Abstract
Transposable elements are endogenous DNA sequences able to integrate into and multiply within genomes. They constitute a major source of genetic innovations, as they can not only rearrange genomes but also spread ready-to-use regulatory sequences able to modify host gene expression, and even can give birth to new host genes. As their evolutionary success depends on their vertical transmission, transposable elements are intrinsically linked to reproduction. In organisms with sexual reproduction, this implies that transposable elements have to manifest their transpositional activity in germ cells or their progenitors. The control of sexual development and function can be very versatile, and several studies have demonstrated the implication of transposable elements in the evolution of sex. In this review, we report the functional and evolutionary relationships between transposable elements and sexual reproduction in animals. In particular, we highlight how transposable elements can influence expression of sexual development genes, and how, reciprocally, they are tightly controlled in gonads. We also review how transposable elements contribute to the organization, expression and evolution of sexual development genes and sex chromosomes. This underscores the intricate co-evolution between host functions and transposable elements, which regularly shift from a parasitic to a domesticated status useful to the host.
Keywords: Transposable element, Sex determination, Sexual development and function, Germline, piRNA, Sex chromosome
Background
Transposable elements (TEs) are major actors of the evolution of genomes and the diversification of species [1]. These DNA sequences have the peculiarity of being able to integrate into and spread within genomes, as well as to recombine and induce genome rearrangements, since they are generally repetitive. First discovered in maize [2], TE families described so far are generally divided into two main classes [3]. Class I TEs (retroelements) spread through a “copy-and-paste” mechanism called retrotransposition, which corresponds to a process of RNA-mediated duplication. They express an RNA intermediate that is reverse-transcribed into a cDNA fragment, which will be inserted somewhere else into the genome. Hence, retrotransposition directly increases the copy number of an element. In contrast, Class II TEs (DNA transposons) move through a “cut-and-paste” mechanism. Most autonomous class II elements encode a transposase that can bind to and excise the transposon from its initial genomic localization, and can subsequently insert it into a new locus [3–5]. This mechanism does not per se duplicate the initial transposon but only changes its location. However, the transposon can be duplicated if the transposition event occurs during the replication process, from an already replicated region to a non-replicated one.
Since they can insert into genomes, recombine and generate different types of rearrangements, TEs are by nature an important source of genomic variability between different species, or between individuals within a given species or population. Most insertions are thought to be deleterious for the host, in particular when they disrupt essential genes, regulatory regions or chromosomal structures, causing negative effects ranging from a slight decrease in host fitness to lethal mutations [6]. When a TE insertion is associated with such a fitness disadvantage, it is generally counter-selected and finally lost. The process of loss can however be modulated by several factors, including the selection coefficient of the insertion, its potential linkage disequilibrium with an advantageous allele, the recombination rate of the region of insertion, and the effective population size of the host [7]. Some insertions, in contrast, can be neutral, for example if they occur in genomic regions that have no crucial impact on host fitness, like gene-poor regions for instance. It is however difficult to classify an insertion as “neutral” once and for all, since it can still induce chromosomal rearrangements through ectopic recombination [8]. Lastly, some TE insertions might bring positively selected changes. In particular, TEs can spread ready-to-use regulatory sequences or trigger epigenetic modifications able to modify the pattern of expression of neighboring genes (for a review see [9]). TEs can also be “domesticated” as new host non-coding RNA genes or genes encoding useful proteins such as the syncytins, which are involved in the development of the placenta in mammals [10–12]. Syncytin genes have been repeatedly derived from envelope genes of endogenous retroviruses during mammalian evolution. Another example of TE-derived host proteins are the Rag proteins, which catalyze the V(D) J recombination responsible for the diversity of immunoglobulins and T cell receptors found in B and T cells, respectively. These proteins were formed from a Transib DNA transposon about 500 million years ago [13]. Many other examples of TE-derived genes have been described in different organisms (for a review see [11, 14]).
Persistence of TEs within a population, which would reflect their evolutionary success, requires their vertical transmission from one generation to the next. In animals with sexual reproduction, i.e. involving the fusion of male and female gametes, this implies transposition in the germline cells that will form the next generation. Sexual reproduction might be instrumental for the propagation of mainly deleterious TEs [15–17]. Indeed, in asexual populations, TEs might not be able to spread and tend to be eliminated if no horizontal transfer occurs [15–17]. Accordingly, experimental studies have shown that TEs are less fit to increase their frequency in asexual populations compared to sexual populations [15, 17–19]. Homologous recombination during meiosis is another feature of sexual reproduction that has an antagonistic impact on the fixation rate of TEs by favoring the elimination of deleterious TE insertions [20, 21]. Recombination triggers the exchange of genetic information between homologous chromosomes belonging to a same chromosome pair. This process has been associated to an increase of purifying selection since it drives the removal of deleterious point mutations and TE insertions [20, 21]. Hence, recombination and sexual reproduction could be considered as a defense mechanism against deleterious TE insertions. Reciprocally, high rates of deleterious mutations such as TE transpositions might favor the maintenance of sexual reproduction as an efficient way to keep these mutations at levels compatible with life [15, 17, 22–24]. In the asexual species Leptopilina clavipes (the wasp), no particularly high TE content is observed, despite the expansion of specific TE families, which could be linked to the switch toward asexuality [25]. The absence of recombination here does not seem to have triggered a massive expansion of TEs, or is counterbalanced by the limited spreading of TEs in the population due to asexuality. Similarly, no difference in TE composition was observed between the genome of an asexual fish of hybrid origin, the amazon molly Poecilia formosa, and the genomes of its parental sexual species, possibly due to the very recent occurrence of the switch from sexuality to asexuality in this lineage [26]. In the more ancient asexual taxa of the bdelloid rotifers, retrotransposons were long thought to be absent [27], supporting the role of sexuality in the genomic maintenance of these TEs [23]. More recent studies somehow challenged this model by highlighting a high diversity of TE families including LTR and non-LTR retrotransposons. However, each of these families presents a very low number of intact copies (one or two for the majority of them) [28]. Such a TE landscape, associated with the relatively low abundance of decayed fragments, the high similarity between LTRs for intact copies, and the localization of TEs in horizontally transmitted regions, led the authors to hypothesize that TEs were mostly acquired by recent horizontal transfers in rotifers [28].
In species with gonochoristic sex, i.e. species in which individuals are either male or female (in contrast to hermaphrodite species, in which individuals produce both male and female gametes), different factors can control sex determination (SD) [29, 30]. Some species undergo environmental sex determination (ESD), while others are subject to genetic sex determination (GSD). In ESD sex is determined by environmental factors, for instance temperature in turtles or crocodilians [31, 32]. Such temperature sex determination seems to be also present, albeit rare, in fish, as it was recently demonstrated for the Southern flounder [33]. In GSD on the contrary, the sex of the individual depends on its genotype. Sex can be determined by several interacting loci in a given species (polygenic sex determinism), but the most prevalent situation appears to be the monogenic GSD. In this situation, the chromosome pair that harbors the master SD gene becomes the sex chromosomes, or gonosomes. Two main sex chromosome configurations exist: the XX/XY system, particularly found in mammals, where males have two types of sex chromosomes (X and Y, male heterogamety), and the ZW/ZZ system, common in birds, where females have two different sex chromosomes (Z and W, female heterogamety) [34, 35]. Many other GSD systems have been reported such as haplodiploidy, where for instance males arise from haploid unfertilized eggs and female from diploid fertilized eggs, like in bees, ants, or some molluscs [36]. In the XX/XY sex determination system in mammals, the Sry gene is the male master sex determining gene for almost all species. Sry is located on the Y but not on the X chromosome and is therefore present in males but not in females. Non-mammalian species such as the fruit fly Drosophila melanogaster or the medaka fish Oryzias latipes also have XX/XY sex determination systems but of independent evolutionary origins. The Sry gene is absent from these species. In O. latipes the Y-linked master gene dmrt1bY, which is a Y-specific duplicate of the dmrt1 gene, drives development toward the male phenotype like Sry in mammals [37, 38]. In D. melanogaster, the X chromosome carries Sxl that has to be in two copies to trigger female differentiation [39]. In this case, the initial choice between the male and female pathways is thus triggered by a dosage effect of the master gene. In birds, a similar process occurs but in a ZW/ZZ system, where ZZ males have two copies of the Z-linked dmrt1 gene and females only one. This creates a gene dosage difference, leading to male or female differentiation [40]. In the nematode C. elegans individuals are either males or hermaphrodites. The presence of two X chromosomes (XX individuals) triggers the differentiation into a hermaphrodite adult that produces both male and female gametes. In contrast, XO individuals differentiate into males as a consequence of the ratio between X chromosomes and autosomes [41, 42].
