromatographic step. This dual reduction approach improved the signals obtained from Cys- or Met-containing peptides by more than 300-fold (Supplementary Table S3).
We then tested the quantitative performance of MANA-SRM, including its linearity and range of detection. For this purpose, various amounts (0.2 μg ~2 μg) of HLA-peptide complexes with different HLA types were spiked into a cell lysate prepared from 35 million SW480 cells prior to IP. In all cases, over 95% of the spiked-in HLA-peptide complexes were recovered, as assessed by subsequent western blot assays (Supplementary Fig. S4D). The amount of 2 μg for HLA-peptide complexes is equivalent to 800,000 HLA molecules per cell, which is far greater than the amount expressed in cells in vivo or in the cells used in the experiments reported below. We concluded that this IP protocol should be able to recover all the HLA-peptide complexes present.
We next tested 33 potential MANA-related peptides covering residues that are frequently mutated in human cancers, each of which was spiked into the eluted antigen solutions following IP from SW480 cell lysates. The purpose of these experiments was to determine whether the peptides could be detected in the context of IP eluates from cell lysates using the MS component of MANA-SRM rather than to test the efficiency of the IP component. We found that all 33 peptides could be detected, and that the signal strength of each of the peptides was at least 70% of the signal strength of the same peptides measured in the absence of IP eluates (Supplementary Fig. S7). This excellent performance was dependent on several aspects of MANA-SRM. For example, in the absence of the HILIC purification step, 16 of the 33 peptides had weak signals (<10% of the expected values) (Supplementary Fig. S6), and in the absence of dual reduction, 11 of the 33 peptides were undetectable (Supplementary Table S3).
To test the efficiency of the entire MANA-SRM process, various amounts of HLA-peptide complexes were spiked into a SW480 cell lysate. Seven different complexes were tested, and an average of 74% of the peptides were recovered as assessed by MS. Additionally, we demonstrated that the same method could be used to quantify peptides presented by HLA class II molecules (see Materials and Methods). In similar spike-in experiments, we recovered 71% of CLIP DR1*0101 complexes loaded with PVSKMRMATPLLMQA and 73% of HIV Integrase DR1*0101 complexes loaded with SGYIEAEVIPAETGQET (Supplementary Table S4).
Based on the success of these validating experiments, we sought to determine whether MANA-related peptides of interest were actually processed and presented on the surface of cells expressing particular HLA alleles. In these experiments, monkey COS-7 cells were transfected with HLA genes as well as oncogenes predicted to generate MANAs that could bind to the HLA gene products. The MANAs tested included those derived from several different common mutant forms of KRAS, as well one from IDH2. In each case, synthetic heavy isotope-labeled peptides were generated that corresponded to the 9- or 10-amino acid peptides predicted to bind to particular HLA alleles.
We found that some of the predicted peptides were processed and presented, whereas others were not (Table 1). We also were able to quantify the number of copies of the peptides bound to HLA on the surfaces of these cells. For example, 196 copies of a KRAS G12D MANA-related peptide of 9-amino acids in length were found on the cell surface, while only 21 copies of the 10-amino acid form of this MANA were presented. Both the 9 and 10-amino acid forms were predicted to bind to HLA-A3 (predicted KD of 1172 nM and 939 nM) respectively. Controls for these experiments included cells that were transfected with HLA but not the KRAS oncogene or vice versa, or cells transfected with HLA genes not expected to bind the MANA-related peptides. In all such cases, there were no peptides detected by MANA-SRM.
Table 1.
Quantification of neo-antigens through MANA-SRM.
