Abstract
Background:
The epidermal growth factor receptor 1 (EGFR1) plays a significant role in cell proliferation and development. Its regulation in humans is very critical and incompletely understood in Non small cell lung cancer (NSCLC).
Methods:
100 newly diagnosed NSCLC (lung adenocarcinoma) patients and 100 healthy controls were included and allele specific (AS) polymerase chain reaction (PCR) was used to genotype and expression was analyzed by quantitative real time PCR. Overall survival of patients was analyzed by Kaplan-Meier method and for prognostic significance ROC curve was plotted.
Results:
A statistically significant difference (p<0.0001) in CC, AA and CA genotypes distribution among patients and healthy controls was observed. Compared to the CC genotype as reference, OR was 30.40 (95%CI 1.75- 524.9, p=0.0002) and 3.97 (95%CI 1.49-10.52, p=0.003) for the homozygous AA and heterozygous CA genotypes respectively. Kaplan-Meier survival analysis was also performed to analyze the relationship of EGFR1 (-191C/A) genotypes with progression free median survival of NSCLC patients and the difference was found to be significantly (p=0.0002) associated with different genotypes. In the ROC curve with respect to TNM stage at optimal cut-off value of 9.88 fold increase in EGFR1 mRNA expression, sensitivity and specificity were 92.9%, 83.3% respectively (AUC=0.95, p<0.0001). ROC curve w.r.t. distant metastases at optimal cut-off value of 13.5 fold change EGFR1 mRNA expression, sensitivity and specificity were 68.2%, 71.4% respectively (AUC=0.81, p<0.0001). In ROC curve w.r.t to presence/ absence of pleural effusion at optimal cut-off value of 14.8 fold change EGFR1 mRNA expression sensitivity and specificity were 66.7%, 68.2% respectively (AUC=0.71, p=0.009).
Conclusions:
Study concluded EGFR1 promoter polymorphism could be a risk factor associated with disease and may be used as prognostic marker for patients’ survival and predictor for disease worseness.
Keywords: Lung adenocarcinoma, EGFR1 (-191C/A) genotype, overall and progression free survival, prognosis
Introduction
Single nucleotide polymorphisms (SNPs) are the most common sources of human genetic variation, and they may contribute to an individuals’ susceptibility to cancer. Several studies have demonstrated that some variants affect either the expression or activities of various enzymes, and that they are therefore associated with the risk of cancer development (Park et al., 2006). The SNP occurs at an important binding site for the transcription factor SP1 that is necessary for activation of EGFR promoter activity and correlates with increased promoter activity and expression of EGFR mRNA (Kobayashi et al., 2005). EGFR is also a possible genetic risk factor for cancer susceptibility. In transgenic mice, overexpression of the EGFR initiates formation of oligodendroglioma and breast cancer (Weiss et al., 2003). Till date, several features of the 5’-regulatory region of EGFR have been described, including a TATA-less, CAAT-less, high GC content promoter with multiple transcriptional start sites (Ishii et al., 1985). EGFR are highly expressed in many tumor types of epithelial origin, including breast, head and neck, bladder cancers, and non small cell lung cancer (Arteaga, 2002). Expression of high levels of EGFR has been associated with a poor prognosis, especially in NSCLC patients (Brabender et al., 2001). Several studies have demonstrated that some variants affect either the expression or activities of various enzymes, and that they are therefore associated with the risk of cancer development (Park et al., 2006). Several polymorphisms in the EGFR gene have been reported (Hsieh et al., 2005) and deposited into public databases (ncbi.nlm.nih.gov/SNP). The variant −191C/A (rs712830) have been associated with increased EGFR promoter activity and gene expression (Liu et al., 2005). This study aimed at characterizing the frequency of EGFR gene polymorphisms in NSCLC patients and determining the correlation with clinicopathological feature and survival of patients.
Materials and Methods
Study population and sample collection
This study included 100 histopathologically confirm newly diagnosed lung adenocarcinoma patients and 100 healthy subjects. Study was approved by institutional ethics committee of Maulana Azad Medical College and Associated Hospitals and All India Institute of Medical Sciences New Delhi. After informed consent, patients’ 3 mL of peripheral blood sample was collected in plain vials from each subject and serum was separated and stored at –80°C until circulating DNA, RNA extraction. Patients included in study were followed for 2 years for overall and progression free survival analysis.