Once sexual development is initiated, the gonad, which comprises both germ cells and somatic cells, differentiates into either a testis or an ovary. A sex-dependent gene regulatory cascade, initiated in the somatic part of the gonad, controls differentiation [30, 43, 44]. Male and female differentiation cascades are often repressing each other, creating a competition between male and female differentiation genes: the most expressed pathway represses the other one [43]. Finally, once the gonad is differentiated, sex is maintained by the expression of specific genes like those encoding the sexual hormone biosynthesis pathways in mammals. It has been shown in mammals and teleost fish that even in adults, de-repressing the opposite pathway can induce sex reversal [45–47]. This demonstrates that expression of at least some of the sexual development network genes is necessary to maintain the differentiated state in sexually mature individuals. Beyond gonads, sex affects many other pathways in the organism, creating a bias in gene expression in several tissues and organs including brain [48–53]. However, gonads remain the most sex-biased organs in terms of gene expression.
Depending on the animal lineage, sexual development and particularly sex determination can show very different evolutionary dynamics. Some SD systems are ancient and at least 100 million years old, such as the mammalian male heterogamety system driven by the Y-linked gene Sry [54] or the avian female heterogametic determination controlled by the Z-linked dmrt1 gene [40]. In other lineages, for instance in teleost fish, sex determination is much more labile, with a frequent switch between and even combination of ESD and GSD, and an important turn-over of sex chromosomes and master sex-determining genes in GSD [55, 56]. For example, the genetic sex determination system is not conserved in the genus Oryzias: while O. latipes, O. curvinotus, O. luzonensis and O. dancena use a XX/XY system, O. javanicus determines sex through ZW/ZZ female heterogamety [57]. Strikingly, Oryzias species with a XX/XY system generally have different sex chromosomes and even different master sex-determining genes: sex is controlled by dmrt1bY (aka dmy) in O. latipes and O. curvinotus, gsdfY in O. luzonensis and sox3Y in O. dancena [57]. Hence, the control of sexual development can be considered as a fast-evolving trait in this clade. Beyond the initiation of sex differentiation, the downstream molecular pathways also appear variable among animals: a comparison of genes expressed in medaka fish and mammalian gonads revealed substantial differences [58]. Very interestingly, the control of sexual development sometimes experiences convergent evolution: in both therian mammals (non-egg-laying placental mammals and marsupials) and Oryzias dancena for instance, the master sex-determining gene evolved from the Sox3 gene [59]. This happened independently in the two lineages, 148 to 166 million years ago in a common ancestor of therian mammals, and less than 20 million years ago in Oryzias dancena. Another striking example is the dmrt1 gene in birds and in the tongue sole. This gene was ancestrally located on the vertebrate linkage group A, which became the Z chromosome independently in both lineages [60].
In this review, we reassess the impact of transposable elements on the structure and expression of genes and genomes through the prism of sex by inventorying the known reciprocal interactions between TEs and sexual development and function in animals. The species sample, however, appears heavily biased towards insects and vertebrates, since most of the studies linking TE and sex have been conducted in classical model organisms commonly used in genetics and development. We first focus on the expression of TEs in germ cells and on the control of their expression. Then, we review how TEs, reciprocally, can impact the expression of sexual development genes. Finally, we document how TEs influence the organization and structural evolution of sexual genes and chromosomes. These diverse and reciprocal influences well illustrate the intricate co-evolution of TEs with their host.
TE expression is tightly controlled in the germline
TEs in the germline: a trade-off between expression and control
Expression and transposition of TEs in the germline are necessary for their vertical transmission to the host progeny, and ultimately for their maintenance within a lineage. The first step of TE transposition consists in the transcription of mRNA to produce enzymes such as a transposase for most DNA transposons, or a reverse transcriptase and an integrase/endonuclease for retroelements. TE mRNAs are expected to be found in cells where TEs are spreading. TE-derived transcripts are indeed found in transcriptomes [61–64], including the germline [65, 66]. In the medaka Oryzias latipes for instance, about 1.2 and 3.5% of the transcriptome of ovaries and testes, respectively, can be assigned to TEs (Dechaud et al. unpublished data).
If evolution fosters TEs that are active in gonads, the putative negative effects of TE insertions, at the same time, require repressive mechanisms. The gonadal activity of a TE results in a trade-off, its own survival depending on the survival of the host, which is needed for vertical transmission and maintenance. This follows the “selfish gene” hypothesis according to which, in a gene-centered view of evolution, some genes can enhance their own transmission, sometimes with a negative effect on the organism fitness [16]. Very interestingly, some TEs like the P element in Drosophila produce different transcripts depending on the organ in which they are expressed [67]. In the gonads, the third intron of the P element is excised allowing its transposition, while in the soma, in addition to a transcriptional control, the P element transcript keeps its third intron and is not able to transpose [67]. Such mechanisms allow the element to limit its impacts on the soma while transposing in the germline.
Germline TE expression is controlled by several mechanisms
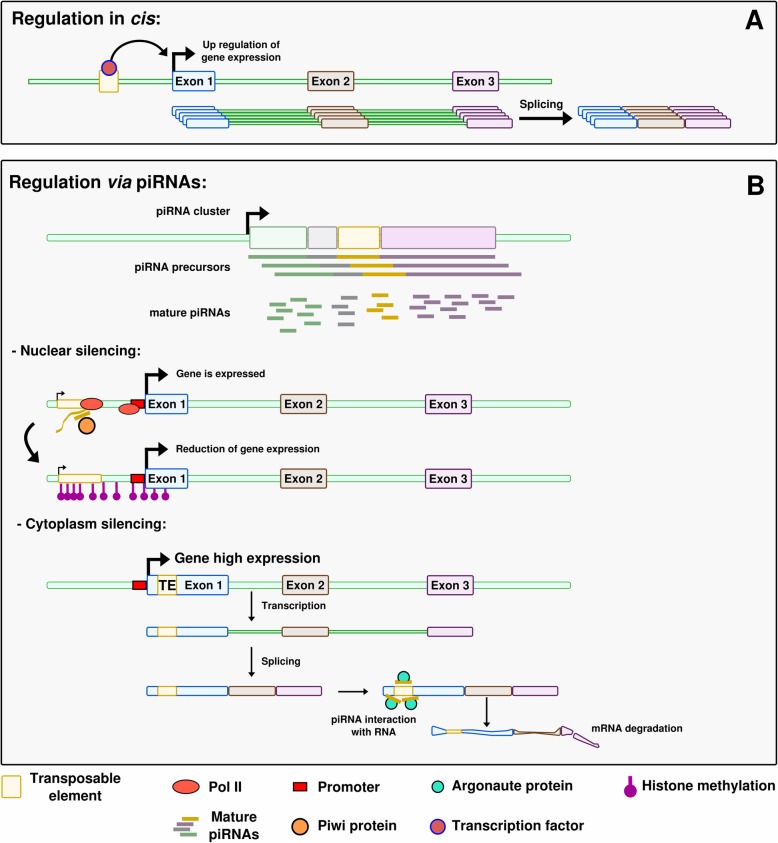
piRNAs (Fig. 1a)
Fig. 1.