| Cell Line | Transfected with HLA + KRAS vector |
Endogenous HLA A alleles | Total # of cells (Million) | Enriched by W6/32 antibody | Enriched by isotype control antibody | Predicted binding peptide by NetMHC 4.0 | Predicted binding affinity by NetMHC 4.0 [unit: nM, the lower value the stroger bidning] | Detected Abundance (femtomole) | Detected Copy Number Per Cell |
|---|---|---|---|---|---|---|---|---|---|
| COS-7 | HLA-A3 + KRAS G12C vector | N/A | 285.3 | YES | NO | VVGACGVGK (9-mer) | 221.43 | N/A | N/A |
| N/A | VVVGACGVGK (10-mer) | 375.53 | N/A | N/A | |||||
| COS-7 | HLA-A3 + KRAS G12D vector | N/A | 270.9 | YES | NO | VVGADGVGK (9-mer) | 1172.08 | 64 | 196 |
| N/A | VVVGADGVGK (10-mer) | 938.8 | 6 | 21 | |||||
| COS-7 | HLA-A3 + KRAS G13D vector | N/A | 286.7 | YES | NO | VVGAGDVGK (9-mer) | 1552.04 | 26 | 75 |
| N/A | VVVGAGDVGK (10-mer) | 936.84 | 2.2 | 7 | |||||
| COS-7 | HLA-A2 + KRAS G12C vector | N/A | 261.9 | YES | NO | N/A | N/A | N/A | N/A |
| COS-7 | HLA-A2 + KRAS G12D vector | N/A | 300.3 | YES | NO | N/A | N/A | N/A | N/A |
| COS-7 | HLA-A2 + KRAS G13D vector | N/A | 276.6 | YES | NO | N/A | N/A | N/A | N/A |
| COS-7 | HLA-A1 + KRAS WT vector | N/A | 290.2 | YES | NO | ILDTAGQEEY | 86.69 | N/A | N/A |
| COS-7 | HLA-A1 + KRAS Q61H vector | N/A | 254.9 | YES | NO | ILDTAGHEEY | 185.91 | 178 | 583 |
| COS-7 | HLA-A2 + KRAS Q61H vector | N/A | 266.2 | YES | NO | N/A | N/A | N/A | N/A |
| COS-7 | HLA-A1 + KRAS Q61L vector | N/A | 267.6 | YES | NO | ILDTAGLEEY | 66.03 | 136 | 512 |
| COS-7 | HLA-A2 + KRAS Q61L vector | N/A | 269.6 | YES | NO | N/A | N/A | N/A | N/A |
| COS-7 | HLA-A1 + KRAS Q61R vector | N/A | 261.9 | YES | NO | ILDTAGREEY | 322.23 | 40 | 127 |
| COS-7 | HLA-B7 + IDH2 R140Q vector | N/A | 273.3 | YES | NO | SPNGTIQNIL | 228.61 | 8 | 25 |
| COS-7 | HLA-B7 + IDH2 R140Q vector | N/A | 289.2 | NO | YES | N/A | N/A | N/A | N/A |
| COS-7 | HLA-A3 + IDH2 R140Q vector | N/A | 268.6 | YES | NO | N/A | N/A | N/A | N/A |
| RPMI-6666 | Not transfected | A*02:01, A*03:01 | 2,100 | YES | NO | N/A | N/A | N/A | N/A |
| HH | Not transfected | A*01:01, A*01:01 | 2,000.0 | YES | NO | N/A | N/A | N/A | N/A |
| CFPAC-1_G12D knockin | Not transfected | A*02:01, A*03:01 | 83.1 | YES | NO | VVGADGVGK (9-mer) | 1172.08 | 4 | 40 |
| HS578.T | Not transfected | A*03:01, A*24:02 | 70.4 | YES | NO | VVGADGVGK (9-mer) | 1172.08 | 6 | 71 |
| HL-60 | Not transfected | A*01:01, A*01:01 | 1,990.1 | YES | NO | ILDTAGLEEY | 66.03 | 10 | 4 |
| SW948 | Not transfected | A*01:01, A*01:01 | 326.1 | YES | NO | ILDTAGLEEY | 66.03 | 6 | 15 |
| RD | Not transfected | A*01:01, A*01:01 | 412.9 | YES | NO | ILDTAGHEEY | 185.91 | 4 | 8 |
| Hs940.T | Not transfected | A*01:01, A*01:01 | 131.3 | YES | NO | ILDTAGREEY | 322.23 | 0.2 | 1 |
Other MANA-related peptides that could be detected on the cell surface were those derived from KRAS mutations at codons 13 and 61, as well as IDH2 R140Q (Table 1). Mutations at Q61 were particularly well-processed and presented, with up to 583 copies per cell. In contrast, mutations at the KRAS codon G12C, encoding peptides that were predicted to bind to HLA-A3, were not detected by MANA-SRM. Because the identical procedures were used to evaluate each of these potential MANAs (transfection into COS-7 cells followed by MANA-SRM using the same antibody) and the heavy isotope labeled G12C peptides were detected successfully, we conclude that the KRAS G12C peptides were processed inefficiently or that the in-silico binding predictions were inaccurate.