Circulating DNA, RNA isolation and cDNA synthesis
Circulating DNA was extracted by using commercially available kit (Epigentech, USA) following manufacturer’s protocol and circulating total RNA was extracted by Trizol reagent according to the manufacturer’s protocol (AMRESCO, USA) of lung adenocarcinoma cases as well as from healthy control’s circulating samples stored at –80 °C. The quality of circulating DNA and total RNA samples were quantified using NanoDrop spectrophotometer. The ratios of the absorbance at 260 and 280nm (A260/280) were used to assess the purity of nucleic acids and for pure DNA, A260/280=1.8 and for pure RNA A260/280 =2.0 were considered.and from total RNA, cDNA was synthesized by using 100 ng total RNA following manufacturers’ protocol (Verso, Thermo Scientific, USA). Briefly, 100 ng of total RNA, 5X cDNA synthesis buffer, dNTPs (5 mM each), RT enhancer, Verso RT enzyme mix and random hexamers/Oligo DT (400 ng/μL) in the total volume of 20 μL incubated for 60 min at 42°C.
Genotyping and quantitative real time polymerase chain reaction (PCR)
The (-191C/A, rs712830) promoter polymorphism of EGFR1 gene was genotyped using allele specific (AS) PCR method using circulating DNA. PCR reaction was performed in 25 μL reaction volume containing 3 μL of 100 ng template circulating DNA, 0.25 μL 25 pmol each primer, 10 μL of mastermix containing 10 mM dNTPs, 20 mM MgCl2, 5 U/μL Taq polymerase with 10× Taq Buffer (Fermantas) and 25 μL reaction volume was maintained by adding nuclease-free ddH2O followed by programme 10 min of initial denaturation at 95°C and 40 cycles at 95 °C for 40 s, 64°C for 40 s and 72°C for 40 s with a final 10 min extension step at 72°C and PCR product of 175 bp was visualized on 2% agarose gel containing ethidium bromide (Figure 1).
Figure 1.
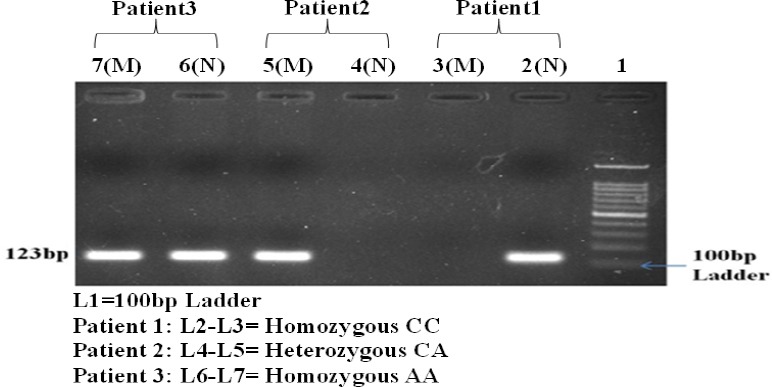
Gel Electrophoresis Image of EGFR 1 Gene Polymorphism in Patients
EGFR-1circulating mRNA expression was studied by QRT-PCR method (SYBR Green I technology) with β-actin gene as internal control. The expression of EGFR-1 and β-actin was performed by PCR programme for 40 cycles, denaturation at 94°C for 40 s, annealing at 60°C for 40 s, extension at 72°C for 40 s and reaction volume was 20 μL. A final extension step at 72°C for 5 min to complete the reaction and melting curve analysis was performed between the range 40 to 90°C to ensure the specific amplification. A control without cDNA was included in each experiment as non template control and all reaction were performed in duplicate. The relative quantification method (2−(ΔΔCT)) was done to analyse the circulating EGFR-1 mRNA expression level by using β-actin as internal control and final results were expressed as mean fold change in circulating EGFR-1 mRNA expression in lung adenocarcinoma patients as compared to control.