Different ways to control TE expression. a piRNAs. piRNAs are produced from piRNA clusters, genomic spots where new TEs can integrate. piRNAs can act through two mechanisms. In the nucleus, piRNAs bind to Piwi proteins. They also bind in anti-sense to TE mRNA being transcribed, triggering histone methylation of TEs and thus inhibiting recruitment of Pol II. This leads to the silencing of TE expression. In the cytoplasm, piRNAs bind to other Argonaute proteins, triggering TE mRNA degradation. b Repressor proteins. A functional P element produces the transposase that triggers its excision and transposition. When repressor proteins are transmitted from the mother through cytoplasm or when the P element is degenerated, it produces an alternatively spliced mRNA. This mRNA encodes a non-functional transposase that will act as a repressor by competing with the functional transposase, and trigger the production of more alternatively spliced mRNA. This positive repression loop, where the repressor protein activates its own production, prevents the transposition of the TE. c DNA methylation. The TE is methylated, preventing its expression
Piwi-interacting RNAs (piRNAs) are 24–31 nucleotides long small non-coding RNAs expressed in the germline and derived from long RNAs that contain TE sequences [68]. They have been described in eukaryotes only, from humans to protozoans [69, 70] and play a large diversity of roles, such as genome rearrangement in ciliates, sex determination in silkworm, telomere protection in Drosophila, long-term memory in sea slug, or oocyte development in human [70]. piRNAs are produced from specific loci called piRNA clusters that regularly integrate new TE-derived sequences and thus extend their target potentialities. They can further be amplified by the so-called “ping-pong” cycle [71].
piRNAs can regulate TE expression by two different mechanisms. The first mechanism occurs in the nucleus, where piRNAs interact with the Piwi proteins, a subfamily of Argonaute nucleases, to target the TE nascent RNAs to which they present sequence similarities, and adds histone repressive marks in the region by interacting with other proteins [68]. This mechanism inhibits the expression of the targeted TEs. The second mechanism happens in the cytoplasm, where piRNAs form a complex with Aubergine (Aub) proteins, which belong to the Piwi subfamily too. This complex post-transcriptionally silences TE expression by interacting with the TE mRNAs. This also triggers a replication of the piRNA, known as the ping-pong cycle [68]. The ubiquitous presence of this regulatory system in the gonads specifically underlines the importance of controlling TE activity in the germline.
As an example, piRNAs are involved in the P-cytotype regulation in Drosophila [72]. In these species, some strains of flies have a DNA transposon, the P element, from which a complementary piRNA is produced. These are called “P strains”, for Paternal contributing strains, in opposition to “M strains”, for Maternal contributing strains. One model proposes that in P strains, P element-derived piRNAs are transmitted from the mother through the oocyte cytoplasm. The transmitted piRNAs then silence the P element both in the nucleus and the cytoplasm by the mechanisms described above. piRNAs are further amplified in the cytoplasm through the ping-pong cycle, maintaining the silencing of the P element. If no piRNA is transmitted from the mother, the P element is not repressed. Consequently, a P male crossed with an M female will have a dysgenic offspring, with increased mutation rates, frequent sterility and abnormally small gonads [73]. This phenomenon, due to the fact that the offspring have the P element but no silencing through maternal piRNA, is known as “hybrid dysgenesis” [67, 72]. In contrast, the offspring of a P female crossed with an M male is fertile, as the P female brings the P element but also some piRNAs to trigger its repression, as well as the ping-pong amplification cycle.
Repressor proteins (Fig. 1b)
TE expression can also be directly controlled by protein factors. In vertebrates, KRAB-ZNF (for Krüppel-associated box domain zing finger) proteins have been shown to play this role ( [74], reviewed in [75]). They constitute a large family of proteins and are able to bind various DNA sequences via the diversity of their ZNF domains. They recruit KAP1 (for KRAB-associated protein 1) to DNA, which in turn mediates transcriptional silencing through histone modifications. KRAB-ZNF proteins were first discovered in mice where they silence genomic insertions of a murine leukemia virus (MLV) [76], but recent studies demonstrated their action on other retroelements [77]. Many KRAB-ZNF proteins are expressed during germline development; however the targeted TE families are still to be discovered for most of the KRAB-ZNF members [77–79]. In Drosophila, a second model of P-element control involves repressor proteins. P strains express a repressor protein that prevents the transposition of the P element in the germline. This mechanism is known as the “protein repressor model” [67, 72]. The repressor is produced from degenerated P elements or from alternatively spliced full P element transcripts. If the precise action mechanism of the repressor protein is unknown, the main hypothesis is a competitive inhibition with the P element transcription [72]. This repressor could also further trigger the production of alternatively spliced transcripts, leading to a feedforward repression loop (Fig. . 1); however this action as a splicing modifier has never been demonstrated. It is inherited from the mother through the cytoplasm. Since the discovery of piRNA however, later demonstrated to repress TEs in the germline [80], an alternative model has been proposed for the P-cytotype regulation (see before). Both models are not mutually exclusive and likely coexist within populations or individuals [72].
Epigenetic modifications (Fig. 1c)
TE activity can be controlled by epigenetic regulations such as DNA methylation [9] or histone modifications [80, 81]. These epigenetic controls however are not specific of the germline. The modifications targeting TEs can sometimes also affect neighboring genes, hence participating in shaping their regulation and influencing genome evolution [82]. Indeed, the epigenetic silencing of TEs is known to be released in cases of stress, for example UV exposure or temperature changes [83]. Thus TEs can be reactivated and expand, influencing genome evolution under stress conditions [82].
TE expression can vary between sexes
Epigenetic modifications and gene expression can differ between sexes. One may wonder, because of these epigenetic differences, whether TE activity would also vary between males and females. Some TE families are expressed at unchanged levels in very different contexts, like SINEs in rats [84]. In this study, 11 organs were tested including testis and uterus, each at 4 developmental stages. Contrary to SINEs, LTR appeared to be more likely to be expressed in specific tissues or conditions, and are also found more differentially expressed between sexes [84, 85].
In mammals, the inactivation of the Piwi regulatory system in the germline of males leads to azoospermia (no production of mature gametes) due to a high rate of illegitimate pairing between non-homologous chromosomes at meiosis that trigger apoptosis [86]. Also, piRNA interacting protein expression was found to be impaired in humans with cryptorchidism (absence of both testes, or location outside the scrotum) [87]. In contrast, Piwi system inactivation in female mice does not lead to over-activation of TEs [86], and neither does a knock-out of dicer, a protein involved in the siRNA degradation system, which would have suggested the involvement of the RNA interference pathway in TE control. One player of this control corresponds instead to the evolutionarily conserved MAEL protein (encoded by the maelstrom gene), found both in mouse and fly [88]. When this factor is mutated, a 2.3-fold excess of L1 mRNA is measured in embryonic day 15.5 mouse oocytes [88]. Although its precise role is still unclear, MAEL intervenes in a silencing step downstream of Piwi [64]. Of note, TEs are hypomethylated in females compared to the male germline. Hence, oocytes seem more resilient to TE transposition than the male germline. It has been suggested that this difference could be linked to the life-long division of spermatogonial cells, in contrast to oocytes, which undergo a long meiotic arrest. Cell division is required for TE transposition, and many more cell divisions occur in the male germline. More cell divisions would allow too many deleterious insertions in the male germline, explaining the need for TE silencing [86].
TEs can regulate the expression of sexual development genes
TEs can have an important impact on gene regulatory networks [89–91]. They can modify the expression of surrounding genes [9, 91] by bringing with them Pol II or III promoters as well as transcription factor binding sites, insulators, splicing sites or epigenetic modifications. TEs could be particularly prone to recruitment into sexual development since they are generally expressed in the gonads.
Regulation in cis (Fig. 2a)
Fig. 2.
Different ways how TEs can affect gene expression. a Regulation in cis. The TE brings a ready-to-use regulatory sequence that carries a transcription factor binding site. The transcription factor can bind on this site and influence expression of the neighboring gene. b Regulation via piRNAs. In the nuclear silencing situation, a TE is present close to the gene of interest. The piRNA, via the Piwi protein, triggers histone modifications that silence the TE but also affect the RNA polymerase binding region of the neighboring gene. Because of the epigenetic modification of the TE, the gene expression is reduced. In the cytoplasm silencing situation, a TE-derived sequence is present in the 5’UTR of the gene. piRNAs specific to this TE bind the transcript in the cytoplasm via an Argonaute protein and trigger the degradation of the transcript
TEs have a strong cis-regulatory potential for host genes through their Pol II or Pol III promoters and binding sites for transcription factors, or other regulatory sequences, which they carry [9]. These regulatory sequences can already exist in the TE sequence, or derive from this sequence by a few point mutations only. Some of the described examples are related to sexual development.