Though not the primary purpose of MANA-SRM, this technique could also be used to assess endogenous processing and presentation of MANAs in cancer cells (rather than in transfected COS-7 cells). We found that Hs578.T lung cancer cells harboring an endogenous KRAS G12D mutation and an HLA-A3 genotype presented 71 copies of the G12D 9-mer by HLA per cell (Table 1). The G12D 10-mer, although also predicted to be present in these cells, was not detected. Controls for these experiments included immuno-precipitation with antibodies that did not bind to HLA but were of the same isotype. Additionally, only the abnormal peptides encoded by the mutant alleles present in these cells were detected by MANA-SRM. For example, G13D peptide-HLA complexes were not detected in cells harboring a G12D mutation, and vice versa.
DISCUSSION
MANAs are critical inducers of T cell-mediated anticancer immune response [1, 24]. Consequently, various immunotherapeutic approaches targeting defined MANAs have been developed [4, 8, 25–28]. Given the efforts and time needed to develop the therapeutic agents against the target MANAs, it is imperative that the presentation on tumor cell surface of the predicted MANAs be verified before such efforts are initiated. Mass spectrometry analysis following immunoprecipitation with relevant anti-HLA antibodies represents a straightforward method to assess whether predicted MANAs are actually expressed, processed correctly, and presented by the HLA molecules [12]. In practice, assessment of MANAs using this method has been challenging, especially when they are presented at low abundance and/or contain reactive amino acids susceptible to side-chain modifications such as cysteine and methionine [23].
We have developed an approach with several critical technical improvements that substantially increased the sensitivity of the published mass spectrometry methods used to identify MANAs. First, we systematically optimized protocols for cell lysis, IP, elution of peptides, as well as crosslinking of anti-HLA on beads to maximize yield of HLA-bound peptides while minimizing co-elution of the antibody which otherwise could lead to signal-suppression and reduced sensitivity of peptide detection. Second, we implemented a new streamlined procedure to purify the eluted peptides, which involved membrane filtration to remove HLA, followed by a hydrophobicity-based and a reverse phase HPLC-based chromatographic steps. Third, we identified derivatives from PEG commonly used in cell lysis and IP buffers as major contaminants and established an efficient method to remove them with the HILIC matrix. Fourth, we developed a dual-reduction strategy to prevent peptides containing methionine and/or cysteine from being modified, which improved the signals derived from these peptides by 300-fold. Together, the new approach with these improvements allowed us to detect endogenous MANAs with frequent driver mutations present at extraordinarily low abundance, likely in the single digit range as suggested by data shown in Table 1.
Massively parallel sequencing coupled with bioinformatics analyses routinely provides a list of candidate MANAs that could potentially be used in immunotherapy. MANA-SRM can determine, at unprecedented sensitivity, which of these candidate MANAs are actually processed and presented on the cell surface. The combination of these technologies thus paves the way for personalized and more effective immune-targeting therapies.
Supplementary Material
ACKNOWLEDGEMENT:
We thank Srona Sengupta and Dr. Robert F. Siliciano for providing us the CLIP DR*0101 and HIV Integrase DR1*0101 tetramers. We thank Agilent Technologies for providing us some of the instrumentation pictures. This work was supported by GI SPORE CA 62924; MSTP GM 07309; ONCOLOGY CORE CA 06973; the Marcus Foundation; the Virginia and D.K. Ludwig Fund for Cancer Research; the Sol Goldman Sequencing Facility at Johns Hopkins; the Conrad R. Hilton Foundation; and the John Templeton Foundation.
Financial support
This work was supported by GI SPORE CA 62924; MSTP GM 07309; ONCOLOGY CORE CA 06973; the Lustgarten Foundation for Pancreatic Cancer Research; the Virginia and D.K. Ludwig Fund for Cancer Research, The Sol Goldman Center for Pancreatic Cancer Research, and the Bloomberg Kimmel Institute at Johns Hopkins.