Statistical analysis
Differences in selected demographic charecteristic and EGFR-1 genotype frequencies between the cases and controls were evaluated by using the Chi-square test. The associations between EGFR-1 variant genotypes and risk of lung adenocarcinoma were estimated by calculating the odds ratios (ORs) with 95% confidence intervals (CIs). Allele frequencies between the cases and controls were evaluated using Hardy-Weinberg equilibrium test. Kruskal Wallis test were used to analyze the association of gene expression with different EGFR 1 genotype. The Kaplan-Meier method was used to calculate the over all and progression free survival of lung adenocarcinoma patients. A P value <0.05 was considered indicative of a statistically significant difference. All statistical analyses were performed using the SPSS 16 and Graph Pad version 6.0.
Results
Demographics
All demographic features of the subjects are depicted (Table 1). In brief, total of 100 lung adenocarcinoma patients were analyzed and healthy controls were age, sex and history of smoking status and type was also matched.
Table 1.
Distribution of Selected Characteristics among NSCLC Patients and Healthy Controls
| Variables | NSCLC patients n (%) | Healthy controls n (%) |
|---|---|---|
| Total no. | 100 (100%) | 100 (100%) |
| Gender | ||
| Males | 71 (71%) | 71 (71%) |
| Females | 29 (29%) | 29 (29%) |
| Age at diagnosis (Years) | ||
| < 55 | 56 (56%) | 56 (56%) |
| > 55 | 44 (44%) | 44 (44%) |
| Mean + SD age (years) | 54.37+10.77 (range 32-75years) | 54.25+10.82 (range 30-70 years) |
| Smoking status | ||
| Non smoker | 55 (55%) | |
| Smoker smokers | 45 (45%) | |
| Current smokers | 24 (24%) | |
| Ex- smokers | 21 (21%) | |
| Smoking type | ||
| Cig | 18 (40%) | |
| Bidi | 16 (35.6%) | |
| Hukka | 11 (24.4%) | |
| Smoking level (pack year) | ||
| Mild (< 10) | 23 (51.1%) | |
| Moderate (< 40) | 18 (40%) | |
| Heavy (> 40) | 4 (8.9%) | |
| TNM Stage | ||
| Early (I&II) | 30 (30%) | |
| Advanced (III&IV) | 70 (70%) | |
| Distant Metastases | ||
| Positive | 44 (44%) | |
| Negative | 56 (56%) | |
| Histopathological Grade | ||
| Grade 1 | 24 (24%) | |
| Grade 2 | 41 (41%) | |
| Grade 3 | 35 (35%) | |
| Pleural effusion | ||
| Yes | 15 (15%) | |
| No | 85 (85%) | |
Case-control genotype distribution
The genotype and allele distribution of EGFR1 (-191C/A) in cases and controls are summarised in Table 2. The genotype and allele frequency distributions of EGFR1 polymorphism (-191C/A) in Lung adenocarcinoma cases and controls was analyzed using circulating DNA. A statistically significant difference (p<0.0001) in CC, AA and CA genotypes distribution among patients and healthy controls was observed. The EGFR1 CC, AA and CA genotypes distribution among NSCLC patients was found to be 71%, 11% and 18%, whereas among controls it was 94%, 0% and 6% respectively.
Table 2.
Frequency of EGFR1 (-191C/A) Polymorphism Genotypes among NSCLC Cases and Healthy Controls
| Genotype | p value | Allele frequency | ||||
|---|---|---|---|---|---|---|
| CC (%) | AA (%) | CA (%) | C allele | A allele | ||
| Cases (n=100) | 71 (71.0) | 11 (11.0) | 18 (18.0) | 0.8 | 0.2 | |
| Controls (n=100) | 94 (94.0) | 0 (0.0) | 6 (6.0) | <0.0001 | 0.97 | 0.03 |
EGFR1 (-191C/A) gene polymorphism and risk of Lung adenocarcinoma
Odds ratio with 95% confidence intervals was calculated for each group to estimate the degree of association between the EGFR1 (-191C/A) genotypes and risk of NSCLC in Indian patients. Compared to the CC genotype as reference, OR was 30.40 (95% CI 1.75- 524.9, p=0.0002) and 3.97 (95% CI 1.49-10.52, p=0.003) for the homozygous AA and heterozygous CA genotypes respectively. This suggested a possible dominant effect of the EGFR1 (-191C/A) mutant A allele on NSCLC risk in Indian population as higher risk was found to be associated with AA homozygous genotype (Table 3).
Table 3.