In Drosophila species, MSL Recognition Elements (MREs) are known to trigger dosage compensation for X chromosomal genes. MSL (for Male Specific Lethal) is a male-specific complex that binds to MREs and increases neighboring gene expression in XY males, hence compensating for the absence of one X chromosome compared to XX females. MREs are found at multiple loci interspersed on the X chromosome. Interestingly, they are carried by Helitron DNA transposons that regulate in cis genes close to their insertion sites [92, 93]. In Drosophila miranda the X chromosome is recent, allowing the detection of the Helitron sequences with alignments methods, while in other Drosophila with older X chromosomes, MREs are present but the Helitrons are not detectable anymore. The authors propose that, on these older chromosomes, selection eroded the Helitron TEs outside of the selected MRE motifs [92, 93]. This example illustrates the efficiency of TEs in the rewiring of gene regulatory networks, as they can spread transcription factor binding sites or other types of regulatory sequences that can then co-regulate several genes. This process appears even more efficient than the birth of transcription factor binding sites “from scratch” by a series of point mutations, which would require much more time to target different genes [89]. More recent studies on MSL in Drosophila show that other mechanisms such as microsatellites expansion also spread MRE motifs on neo-X chromosomes [94]. In Drosophila melanogaster, the promoter of the Su (Ste) piRNA – one of the most abundant piRNA in the testes – derives from a 1360 transposon [95, 96]. Su (Ste) silences the Stellate genes, hindering the accumulation of Stellate proteins, which causes formation of crystals and results in male sterility [97].
Other cases of TE-controlled genes have been described in other organisms. In the medaka fish Oryzias latipes, the master sex determining gene dmrt1bY has been formed through the duplication of the autosomal gene dmrt1a, which has a downstream position in the male sex differentiation cascade in vertebrates. Dmrt1bY is controlled by different transcription factors including itself, its paralog Dmrt1a and Sox5. Binding sites for these transcription factors are located in the upstream region of dmrt1bY, which corresponds to a non-autonomous P element called Izanagi, in which a LINE/Rex1 retroelement was inserted later (Fig. 3a) [98]. The binding sites for Dmrt1A and Dmrt1bY are located within Izanagi, while the binding site for Sox5 lies within the Rex1-derived sequence [47, 98]. Here, the TEs directly brought the cis-regulatory elements that conferred to dmrt1bY an expression pattern compatible with a function as a master sex-determining gene. This makes a convincing case for TEs being actors of sex determination evolution (Fig. 3b) [98]. Accordingly, it has also been suggested that recent TE insertions in humans (like Izanagi in medaka) usually bring context-specific gene activities, while older TE insertions are more likely to correspond to broad enhancers [99]. In human, enhancers are globally depleted in recent TE insertions. However, enrichment of young TE families is observed in enhancers of genes specifically expressed in testis [99].
Fig. 3.

dmrt1bY evolution and regulation in Oryzias latipes. a In the ancestor, the dmrt1 gene existed in a single copy on a pair of autosomes. dmrt1 was then duplicated into dmrt1a and dmrt1b. Later, two TEs inserted upstream of dmrt1b: Izanagi, a DNA/P-element, followed by Rex1, a LINE retrotransposon [98]. After the insertion of these TEs, dmrt1b became the master sex-determining gene dmrt1bY and the chromosome harboring it became the Y chromosome (the gene is absent from the X). b dmrt1bY is expressed during sex determination in the prospective males. Its product triggers sex determination towards the male phenotype. It also binds on its own binding site in Izanagi, down-regulating its own expression. After sex determination and in adults, dmrt1a, the ancestral paralog of dmrt1bY, is expressed. It binds to Izanagi, down-regulating and silencing dmrt1bY once sex determination has occurred. This silencing is also ensured by the binding of Sox5 to a motif encompassed in the Rex1 sequence
Regulation by piRNAs (Fig. 2b)
TEs can affect the regulation of genes in trans via piRNAs. If piRNAs are originally devoted to the down-regulation of TEs, there is now accumulating evidence that piRNAs regulate host developmental genes and maternal mRNA decay [100]. As an example, TE-derived piRNAs can target maternally-deposited copies of the Drosophila embryo nos mRNA for degradation, which is required for a proper development of the head [101]. The region of the nos 3′ untranslated region that is recognized by the piRNAs originates from two different TEs [101]. We can find some evidence of such regulation in gonads. In Drosophila ovarian somatic sheet cells a piRNA knock-down affects the expression of about 100 transcripts [102]. Most of these deregulated transcripts originate from TEs, but a significant part of them still corresponds to host protein-coding genes, with different genes being affected according to cell lineage. Some of these genes presented de novo inserted TEs in their introns or UTRs that induced suppression by the PIWI machinery at the nascent RNA level [102]. In mouse spermatocytes, piRNAs derived from TEs were shown to mediate the degradation of numerous mRNAs and lncRNAs [103]. This regulation involves PIWIL1, a major actor of the piRNA pathway, the knockdown of which leads to the upregulation of 172 genes. piRNAs were shown to target in particular retrotransposon sequences located in the 3′ UTR of mRNAs [103]. TE-derived sequences thus play a role in the control of germline expressed genes through piRNAs.
Some piRNAs have been demonstrated to trigger sex determination. In Bombyx mori, a species where the sex determining system is ZW/ZZ, the master sex-determining region is localized on the W chromosome and produces female enriched piRNAs deriving from TEs and repetitive sequences. The Fem piRNA encoded in this sex-determining region of the W chromosome derives from a non-TE repetitive region and forms a complex with a silkworm equivalent of the Piwi protein. The complex targets and cleaves a masculinizing protein-coding mRNA transcribed from the Z chromosome, triggering feminization [104, 105]. A similar example has been described in C. elegans, where the 21ux-1 piRNA downregulates the xol-1 gene involved in X chromosome dosage compensation and sex determination [42]. This piRNA control of xol-1 appears to be conserved in the related nematode C. briggsae, suggesting a robust involvement of piRNA in controlling gene expression [42]. In these two examples however, neither the piRNA nor its target were shown to be derived from TEs. In mammals, as described before, the inactivation of the epigenetic control of TEs in male gonads leads to azoospermia and thus infertility [86]. However, a certain relaxation of epigenetic control is observed in the germline, leading to demethylation of TEs and their reactivation. At a first look, this could be considered as deleterious for the host. The relaxation happening in the germline leads to a low level of TE activity that is actually thought to allow the host to sense the TEs present in the genome [86]. Such sensing would help to better control TE transposition. According to the authors, this sensing could be ensured by piRNAs. Relaxation of the epigenetic control allows TE expression that itself triggers piRNA production. piRNAs could then limit the impact of TEs but also regulate the expression of other genes, and through these possibly participate in sexual development. Taken together, the presence of TEs in genomes could be linked to the fact that they have an indirect effect, via piRNAs, on the control of specific genes, and sometimes on critical event such as sexual development.
TEs are involved in sex chromosome structure and evolution
We have described how sex can influence TEs expression, and reciprocally how TEs can modulate expression of genes involved in sexual development. In addition to effects of TE on host gene expression, genomic differences can exist between males and females in terms of TE and gene position and content. These differences can impact sexual development.
In mammals, the X and Y chromosomes are derived from a same pair of autosomes. Accordingly, even if the Y chromosome has lost many of its genes due to suppression of recombination, most genes carried on the Y chromosome have homologs on the X chromosome. This scenario of gene loss, however, does not appear universal, since in certain cases, like in Drosophila melanogaster, sex chromosomes evolved more through gene gain [106]. In the platyfish (Xiphophorus maculatus), an accumulation of Texim genes is observed on the Y chromosome [107]. These genes are physically associated to a Helitron transposon, which might have spread the Texim sequences on the Y chromosome but not on the X. In salmonids, recent findings on SD showed that the master sex-determining gene, sdY, is conserved in many species. However, it does not always locate on the same chromosome, but instead seems to behave like a “jumping gene” [108, 109]. An analysis of the boundaries of the moving region that carries sdY revealed the presence of several TE sequences, leading authors to propose a mechanism of TE-associated transduction [108, 109]. This phenomenon could be linked to a rapid turnover of sexual chromosomes in this clade. Other examples of such sex determining “jumping genes” have been described in animals, such as in the house fly [110] or in Chironomus species [111]. In these cases the possible involvement of TEs in the translocation of the determining cassette has not been investigated, but we can notice that, in the case of the house fly, about two thirds of the Y-linked scaffolds present sequence similarities with TEs [110].