Footnotes
Conflict of Interest Statement
Bert Vogelstein and Kenneth W. Kinzler are founder of Personal Genome Diagnostics and Thrive and advisors of Sysmex, Eisai, CAGE, Neophore. BV is also an advisor to Nexus. Nickolas Papadopolous and Shibin Zhou are founders of Personal Genome Diagnostics and Thrive. These companies and others have licensed technologies related to the work described in this paper from Johns Hopkins University. Some of these licenses are associated with equity or royalty payments to the afore-mentioned individuals. Additional patent applications on the work described in this paper may be filed by Johns Hopkins University. The terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
REFERENCE:
- 1.Schumacher TN and Schreiber RD, Neoantigens in cancer immunotherapy. Science, 2015. 348(6230): p. 69–74. [DOI] [PubMed] [Google Scholar]
- 2.Riaz N, et al. , The role of neoantigens in response to immune checkpoint blockade. Int Immunol, 2016. 28(8): p. 411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauss M, et al. , Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat Commun, 2017. 8(1): p. 1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott PA, et al. , An immunogenic personal neoantigen vaccine for patients with melanoma. Nature, 2017. 547(7662): p. 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz LA Jr. and Le DT, PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med, 2015. 373(20): p. 1979. [DOI] [PubMed] [Google Scholar]
- 6.Larkin J, Hodi FS, and Wolchok JD, Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med, 2015. 373(13): p. 1270–1. [DOI] [PubMed] [Google Scholar]
- 7.Vitiello A and Zanetti M, Neoantigen prediction and the need for validation. Nat Biotechnol, 2017. 35(9): p. 815–817. [DOI] [PubMed] [Google Scholar]
- 8.Sahin U, et al. , Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature, 2017. 547(7662): p. 222–226. [DOI] [PubMed] [Google Scholar]
- 9.The problem with neoantigen prediction. Nat Biotechnol, 2017. 35(2): p. 97. [DOI] [PubMed] [Google Scholar]
- 10.Lu YC, et al. , Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res, 2014. 20(13): p. 3401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danilova L, et al. , The Mutation-Associated Neoantigen Functional Expansion of Specific T Cells (MANAFEST) Assay: A Sensitive Platform for Monitoring Antitumor Immunity. Cancer Immunol Res, 2018. 6(8): p. 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassani-Sternberg M, et al. , Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun, 2016. 7: p. 13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creech AL, et al. , The Role of Mass Spectrometry and Proteogenomics in the Advancement of HLA Epitope Prediction. Proteomics, 2018. 18(12): p. e1700259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan C, et al. , Accurate quantitation of MHC-bound peptides by application of isotopically labeled peptide MHC complexes. J Proteomics, 2014. 109: p. 240–4. [DOI] [PubMed] [Google Scholar]
- 15.Bassani-Sternberg M, et al. , Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc Natl Acad Sci U S A, 2010. 107(44): p. 18769–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ternette N, et al. , Early Kinetics of the HLA Class I-Associated Peptidome of MVA.HIVconsv-Infected Cells. J Virol, 2015. 89(11): p. 5760–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson CD, et al. , Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol, 2016. 34(3): p. 339–44. [DOI] [PubMed] [Google Scholar]
- 18.Agudelo D, et al. , Marker-free coselection for CRISPR-driven genome editing in human cells. Nat Methods, 2017. 14(6): p. 615–620. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, et al. , Selected reaction monitoring approach for validating peptide biomarkers. Proc Natl Acad Sci U S A, 2017. 114(51): p. 13519–13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreatta M and Nielsen M, Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics, 2016. 32(4): p. 511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, et al. , Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci U S A, 2011. 108(6): p. 2444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Englard S and Seifter S, Precipitation techniques. Methods Enzymol, 1990. 182: p. 285–300. [DOI] [PubMed] [Google Scholar]
- 23.Lange V, et al. , Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol, 2008. 4: p. 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarchoan M, Hopkins A, and Jaffee EM, Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med, 2017. 377(25): p. 2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skora AD, et al. , Generation of MANAbodies specific to HLA-restricted epitopes encoded by somatically mutated genes. Proc Natl Acad Sci U S A, 2015. 112(32): p. 9967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stronen E, et al. , Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science, 2016. 352(6291): p. 1337–41. [DOI] [PubMed] [Google Scholar]
- 27.Tran E, et al. , T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med, 2016. 375(23): p. 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran E, et al. , Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science, 2014. 344(6184): p. 641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.