Risk of Developing NSCLC Associated with EGFR1 (-191C/A) Genotypes
| Genotypes | Controls (n=100) | Cases (n=100) | OR (95% CI) | p value |
|---|---|---|---|---|
| CC | 94 | 71 | 1 (ref) | |
| AA | 0 | 11 | 30.40 (1.76-524.9) | 0.0002 |
| CA | 6 | 18 | 3.97 (1.49-10.52) | 0.003 |
| CA+AA | 6 | 29 | 6.39 (2.52-16.25) | <0.0001 |
EGFR1 (-191C/A) genotypes and circulating EGFR1 m RNA expression
Real time relative quantification analysis showed increased circulating EGFR1 mRNA expression among NSCLC patients than healthy controls but no significant difference was observed in circulating mRNA expression with respect to EGFR1 genotype in NSCLC patients. The fold change was calculated with respect to the EGFR1 CC wild type genotype NSCLC patient taken as the reference value. It was observed that there was more than 15 fold (mean) increase circulating EGFR1 mRNA expression among patients with homozygous AA and heterozygous CA genotype while homozygous CC wild type genotype showed more than 12 fold of circulating EGFR1 mRNA expression (Table 4).
Table 4.
Fold Change Increase in Circulating EGFR1 mRNA Expression Associated with Different EGFR1 (-191C/A) Genotypes among Lung Adenocarcinoma Patients.
| Genotypes | Mean + SD | Range | Median | p value |
|---|---|---|---|---|
| CC | 12.66+7.6 | 2.07-31.77 | 12.55 | |
| AA | 15.89+8.27 | 3.78-29.65 | 15.56 | 0.22 |
| CA | 15.91+8.35 | 4.65-37.53 | 13.64 |
EGFR1 (-191C/A) genotypes and survival outcome
Overall survival
Kaplan-Meier survival analysis was performed to analyze the relationship of EGFR1 (-191C/A) polymorphism with overall survival of NSCLC patients. No significant difference was observed in overall survival of NSCLC patients with respect to different genotypes, though patients with EGFR1 AA genotype (8.9 months) and CA genotype (8.0 months) showed reduced overall median survival time compared to wild type CC homozygous (11.2 months) genotype (Figure 2a).
Figure 2.
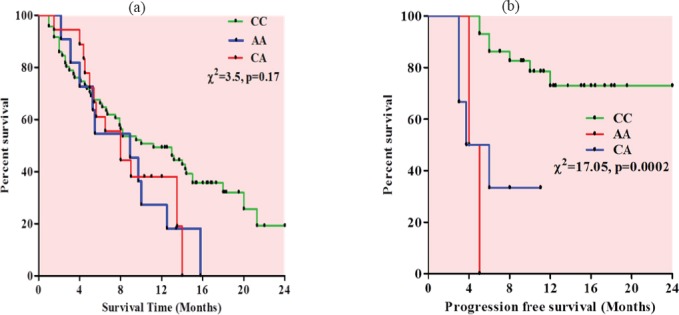
Kaplan-Meier Survival Curve (a) Overall survival curve with respect to EGFR1 genotypes (b) Progression free survival curve with respect to EGFR1 genotypes.
Progression free survival
Kaplan-Meier survival analysis was also performed to analyze the relationship of EGFR1 (-191C/A) genotypes with progression free median survival of NSCLC patients. Patients with EGFR1 AA genotype (4.5months) and CA genotype (4.8 months) showed reduced progression free survival compared to CC genotype (11.3months) and the difference was found to be significant (p=0.0002) (Figure 2b).
Circulating EGFR1 mRNA expression as predictive/prognostic marker in Lung adenocarcinoma patients
To evaluate the role of circulating EGFR1 mRNA as predictive/prognostic marker for NSCLC patients, different clinical parameters were dichotomised into two groups and ROC curves were plotted between early and advanced stage NSCLC patients. In the ROC curve with respect to TNM stage at optimal cut-off value of 9.88 fold increase in EGFR1 mRNA expression, sensitivity and specificity were 92.9%, 83.3% respectively (AUC=0.95, p<0.0001). ROC curves were also plotted with respect to presence/absence of distant metastases and presence/absence of pleural effusion positive. In ROC curve w.r.t. + distant metastases at optimal cut-off value of 13.5 fold change EGFR1 mRNA expression, sensitivity and specificity were 68.2%, 71.4% respectively (AUC=0.81, p<0.0001). In ROC curve w.r.t to presence/ absence of pleural effusion at optimal cut-off value of 14.8 fold change EGFR1 mRNA expression sensitivity and specificity were 66.7%, 68.2% respectively (AUC=0.71, p=0.009) (Figure 3).