TEs can also themselves present sex-specific localizations. As described before, in Drosophila miranda the recently formed X chromosome, called “neo-X”, accumulates Helitron DNA transposons [92]. The success of fixation of this TE on this specific sex chromosome is probably linked to its role in the expression of X-chromosomal genes, bringing an evolutionary advantage (see part 2A) [92]. Sex chromosomes are actually often enriched in TEs [112–115]. This accumulation might be in some cases the consequence of the impossibility for sex chromosomes to recombine and thus eliminate deleterious insertions. In the genome of the African clawed frog Xenopus laevis, recombination between W and Z sex chromosomes stopped recently, and a large accumulation of TEs already started in the W specific regions [115]. Such accumulation has also been observed on several young sex chromosomes of teleost fishes [112]. The higher density of TEs on these chromosomes might increase their probability to regulate some key sexual development genes and consequently to impact sexual development. In birds, such as woodpeckers for instance, the female specific chromosome W is enriched in CR1 insertions, which is a retrotransposon [116, 117]. In human, the Y chromosome is a hot spot for specific TE insertions [118]. All TE types show a higher density on the Y compared to autosomes, except for SVA short retrotransposons. In particular, density is 30 times higher than the genome average for LTR elements, and four times higher for Alu and L1 elements. The authors assume that this cannot be due to a genome assembly artifact, since the enrichment varies according to TE families. Nevertheless, they do not provide any explanation for the insertion rate differences between TE types on the Y chromosome. This high TE density on the Y chromosome is not explainable by low gene density as human chromosome 13 has a lower gene density and is not enriched for TEs [118]. This accumulation of active elements suggests that the Y chromosome is not shrinking in man, but still expanding through new insertions [119]. Of note, in contrast to what is observed in mammals and birds, the heterogametic sex chromosome (W or Y), in many fish, reptiles and amphibians, is much larger than the Z or X, and often the largest chromosome of the complement. In these groups, sex chromosomes are usually younger than in mammals and birds, with frequent turnover. In addition to bringing additional DNA material, it has been hypothesized that TE insertions could favor, in a fast and effective manner, structural differences between gonosomes, that in turn help the expansion of the region of suppressed recombination [120]. This could thus lead to an increase in sex chromosome size during the early phase of their differentiation, while size diminishing would occur later in their evolution [120]. The accumulation of TEs and other repetitive sequences on the Y chromosome has been hypothesized to globally impact the chromatin landscape of the genome [121, 122]. Indeed, polymorphic Y chromosomes that differ only by their quantity of repeats are associated to different levels of chromatin repression on autosomes [122]. The high density of TEs and satellite DNA on the Y chromosome could function as a sink for heterochromatin marks, leading to a dilution of these marks in the rest of the genome, and hence to differential expression between males and females [122].
The X chromosome inactivation in mammals, also called Lyonisation, is a dosage compensation process in which one of the two X chromosomes is inactivated in XX females, preventing gene overexpression compared to males, which have a single X [123, 124]. The enrichment of LINE retrotransposons on the X chromosomes of human and mice led to the hypothesis of an involvement of LINEs in this process [114, 124]. This hypothesis has been tested in the spiny rat Tokudaia osimensis, where males and females are XO [125]. No dosage compensation by X inactivation is required here, suggesting that LINEs would not be required on this X chromosome. Interestingly, the authors describe a similar high concentration of LINEs on this X chromosome compared to humans or mice. They conclude that the accumulation of TEs on X chromosomes might be only a by-product of reduced recombination [125]. This idea was also reviewed later by Lyon, leading to the same conclusion [126]. Further investigations on the role of LINEs in X chromosome inactivation have been conducted in mammals. On the human X chromosome, regions poor in L1 elements contain genes escaping X inactivation [127]. In placental mammals, the inactivated X chromosome is coated with Xist (X-inactive specific transcript) RNAs, which have a silencing effect. These regions are composed of silent LINEs that are closed in chromatin 3D structure, and are formed prior to gene inactivation [128, 129]. As genes “move” in the Xist silenced region via a modification of the 3D conformation of the chromosome, they become inactivated. Conversely, LINE poor regions are physically distant from Xist silenced regions [123, 129]. In these studies, the authors show that LINEs play a role in the spread of X chromosome silencing by recruiting Xist RNAs, suggesting a general role in the regulation of X-chromosomal gene expression. This phenomenon also exemplifies that for understanding chromosomal organization the intricate structure and function relationships have to be considered.
Conclusions
Sex is an important parameter to take into account when performing experiments, in particular when analyzing gene expression [130]. Many studies, including genome sequencing, are conducted in individuals of only one sex, and results observed might not be generalizable to the other [131]. We presented in this review the many facets linking sex with TEs, both influencing each other in a co-evolutionary process. TE expression in germlines is essential for them to get fixed in the genome and be transmitted vertically. Conversely, TEs have an influence on sex differentiation mechanisms, for example through the intermediary of piRNAs. They could also influence sex evolution by the regulatory novelties they create. TEs are indeed great tools for evolution as they can rapidly propagate regulatory elements and thus provide the necessary rewiring of the genetic network. The high density of TEs on sex chromosomes, linked to the absence of recombination of these chromosomes, could increase the probability for TEs to locate in the vicinity of sexual development genes and interact with them. They can influence and be influenced by sex depending on the process studied.
Another way TEs can influence gene expression is by triggering alternative splicing, via the new splicing sites they sometimes bring with them [9]. In the case of sexual development gene regulation, however, such involvement of TEs has yet to be demonstrated. In Drosophila melanogaster, some intron retention events are known to be linked to sex [132]. Although the exact trigger of the alternative splicing is not clearly elucidated for now, a hypothesis proposed that the high coverage of repetitive sequences on the Y chromosome could be involved in the process, as presented earlier in this review: the Y chromosome would attract on its repeats high quantities of chromatin-modifying proteins, which would in turn lead to a global modification of the chromatin state on other chromosomes, and in the end would affect the accessibility of splicing factors to the nascent transcripts. Here, the impact of TEs on the splicing machinery would thus be indirect and not specific to particular genes.
Finally, genes involved in sexual development and sexual functions seem to evolve faster than other genes [133, 134]. These observations of positive selection and rapid evolution are not really consistent with earlier observations of the sex determination and differentiation cascade. Indeed, a popular model, formulated by Graham in 2003, states that “masters change, slaves remain” [135], where “masters” refer to genes at the top of the sex determination cascade, and “slaves” to genes acting at the end of the cascade. A renewal of this initial proposition has been proposed by Herpin et al.: “When masters change, some slaves remain, others are dismissed or acquire new tasks, and new ones can be hired” [34, 55]. Knowing that TEs are a source of genomic diversification, studying the evolution of sexual development genes in the perspective of TEs, just as the evolution of their regulation, could reveal interesting trends. A perspective could be to investigate RNA-seq dataset for species-specific sex-biased genes associated to TE location variation between closely related species to reveal candidate genes recently controlled by TEs. Global approaches by sequencing piRNAs and mapping them to sex-biased genes could also give more clues about the regulation and evolution of genes involved in sexual development and function.
Acknowledgments
The authors sincerely thank Joanne Burden for her diligent proofreading of the article.
Abbreviations
- ESD
Environmental Sex Determination
- GSD
Genetic Sex Determination
- KAP1
KRAB-associated protein 1
- KRAB-ZNF
Krüppel-associated box domain zinc finger
- MRE
MSL Recognition Element
- MSL
Male Specific Lethal
- piRNA
Piwi-Interacting RNA
- SD
Sex Determination
- TE
Transposable Element
Authors’ contributions
CD has drafted the initial version of the review and designed the figures; JNV, MS and MN have contributed to the writing of the manuscript. All authors have approved the final version.
Funding
This work was supported by a PRCI (International Collaborative Research Project) grant overseen by the French National Research Agency (ANR) together with the German Research Agency (Deutsche Forschungsgemeinschaft) (ANR-16-CE92–0019 – EVOBOOSTER). This publication was funded by the German Research Foundation (DFG) and the University of Würzburg in the funding program Open Access Publishing.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Manfred Schartl, Email: phch1@biozentrum.uni-wuerzburg.de.
Magali Naville, Email: magali.naville@ens-lyon.fr.
References
- 1.Biémont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443(7111):521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 2.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci. 1950;36(6):344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 4.Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9(5):411–412. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- 5.Seberg O, Petersen G. A unified classification system for eukaryotic transposable elements should reflect their phylogeny. Nat Rev Genet. 2009;10(4):276. doi: 10.1038/nrg2165-c3. [DOI] [PubMed] [Google Scholar]
- 6.Rishishwar L, Tellez Villa CE, Jordan IK. Transposable element polymorphisms recapitulate human evolution. Mob DNA. 2015;6:21. doi: 10.1186/s13100-015-0052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgeois Yann, Boissinot Stéphane. On the Population Dynamics of Junk: A Review on the Population Genomics of Transposable Elements. Genes. 2019;10(6):419. doi: 10.3390/genes10060419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrov DA, Fiston-Lavier A-S, Lipatov M, Lenkov K, Gonzalez J. Population genomics of transposable elements in Drosophila melanogaster. Mol Biol Evol. 2011;28(5):1633–1644. doi: 10.1093/molbev/msq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46(1):21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 10.Dupressoir A, Marceau G, Vernochet C, Bénit L, Kanellopoulos C, Sapin V, et al. Syncytin-a and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A. 2005;102(3):725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volff J-N. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays News Rev Mol Cell Dev Biol. 2006;28(9):913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert C, Feschotte C. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr Opin Genet Dev. 2018;49:15–24. doi: 10.1016/j.gde.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapitonov Vladimir V, Jurka Jerzy. RAG1 Core and V(D)J Recombination Signal Sequences Were Derived from Transib Transposons. PLoS Biology. 2005;3(6):e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naville M, Warren IA, Haftek-Terreau Z, Chalopin D, Brunet F, Levin P, et al. Not so bad after all: retroviruses and long terminal repeat retrotransposons as a source of new genes in vertebrates. Clin Microbiol Infect. 2016;22(4):312–323. doi: 10.1016/j.cmi.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Dolgin ES, Charlesworth B. The fate of transposable elements in asexual populations. Genetics. 2006;174(2):817–827. doi: 10.1534/genetics.106.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickey DA. Selfish DNA: a sexually-transmitted nuclear parasite. Genetics. 1982;101(3–4):519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright S, Finnegan D. Genome evolution: sex and the transposable element. Curr Biol. 2001;11(8):R296–R299. doi: 10.1016/S0960-9822(01)00168-3. [DOI] [PubMed] [Google Scholar]
- 18.Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25(9):537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeyl C, Bell G, Green DM. Sex and the spread of retrotransposon Ty3 in experimental populations of Saccharomyces cerevisiae. Genetics. 1996;143(4):1567–1577. doi: 10.1093/genetics/143.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78(2):737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8(3):269–294. doi: 10.1017/S0016672300010156. [DOI] [PubMed] [Google Scholar]
- 22.Arkhipova IR. Mobile genetic elements and sexual reproduction. Cytogenet Genome Res. 2005;110(1–4):372–382. doi: 10.1159/000084969. [DOI] [PubMed] [Google Scholar]
- 23.Barsoum E, Martinez P, Astrom SU. Alpha 3, a transposable element that promotes host sexual reproduction. Genes Dev. 2010;24(1):33–44. doi: 10.1101/gad.557310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bestor TH. Sex brings transposons and genomes into conflict. Genetica. 1999;107(1–3):289–295. doi: 10.1023/A:1003990818251. [DOI] [PubMed] [Google Scholar]
- 25.Kraaijeveld K, Zwanenburg B, Hubert B, Vieira C, De Pater S, Van Alphen JJM, et al. Transposon proliferation in an asexual parasitoid. Mol Ecol. 2012;21(16):3898–3906. doi: 10.1111/j.1365-294X.2012.5582.x. [DOI] [PubMed] [Google Scholar]
- 26.Warren WC, García-Pérez R, Xu S, Lampert KP, Chalopin D, Stöck M, et al. Clonal polymorphism and high heterozygosity in the celibate genome of the Amazon molly. Nat Ecol Evol. 2018;2(4):669–679. doi: 10.1038/s41559-018-0473-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arkhipova I, Meselson M. Transposable elements in sexual and ancient asexual taxa. Proc Natl Acad Sci U S A. 2000;97(26):14473–14477. doi: 10.1073/pnas.97.26.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flot J-F, Hespeels B, Li X, Noel B, Arkhipova I, Danchin EGJ, et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 2013;500(7463):453–457. doi: 10.1038/nature12326. [DOI] [PubMed] [Google Scholar]
- 29.Capel B. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat Rev Genet. 2017;18(11):675–689. doi: 10.1038/nrg.2017.60. [DOI] [PubMed] [Google Scholar]
- 30.Pan Q, Anderson J, Bertho S, Herpin A, Wilson C, Postlethwait JH, et al. Vertebrate sex-determining genes play musical chairs. C R Biol. 2016;339(7–8):258–262. doi: 10.1016/j.crvi.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bull JJ, Vogt RC. Temperature-dependent sex determination in turtles. Science. 1979;206(4423):1186–1188. doi: 10.1126/science.505003. [DOI] [PubMed] [Google Scholar]
- 32.Lang JW, Andrews HV. Temperature-dependent sex determination in crocodilians. J Exp Zool. 1994;270(1):28–44. doi: 10.1002/jez.1402700105. [DOI] [PubMed] [Google Scholar]
- 33.Honeycutt JL, Deck CA, Miller SC, Severance ME, Atkins EB, Luckenbach JA, et al. Warmer waters masculinize wild populations of a fish with temperature-dependent sex determination. Sci Rep. 2019;9(1):6527. doi: 10.1038/s41598-019-42944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L, et al. Sex determination: why so many ways of doing it? PLoS Biol. 2014;12(7):e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schartl M. Sex chromosome evolution in non-mammalian vertebrates. Curr Opin Genet Dev. 2004;14(6):634–641. doi: 10.1016/j.gde.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Koene JM. Sex determination and gender expression: reproductive investment in snails. Mol Reprod Dev. 2017;84(2):132–143. doi: 10.1002/mrd.22662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuda M. Sex determination in the teleost medaka, Oryzias latipes. Annu Rev Genet. 2005;39:293–307. doi: 10.1146/annurev.genet.39.110304.095800. [DOI] [PubMed] [Google Scholar]
- 38.Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, Shimizu A, et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci. 2002;99(18):11778–11783. doi: 10.1073/pnas.182314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert SF. Chromosomal sex determination in drosophila. Dev Biol 6th Ed. 2000.