Figure 3.
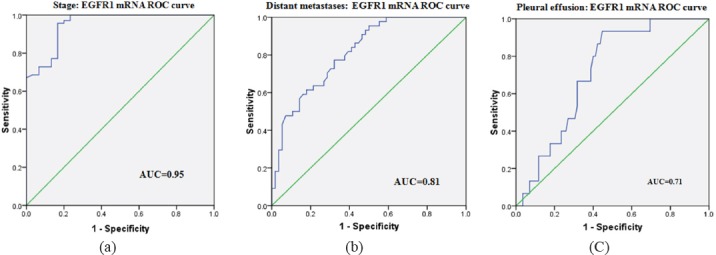
ROC curve for circulating EGFR1 mRNA expression: (a) between early and advanced stage patients, (b) between presence/absence of distant metastases, (c) between patients with/without pleural effusion.
Discussion
EGFR1 essential promoter region was associated with altered promoter activity and gene expression both in vitro and in vivo. The SNP occurs at binding site for the transcription factor SP1 that is necessary for activation of EGFR1 promoter activity and correlates with increased promoter activity and expression of EGFR1 mRNA (Kobayashi et al., 2005). Several polymorphisms in the EGFR1 gene have been reported (Hsieh et al., 2005) and deposited into public databases (Liu et al., 2005). The variant -191C/A have been associated with increased EGFR1 promoter activity and gene expression (Moriai et al., 1994) and Costa BM et al in 2011 found that the heterozygous -191C/A genotype EGFR1 could be used as predictive marker in glioblastoma (Costa et al., 2018). EGFR1 (-191 C/A) promoter polymorphism was found to be associated with increased risk and poor prognosis of NSCLC in Indian population and significant difference was observed in genotype distribution of EGFR1 -191C/A among NSCLC cases and healthy controls. It has been observed that the risk of developing NSCLC was more than 30.0 fold higher in association with homozygous EGFR1 -191AA genotype than homozygous EGFR1 -191CC genotype. Genome-wide studies (GWAS) showed that EGFR1 polymorphisms found to be associated with increased glioma risk (Sanson et al., 2011). Higher expression of circulating EGFR1 mRNA expression was observed with AA mutant homozygous and CA heterozygous compare to CC homozygous wild type genotype. It has been observed that the overall survival was reduced with EGFR1 AA mutant homozygous genotype compare to EGFR1 CC wild type genotype, however progression free survival was also found to be reduced with EGFR1 AA mutant homozygous genotype and CA heterozygous genotype. Ligands binding to EGFR1 triggers different downstream signalling pathways and results in several biological responses varying from proliferation, differentiation, migration and apoptosis (Wells, 1999; Pu et al., 2009). EGFR1 was found upregulated in human carcinomas and often related to poor prognosis or advanced pathological stages (Fischer-Colbrie et al., 1997). The switch from greater cytoplasmic EGFR1 to greater membranous EGFR1 expression occurs in progression in cancers (Piyathilake et al., 2002). Increased expression of EGFR1 mRNA in plasma of NSCLC patients provides a potential tool for molecular approach in diagnosis (Radostina et al., 2010). Higher AUC in ROC curves of advanced vs early stage patients for serum EGFR1 mRNAs expression and very high sensitivity and specificity suggested to be useful as good predictive marker for disease progression in NSCLC. Scagliotti et al., (2004) also have analysed that EGFR1 expression in NSCLC is associated with reduced survival. EGFR1 is overexpressed in the NSCLC patients, and is associated with the poor survival and EGFR1 expression is clearly involved in the lung cancer pathogenesis (Dowell and Minna, 2005). Expression study previously published and it was found that circulating EGFR1 mRNA over expression may be a predictive marker in patients’ poor prognosis, overall survival and metastatic behaviour of NSCLC patients (Mirza et al., 2015). Current study suggest that circulating EGFR1 gene polymorphism could be the indicative marker to be associated with risk for Lung adenocarcinoma, poor progression free survival and expression could be a prognostic indicator for disease advancement and metastatic behaviour of disease.