- 40.Sanchez L, Chaouiya C. Logical modelling uncovers developmental constraints for primary sex determination of chicken gonads. J R Soc Interface. 2018;15(142):20180165. doi: 10.1098/rsif.2018.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stothard P, Pilgrim D. Sex-determination gene and pathway evolution in nematodes. BioEssays News Rev Mol Cell Dev Biol. 2003;25(3):221–231. doi: 10.1002/bies.10239. [DOI] [PubMed] [Google Scholar]
- 42.Tang W, Seth M, Tu S, Shen E-Z, Li Q, Shirayama M, et al. A Sex Chromosome piRNA Promotes Robust Dosage Compensation and Sex Determination in C. elegans. Dev Cell. 2018;44(6):762–770.e3. doi: 10.1016/j.devcel.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eggers S, Ohnesorg T, Sinclair A. Genetic regulation of mammalian gonad development. Nat Rev Endocrinol. 2014;10(11):673–683. doi: 10.1038/nrendo.2014.163. [DOI] [PubMed] [Google Scholar]
- 44.Hsu C, Pan Y-J, Wang Y-W, Tong S-K, Chung B. Changes in the morphology and gene expression of developing zebrafish gonads. Gen Comp Endocrinol. 2018;265:154–159. doi: 10.1016/j.ygcen.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 45.Croft B, Ohnesorg T, Hewitt J, Bowles J, Quinn A, Tan J, et al. Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9. Nat Commun. 2018;9(1):5319. doi: 10.1038/s41467-018-07784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonen N, Futtner CR, Wood S, Garcia-Moreno SA, Salamone IM, Samson SC, et al. Sex reversal following deletion of a single distal enhancer of Sox9. Science. 2018;360(6396):1469–1473. doi: 10.1126/science.aas9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schartl M, Schories S, Wakamatsu Y, Nagao Y, Hashimoto H, Bertin C, et al. Sox5 is involved in germ-cell regulation and sex determination in medaka following co-option of nested transposable elements. BMC Biol. 2018;16:16. [DOI] [PMC free article] [PubMed]
- 48.Assis R, Zhou Q, Bachtrog D. Sex-biased transcriptome evolution in drosophila. Genome Biol Evol. 2012;4(11):1189–1200. doi: 10.1093/gbe/evs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Böhne A, Sengstag T, Salzburger W. Comparative transcriptomics in east african cichlids reveals sex- and species-specific expression and new candidates for sex differentiation in fishes. Genome Biol Evol. 2014;6(9):2567–2585. doi: 10.1093/gbe/evu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catalan A, Hutter S, Parsch J. Population and sex differences in Drosophila melanogaster brain gene expression. BMC Genomics. 2012;13:1–12. doi: 10.1186/1471-2164-13-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ledón-Rettig CC, Zattara EE, Moczek AP. Asymmetric interactions between doublesex and tissue- and sex-specific target genes mediate sexual dimorphism in beetles. Nat Commun. 2017;8:14593. doi: 10.1038/ncomms14593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu H, Lamm MS, Rutherford K, Black MA, Godwin JR, Gemmell NJ. Large-scale transcriptome sequencing reveals novel expression patterns for key sex-related genes in a sex-changing fish. Biol Sex Differ. 2015;6:26. doi: 10.1186/s13293-015-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi L, Zhang Z, Su B. Sex biased gene expression profiling of human brains at major developmental stages. Sci Rep. 2016;6:21181. [DOI] [PMC free article] [PubMed]
- 54.Waters PD, Wallis MC, Graves JAM. Mammalian sex—origin and evolution of the Y chromosome and SRY. Semin Cell Dev Biol. 2007;18(3):389–400. doi: 10.1016/j.semcdb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Herpin A, Schartl M. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 2015;16(10):1260–1274. doi: 10.15252/embr.201540667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schartl M. A comparative view on sex determination in medaka. Mech Dev. 2004;121(7–8):639–645. doi: 10.1016/j.mod.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 57.Matsuda M, Sakaizumi M. Evolution of the sex-determining gene in the teleostean genus Oryzias. Gen Comp Endocrinol. 2016;239:80–88. doi: 10.1016/j.ygcen.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 58.Herpin A, Adolfi MC, Nicol B, Hinzmann M, Schmidt C, Klughammer J, et al. Divergent expression regulation of gonad development genes in medaka shows incomplete conservation of the downstream regulatory network of vertebrate sex determination. Mol Biol Evol. 2013;30(10):2328–2346. doi: 10.1093/molbev/mst130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takehana Y, Matsuda M, Myosho T, Suster ML, Kawakami K, Shin T, et al. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat Commun. 2014;5:4157. doi: 10.1038/ncomms5157. [DOI] [PubMed] [Google Scholar]
- 60.Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 2014;46(3):253–260. doi: 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- 61.Deloger M, Cavalli FMG, Lerat E, Biémont C, Sagot M-F, Vieira C. Identification of expressed transposable element insertions in the sequenced genome of Drosophila melanogaster. Gene. 2009;439(1–2):55–62. doi: 10.1016/j.gene.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 62.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lipatov M, Lenkov K, Petrov DA, Bergman CM. Paucity of chimeric gene-transposable element transcripts in the Drosophila melanogaster genome. BMC Biol. 2005;3(1):24. doi: 10.1186/1741-7007-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151(5):964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brunet F, Roche A, Chalopin D, Naville M, Klopp C, Vizziano-Cantonnet D, et al. Analysis of transposable elements expressed in the gonads of the siberian sturgeon. In: Williot P, Nonnotte G, Vizziano-Cantonnet D, Chebanov M, et al., editors. The Siberian Sturgeon (Acipenser baerii, Brandt, 1869) Volume 1 - Biology. Cham: Springer International Publishing; 2018. pp. 115–130. [Google Scholar]
- 66.Esteve-Codina A, Kofler R, Palmieri N, Bussotti G, Notredame C, Perez-Enciso M. Exploring the gonad transcriptome of two extreme male pigs with RNA-seq. BMC Genomics. 2011;12:552. doi: 10.1186/1471-2164-12-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laski FA, Rio DC, Rubin GM. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986;44(1):7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 68.Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its biogenesis and functions. Annu Rev Biochem. 2015;84(1):405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 69.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sarkar A, Volff J-N, Vaury C. piRNAs and their diverse roles: a transposable element-driven tactic for gene regulation? FASEB J. 2017;31(2):436–446. doi: 10.1096/fj.201600637RR. [DOI] [PubMed] [Google Scholar]
- 71.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 72.Kelleher ES. Reexamining the P-element invasion of Drosophila melanogaster through the lens of piRNA silencing. Genetics. 2016;203(4):1513–1531. doi: 10.1534/genetics.115.184119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hill Tom, Schlötterer Christian, Betancourt Andrea J. Hybrid Dysgenesis in Drosophila simulans Associated with a Rapid Invasion of the P-Element. PLOS Genetics. 2016;12(3):e1005920. doi: 10.1371/journal.pgen.1005920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ecco G, Imbeault M, Trono D. KRAB zinc finger proteins. Dev. 2017;144(15):2719–2729. doi: 10.1242/dev.132605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Molaro A, Malik HS. Hide and seek: how chromatin-based pathways silence retroelements in the mammalian germline. Curr Opin Genet Dev. 2016;37:51–58. doi: 10.1016/j.gde.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458(7242):1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516(7530):242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan X, Xu X, Elkenani M, Smorag L, Zechner U, Nolte J, et al. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res. 2013;11(3):1045–1059. doi: 10.1016/j.scr.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 79.Helleboid P-Y, Heusel M, Duc J, Piot C, Thorball CW, Coluccio A, et al. The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification. EMBO J. 2019;38(18):e101220. doi: 10.15252/embj.2018101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322(5906):1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zempleni J, Chew YC, Bao B, Pestinger V, Wijeratne SSK. Repression of transposable elements by histone biotinylation. J Nutr. 2009;139(12):2389–2392. doi: 10.3945/jn.109.111856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 83.Capy P, Gasperi G, Biémont C, Bazin C. Stress and transposable elements: co-evolution or useful parasites? Heredity. 2000;85(2):101. doi: 10.1046/j.1365-2540.2000.00751.x. [DOI] [PubMed] [Google Scholar]
- 84.Dong Y, Huang Z, Kuang Q, Wen Z, Liu Z, Li Y, et al. Expression dynamics and relations with nearby genes of rat transposable elements across 11 organs, 4 developmental stages and both sexes. BMC Genomics. 2017;18:666. [DOI] [PMC free article] [PubMed]
- 85.Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, et al. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 2017;27(10):1623–1633. doi: 10.1101/gr.218149.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zamudio N, Bourc’his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity. 2010;105(1):92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 87.Hadziselimovic F, Hadziselimovic NO, Demougin P, Krey G, Oakeley E. Piwi-pathway alteration induces LINE-1 transposon derepression and infertility development in cryptorchidism. Sex Dev. 2015;9(2):98–104. doi: 10.1159/000375351. [DOI] [PubMed] [Google Scholar]
- 88.Malki S, van der Heijden GW, O’Donnell KA, Martin SL, Bortvin A. A role for Retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell. 2014;29(5):521–533. doi: 10.1016/j.devcel.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sundaram Vasavi, Wang Ting. Transposable Element Mediated Innovation in Gene Regulatory Landscapes of Cells: Re-Visiting the “Gene-Battery” Model. BioEssays. 2017;40(1):1700155. doi: 10.1002/bies.201700155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sundaram V, Choudhary MNK, Pehrsson E, Xing X, Fiore C, Pandey M, et al. Functional cis-regulatory modules encoded by mouse-specific endogenousretrovirus. Nat Commun. 2017;8:14550. [DOI] [PMC free article] [PubMed]
- 92.Ellison CE, Bachtrog D. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science. 2013;342(6160):846–850. doi: 10.1126/science.1239552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ellison CE, Bachtrog D. Non-allelic gene conversion enables rapid evolutionary change at multiple regulatory sites encoded by transposable elements. Elife. 2015;4:e05899. doi: 10.7554/eLife.05899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ellison C, Bachtrog D. Contingency in the convergent evolution of a regulatory network: dosage compensation in Drosophila. PLoS Biol. 2019;17(2):e3000094. doi: 10.1371/journal.pbio.3000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA. 2010;16(12):2503–2515. doi: 10.1261/rna.2270710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Malone CD, Lehmann R, Teixeira FK. The cellular basis of hybrid dysgenesis and stellate regulation in Drosophila. Curr Opin Genet Dev. 2015;34:88–94. doi: 10.1016/j.gde.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kotelnikov RN, Klenov MS, Rozovsky YM, Olenina LV, Kibanov MV, Gvozdev VA. Peculiarities of piRNA-mediated post-transcriptional silencing of stellate repeats in testes of Drosophila melanogaster. Nucleic Acids Res. 2009;37(10):3254–3263. doi: 10.1093/nar/gkp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Herpin A, Braasch I, Kraeussling M, Schmidt C, Thoma EC, Nakamura S, et al. Transcriptional Rewiring of the Sex Determining dmrt1 Gene Duplicate by Transposable Elements. PLoS Genet. 2010;6(2):e1000844. doi: 10.1371/journal.pgen.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Simonti CN, Pavličev M, Capra JA. Transposable element exaptation into regulatory regions is rare, influenced by evolutionary age, and subject to pleiotropic constraints. Mol Biol Evol. 2017;34(11):2856–2869. doi: 10.1093/molbev/msx219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rojas-Ríos Patricia, Simonelig Martine. piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development. 2018;145(17):dev161786. doi: 10.1242/dev.161786. [DOI] [PubMed] [Google Scholar]
- 101.Rouget C, Papin C, Boureux A, Meunier A-C, Franco B, Robine N, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467(7319):1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sytnikova YA, Rahman R, Chirn G, Clark JP, Lau NC. Transposable element dynamics and PIWI regulation impacts lncRNA and gene expression diversity in Drosophila ovarian cell cultures. Genome Res. 2014;24(12):1977–1990. doi: 10.1101/gr.178129.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Watanabe T, Cheng E, Zhong M, Lin H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015;25(3):368–380. doi: 10.1101/gr.180802.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Katsuma S, Kawamoto M, Kiuchi T. Guardian small RNAs and sex determination. RNA Biol. 2014;11(10):1238–1242. doi: 10.1080/15476286.2014.996060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature. 2014;509(7502):633–636. doi: 10.1038/nature13315. [DOI] [PubMed] [Google Scholar]
- 106.Carvalho AB, Vicoso B, Russo CAM, Swenor B, Clark AG. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2015;112(40):12450–12455. doi: 10.1073/pnas.1516543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tomaszkiewicz M, Chalopin D, Schartl M, Galiana D, Volff J-N. A multicopy Y-chromosomal SGNH hydrolase gene expressed in the testis of the platyfish has been captured and mobilized by a Helitron transposon. BMC Genet. 2014;15:44. doi: 10.1186/1471-2156-15-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Faber-Hammond JJ, Phillips RB, Brown KH. Comparative analysis of the shared sex-determination region (SDR) among salmonid fishes. Genome Biol Evol. 2015;7(7):1972–1987. doi: 10.1093/gbe/evv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lubieniecki KP, Lin S, Cabana EI, Li J, Lai YYY, Davidson WS. Genomic Instability of the Sex-Determining Locus in Atlantic Salmon (Salmo salar) G3. 2015;5(11):2513–2522. doi: 10.1534/g3.115.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meisel RP, Gonzales CA, Luu H. The house fly Y chromosome is young and minimally differentiated from its ancient X chromosome partner. Genome Res. 2017;27(8):1417–1426. doi: 10.1101/gr.215509.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martin J, Kuvangkadilok C, Peart DH, Lee BTO. Multiple sex determining regions in a group of related Chironomus species (Diptera:Chironomidae) Heredity. 1980;44(3):367–382. doi: 10.1038/hdy.1980.34. [DOI] [Google Scholar]
- 112.Chalopin D, Naville M, Plard F, Galiana D, Volff J-N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol. 2015;7(2):567–580. doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Erlandsson R, Wilson JF, Pääbo S. Sex chromosomal transposable element accumulation and male-driven substitutional evolution in humans. Mol Biol Evol. 2000;17(5):804–812. doi: 10.1093/oxfordjournals.molbev.a026359. [DOI] [PubMed] [Google Scholar]
- 114.Lyon MF. The Lyon and the LINE hypothesis. Semin Cell Dev Biol. 2003;14(6):313–318. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 115.Mawaribuchi S, Takahashi S, Wada M, Uno Y, Matsuda Y, Kondo M, et al. Sex chromosome differentiation and the W- and Z-specific loci in Xenopus laevis. Dev Biol. 2017;426(2):393–400. doi: 10.1016/j.ydbio.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 116.Bertocchi NA, de Oliveira TD, Del Valle GA, Coan RLB, Gunski RJ, Martins C, et al. Distribution of CR1-like transposable element in woodpeckers (Aves Piciformes): Z sex chromosomes can act as a refuge for transposable elements. Chromosom Res. 2018;26(4):333–343. doi: 10.1007/s10577-018-9592-1. [DOI] [PubMed] [Google Scholar]
- 117.Śliwińska EB, Martyka R, Tryjanowski P. Evolutionary interaction between W/Y chromosome and transposable elements. Genetica. 2016;144(3):267–278. doi: 10.1007/s10709-016-9895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tang W, Mun S, Joshi A, Han K, Liang P. Mobile elements contribute to the uniqueness of human genome with 15,000 human-specific insertions and 14 Mbp sequence increase. DNA Res. 2018;25(5):521–533. doi: 10.1093/dnares/dsy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Griffin DK. Is the Y chromosome disappearing?—both sides of the argument. Chromosom Res. 2012;20(1):35–45. doi: 10.1007/s10577-011-9252-1. [DOI] [PubMed] [Google Scholar]
- 120.Schartl M, Schmid M, Nanda I. Dynamics of vertebrate sex chromosome evolution: from equal size to giants and dwarfs. Chromosoma. 2016;125(3):553–571. doi: 10.1007/s00412-015-0569-y. [DOI] [PubMed] [Google Scholar]
- 121.Brown EJ, Bachtrog D. The chromatin landscape of Drosophila: comparisons between species, sexes, and chromosomes. Genome Res. 2014;24(7):1125–1137. doi: 10.1101/gr.172155.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci. 2010;107(36):15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21(3):359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 124.Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80(1–4):133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 125.Scott LA, Kuroiwa A, Matsuda Y, Wichman HA. X accumulation of LINE-1 retrotransposons in Tokudaia osimensis, a spiny rat with the karyotype XO. Cytogenet Genome Res. 2006;112(3–4):261–269. doi: 10.1159/000089880. [DOI] [PubMed] [Google Scholar]
- 126.Lyon MF. Do LINEs have a role in X-chromosome inactivation? J Biomed Biotechnol. 2006;2006(1):59746. doi: 10.1155/JBB/2006/59746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A. 2000;97(12):6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20(16):2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141(6):956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 130.Jazin E, Cahill L. Sex differences in molecular neuroscience: from fruit flies to humans. Nat Rev Neurosci. 2010;11(1):9–17. doi: 10.1038/nrn2754. [DOI] [PubMed] [Google Scholar]
- 131.Shansky RM. Are hormones a “female problem” for animal research? Science. 2019;364(6443):825–826. doi: 10.1126/science.aaw7570. [DOI] [PubMed] [Google Scholar]
- 132.Wang M, Branco AT, Lemos B. The Y chromosome modulates splicing and sex-biased intron retention rates in Drosophila. Genetics. 2018;208(3):1057–1067. doi: 10.1534/genetics.117.300637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8(9):689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 134.Yang L, Zhang Z, He S. Both male-biased and female-biased genes evolve faster in fish genomes. Genome Biol Evol. 2016;8(11):3433–3445. doi: 10.1093/gbe/evw239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Graham P, Penn JKM, Schedl P. Masters change, slaves remain. BioEssays. 2003;25(1):1–4. doi: 10.1002/bies.10207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.