Conflict of interest
None.
Acknowledgements
The authors thank to all the study subjects and All India Institute of Medical Sciences, New Delhi, for assistance in recruiting the subjects.
References
- 1.Arteaga CL. Epidermal growth factor receptor dependence in human tumors: more than just expression? Oncologist. 2002;4:31–9. doi: 10.1634/theoncologist.7-suppl_4-31. [DOI] [PubMed] [Google Scholar]
- 2.Brabender J, Danenberg KD, Metzger R, et al. Epidermal growth factor receptor and HER2-neu mRNA expression in non-small cell lung cancer is correlated with survival. Clin Cancer Res. 2001;7:1850–5. [PubMed] [Google Scholar]
- 3.Costa BM, Marta VP, Ricardo F, et al. Impact of EGFR genetic variants on Glioma risk and patient outcome. Cancer Epidemiol Biomarkers Prev. 2011;20:2610–7. doi: 10.1158/1055-9965.EPI-11-0340. [DOI] [PubMed] [Google Scholar]
- 4.Dowell JE, Minna JD. Chasing mutations in the epidermal growth factor in lung cancer. N Engl J Med. 2005;352:830–2. doi: 10.1056/NEJMe058033. [DOI] [PubMed] [Google Scholar]
- 5.Fischer-Colbrie J, Witt A, Heinzl H, et al. EGFR and steroid receptors in ovarian carcinoma: comparison with prognostic parameters and outcome of patients. Anticancer Res. 1997;17:613–9. [PubMed] [Google Scholar]
- 6.Hsieh YY, Chang CC, Tsai FJ, et al. T homozygote and allele of epidermal growth factor receptor 2073 gene polymorphism are associated with higher susceptibility to endometriosis and leiomyomas. Fertil Steril. 2005;83:796–9. doi: 10.1016/j.fertnstert.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Ishii S, Xu YH, Stratton RH, et al. Characterization and sequence of the promoter region of the human epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1985;82:4920–4. doi: 10.1073/pnas.82.15.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 9.Liu W, Innocenti F, Wu MH, et al. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- 10.Mirza M, Rashid M, Jamsheed J, et al. Cell free EGFR mRNA expression and implications for survival and metastasis in non-small cell lung cancer cases. Asian Pac J Cancer Prev. 2015;16:6445–9. doi: 10.7314/apjcp.2015.16.15.6445. [DOI] [PubMed] [Google Scholar]
- 11.Moriai T, Kobrin MS, Hope C, et al. A variant epidermal growth factor receptor exhibits altered type a transforming growth factor binding and transmembrane signaling. Proc Natl Acad Sci U S A. 1994;91:10217–21. doi: 10.1073/pnas.91.21.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park JY, Park JM, Jang JS, et al. Caspase 9 promoter polymorphisms and risk of primary lung cancer. Hum Mol Genet. 2006;15:1963–71. doi: 10.1093/hmg/ddl119. [DOI] [PubMed] [Google Scholar]
- 13.Piyathilake CJ, Frost AR, Manne U, et al. Differential expression of growth factors in squamous cell carcinoma and precancerous lesions of the lung. Clin Cancer Res. 2002;8:734–4. [PubMed] [Google Scholar]
- 14.Pu YS, Huang CY, Kuo YZ, et al. Characterization of membranous and cytoplasmic EGFR expression in human normal renal cortex and renal cell carcinoma. J Biomed Sci. 2009;16:82. doi: 10.1186/1423-0127-16-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radostina C, Ognian G, Ivanka D, et al. EGFR and hTERT expression as a diagnostic approach for non-small cell lung cancer in high risk groups. Biomark Cancer. 2010;2:57–63. doi: 10.4137/BIC.S3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanson M, Hosking FJ, Shete S, et al. Chromosome 7p11.2 (EGFR) variation influences gliomarisk. Hum Mol Genet. 2011;20:2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scagliotti GV, Selvaggi G, Novello S, et al. The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res. 2004;10:4227–32. doi: 10.1158/1078-0432.CCR-040007. [DOI] [PubMed] [Google Scholar]
- 18.Weiss WA, Burns MJ, Hackett C, et al. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res. 2003;63:1589–95. [PubMed] [Google Scholar]
- 19.Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–43. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]


